Abstract
Traditional pest and disease identification methods mainly rely on manual detection or traditional machine learning techniques, but they have obvious deficiencies in terms of their accuracy and generalisation ability. In recent years, deep learning has gradually become the preferred solution for the intelligent identification of pests and diseases by virtue of its powerful automatic feature extraction and complex data-processing capabilities. In this paper, we systematically present the application of traditional machine learning methods in pest and disease identification and their limitations, and focus on the research progress of deep learning methods, covering three mainstream architectures: convolutional neural network (CNN), Vision Transformer model and CNN–Transformer hybrid model. In addition, this paper provides an in-depth analysis of the key challenges currently faced in the field of pest recognition, including the problems of small-sample learning, complex background interference and model lightweighting, and further propose solutions for future research to provide theoretical references and technical guidance for the development of related fields.
1. Introduction
Food security and agricultural sustainable development are seriously threatened by the issue of crop pests and diseases, which is growing more and more prevalent in the context of global climate change and rising agricultural output. The Food and Agriculture Organization of the United Nations (FAO) estimates that global crop losses due to pests and diseases amount to 40% per year [1]. In order to control pests and diseases, many farmers have to increase the application of pesticides and fungicides. However, the abuse of pesticides in large doses and at high frequencies not only results in increasing the resistance of pests and diseases but also poses a serious threat to the environment and human health. How to identify crop pests and diseases in a timely and accurate manner and to control them scientifically according to their type and degree has become an urgent problem.
The early identification of pests and diseases mainly relies on manual identification methods, i.e., subjective judgement by professionals through on-site observation or laboratory testing [2]. However, this method has problems such as strong subjectivity, poor accuracy and low efficiency, which make it difficult to meet the needs of modern agricultural development. With the development of computer vision technology, methods based on conventional machine learning are used in the identification of pests and diseases. The methods involve manually designing feature detection algorithms to extract image features and then using classification or clustering algorithms, such as K-means and SVM, to identify and classify features. These methods improve the identification efficiency and accuracy to a certain extent, but there are still problems, such as a poor model generalisation ability.
In recent years, the rise of deep learning techniques has brought new opportunities for pest and disease identification. The introduction of convolutional neural network (CNN) and Transformer architectures has greatly improved the performance of pest and disease identification and pushed the development of the field. And with the increasing demand for agricultural intelligence and precision, multimodal data fusion and advanced algorithm design have gradually become the key technical means to improve the efficiency of agricultural production. In the field of target detection, the novel embedded cross-framework [3] solves the real-time and accuracy problems of detecting significant targets in high-resolution images by fusing multi-scale features and lightweight design, which is especially suitable for the identification and localisation tasks regarding pests and diseases in complex farmland environments. In addition, in the face of continuous changes in data distribution in dynamic agricultural scenarios, the dual-channel collaborative Transformer [4] effectively mitigates the forgetting problem of the model in incremental learning through the dual-stream feature interaction and continuous learning mechanism, providing an innovative solution for the long-term adaptability of agricultural intelligent systems. Although these technologies have their own focus on methods and application scenarios, they collectively reflect the whole chain of innovation from data acquisition and feature modelling to algorithm optimisation, and provide a multi-dimensional technical path for the intelligent identification of pests and diseases.
In this paper, we present the research on the application of models such as a CNN and Transformer in the field of pests and diseases, discuss their specific methods and research progress in pest and disease identification, show the potential and application prospects of deep learning technology in crop pest and disease identification, and point out the challenges of current research and future development directions.
2. Methods Based on Conventional Machine Learning
The pest and disease identification methods based on conventional machine learning combine feature extraction and conventional machine learning algorithms by extracting the global and local features of the image using designed feature detection algorithms (e.g., edge detection, corner detection, texture features) on the preprocessed pest and disease images, and then designing and training classifiers to achieve the identification and classification of pests and diseases. The steps are shown in Figure 1. First, the image data of crop pests and diseases are collected through on-site collection and public datasets to make them more diversified and representative, and then preprocessing operations, such as denoising, filtering and resizing, are carried out to facilitate the subsequent feature extraction based on the relevant features. Conventional machine learning algorithms, such as K-means, Least Squares, Random Forest Algorithm (RF) and Support Vector Machine Algorithm (SVM), are selected for classifier design and training; the model is evaluated and validated according to indicators, such as precision, recall, accuracy and F1 value; and then, finally, the model is deployed and applied.
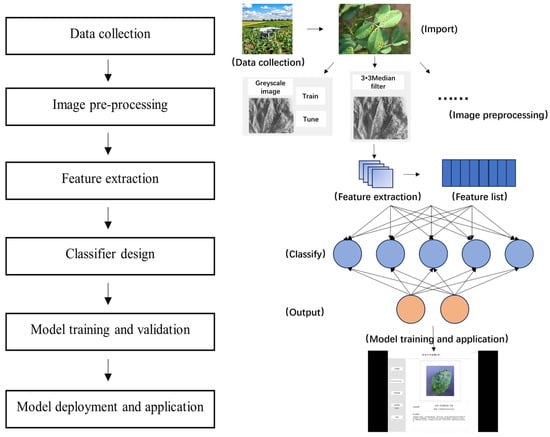
Figure 1.
Steps of pest and disease identification based on conventional machine learning.
The usual flow of the pest and disease identification method based on machine learning is shown in Figure 1.
Step 1: The collection methods include public datasets, agricultural field collection and drone photography, and the data should be diverse and representative.
Step 2: The purpose is to improve image quality and reduce noise for subsequent feature extraction, and the operations include denoising, image resizing, filtering, colour space conversion and histogram equalisation.
Step 3: The relevant features include colour, texture and shape, and the extraction methods include colour co-occurrence matrix (CCM), greyscale co-occurrence matrix (GLCM) and geometric parameters.
Step 4: Traditional machine learning algorithms are chosen as classifiers, such as K-means [5], Least Squares [6], Random Forest Algorithm (RF) [7], Support Vector Machine Algorithm (SVM) [8] and principal component analysis (PCA) [9].
Step 5: The following metrics:
among others, are used for the modelling assessment. Here, TP (True Positive) is the number of samples correctly predicted by the model to be in the positive category, FP (False Positive) is the number of samples incorrectly predicted by the model to be in the positive category, TN (True Negative) is the number of samples correctly predicted by the model to be in the negative category and FN (False Negative) is the number of samples incorrectly predicted by the model to be in the negative category. Precision indicates the proportion of samples predicted by the model to be in the positive category that are actually in the positive category. Recall indicates the proportion of samples that are actually in the positive category that are correctly predicted by the model to be in the positive category. Accuracy represents the proportion of samples correctly predicted by the model to the total number of samples. F1 is the harmonic mean of the precision rate and the recall rate.
Step 6: The model is applied to a practical pest and disease identification system to provide pest and disease monitoring and early warning services for agricultural production.
Different identification models have specific strengths and weaknesses for specific objects, and some applications of pest and disease identification models based on traditional machine learning and their characteristics in recent years are listed in Table 1.

Table 1.
Characteristics of pest and disease identification models based on conventional machine learning.
Compared with the manual identification methods, the pest and disease identification techniques based on conventional machine learning provide obvious improvements in identification efficiency and accuracy, but there are still some limitations: (1) The selection of manual features relies heavily on the experience and intuition of the experts, which can lead to subjectivity and instability. (2) The manual features cannot be adapted well to the new data, resulting in a poor model generalisation ability. For example, environmental changes, such as different light conditions, weather conditions or shading, as well as different pests, diseases or crop species, can affect the accuracy of the identification.
3. Methods Based on Deep Learning
Deep learning can process more complicated data, learn features automatically and greatly enhance the model’s capacity for generalisation as compared with traditional machine learning.
The basic idea is that multiple hidden layers of neural networks are used for data analysis and feature learning, each hidden layer can be regarded as a perceptron for extracting local features and these local features are subsequently abstracted into global features [27]. By this approach, deep neural networks not only significantly alleviate the problem of local minima but also perform well in a variety of complex tasks, such as image identification and natural language processing [28].
The basic flow of the pest and disease identification method based on deep learning is shown in Figure 2. First, the dataset is preprocessed, including dataset labelling and enhancement. Then, the dataset is split into a training set and a test set, the training set is used to train the deep neural network, the trained model is tested on the test data to evaluate the performance of the deep learning model, and the model is validated and optimised to output the results and applications. In the following, we describe the current status and problems faced by the application of deep learning in crop pest and disease identification in terms of the data collection and preprocessing and the model design.
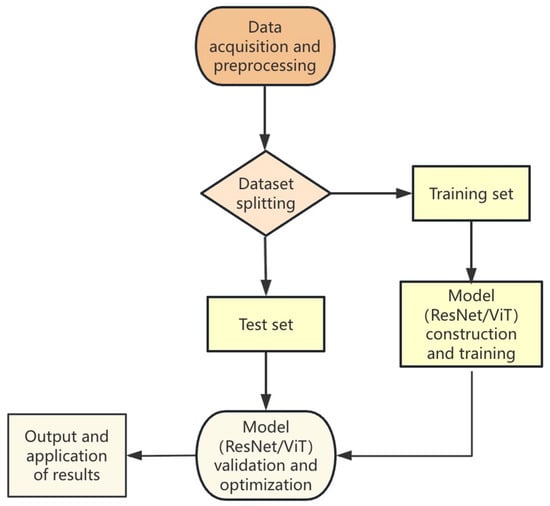
Figure 2.
Flow of pest and disease identification based on deep learning.
3.1. Data Collection and Preprocessing
In the research of pest and disease identification, data collection usually adopts both public datasets and self-constructed datasets. Commonly used public datasets for pests and diseases include the Agricultural Pest and Disease Research Atlas (IDADP), Plant Village dataset, AI Challenger 2018 Pest and Disease Classification dataset, Apple Leaf Pathology dataset and Rice Leaf Pest dataset; public datasets for insects and pests include the IP102 dataset, the OIDS-45 dataset, the YOLO Crop Insect Pest Recognition Dataset (Insect Pest Recognition Dataset, IPRD) and the D0 dataset. By crop species, there are fruit tree pest and disease open datasets, such as the Agricultural Pest and Disease Research Atlas (IDADP), Plant Village dataset, Apple Leaf Pathology Dataset, YOLO Crop Pest Recognition Dataset, OIDS-45 dataset and D0 dataset, and vegetable pest and disease open datasets, such as the Plant Village dataset, AI Challenger 2018 pest classification dataset and D0 dataset. Public datasets have the advantages of high data quality, easy access, and easy cooperation and communication between researchers, but they also have the problems of insufficient data relevance, variable data quality, and limited data volume and diversity, so self-built datasets are still used in many studies in pest and disease identification. The self-built dataset can be targeted to collect and process pest and disease images according to specific needs, i.e., meet the needs of specific application scenarios and be targeted and scalable.
The dataset needs to be preprocessed before model training, such as data labelling and data enhancement. Commonly used data annotation methods are image classification annotation, object detection annotation, semantic segmentation annotation and so on.
Image classification annotation refers to tagging each image with a specific ‘category’, such as mildew and late blight, which can clearly classify the image; object detection annotation focuses on identifying and accurately locating specific areas in the image, for example, for wheat infected with brown rot, this annotation method can accurately identify and locate the area where the rust spots are located. Semantic segmentation annotation focuses on classifying each pixel in the image in a more detailed way, for example, in the case of apple, some leaves are affected by black rot disease, and this method can accurately differentiate between diseased leaves and healthy leaves.
Data enhancement refers to expanding the data samples by changing the training data so as to enhance the training effect of the dataset and improve the generalisation ability and robustness of the model. Commonly used data enhancement methods include rotation, scaling, panning, cropping, flipping and adding noise. Among them, image flipping and image panning mainly enhance the model performance by increasing the amount of data, as well as enhancing the panning isotropy of the model [29]. These changes simulate to a certain extent the various changing situations in the real environment, which can enable the model to adapt to more different application scenarios. The geometric transformation type of image enhancement technique has strong versatility and relatively low computational cost, which can play an effective role in assisting the training of deep learning models [30].
However, in practice, relying only on the above data enhancement methods may still make it difficult to meet the training needs of complex pest and disease identification models. Introducing generative adversarial networks (GANs) to extend the dataset is also a common choice when the sample size is insufficient.
Abbas A et al. [31] expanded the pest and disease dataset using a GAN. The process is shown in Figure 3, where real pest and disease image data are first input into the discriminator model, and at the same time, a random photo is given to the generator model. Then, the generator model generates a fake image of pests and diseases based on the feature labels in the image, and at the same time, inputs this fake image into the discriminator model. Finally, the discriminator is used to differentiate between the true and false data. In the whole training process, the generator and the discriminator confront each other, improve each other, and continuously improve their own abilities until the new data formed by the generator can no longer be recognised by the discriminator and a state of equilibrium is reached. By virtue of the mutual confrontation between the generator and the discriminator, this method can generate high-quality and highly realistic new data, which effectively improves the generalisation ability and robustness of the model. Studies in the literature [31] showed that a significant classification accuracy can be achieved through the above processing, which effectively alleviates the dependence of the deep learning model on a large amount of high-quality labelled data and provides strong support for the timely detection and control of tomato diseases. Kui Hu et al. [32] addressed the problems of small datasets and complex backgrounds in the natural environments of rice pests by using conventional data enhancement methods, such as Gaussian noise, random masking, random brightness and motion blur; the model also adopts a GAN to further expand the dataset and proposes a multi-scale two-branch structure model based on a GAN and improved ResNet network, and the experimental results show that by introducing the GAN for data enhancement, the model identification accuracy was improved by 2.47% compared with the original ResNet-50 model. GANs significantly improve the accuracy and adaptability of deep learning models in pest and disease identification through local feature enhancement, environment diversity extension and cross-modal synthesis in fine-grained data generation.
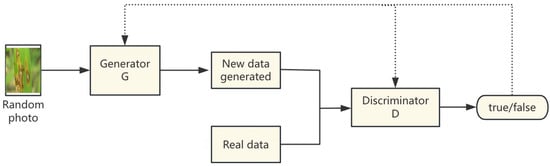
Figure 3.
Flowchart of data enhancement based on GAN.
In addition, several studies have used simulation tools, such as Blender and Unity, to generate new data to generate new images to expand the dataset for pest and disease identification. Bates E et al. [33] used the Unity simulator to construct a dataset to design a synthetic forest environment for generating images of ash leaves in their study of leaf disease sensing in ash trees.
The application of generative adversarial networks (GANs) in data enhancement is not limited to agricultural image generation. For example, in the field of traffic flow prediction, reference [34] proposes a heterogeneous traffic flow prediction framework by fusing visual quantitative features with spatio-temporal data, and its multimodal feature fusion strategy inspires the modelling of complex backgrounds and illumination changes in agricultural images. Similarly, the dynamic trend fusion module proposed in reference [35] optimises time-series features through the adaptive weight assignment for the time-series analysis of agricultural pest and disease data. Furthermore, the bilinear spatio-temporal fusion network in reference [36] demonstrated the potential of lightweight model design by efficiently combining spatial and temporal information, which has implications for edge deployment for real-time pest and disease identification in agricultural scenarios.
3.2. Models
There are a variety of deep learning network models for pest and disease identification, which are discussed in this paper in three categories, CNN, Transformer and CNN–Transformer hybrid models, according to the infrastructure of the models.
3.2.1. CNN-Based Models
Unlike the pest and disease identification models based on traditional machine learning, a convolutional neural network (CNN) has strong feature extraction capability, which can automatically learn and extract features from the original image, and the model has better robustness and generalisation capability. Therefore, CNN-based network models have become the most commonly used in pest and disease classification [37].
In the object identification task, CNN-based identification models can be divided into two-stage models and single-stage models. The two-stage model first needs to form a candidate region and then accurately locate and classify it, so the detection speed is slower, but its accuracy is higher; the single-stage model does not need to form a candidate region, and directly identifies and locates the object in the image, so the detection speed is faster, but the accuracy is lacking. Typical object identification algorithms include AlexNet, VGG, YOLO, SSD, Faster R-CNN and Mask R-CNN, and Figure 4 lists the research progress of some representative object identification algorithms.
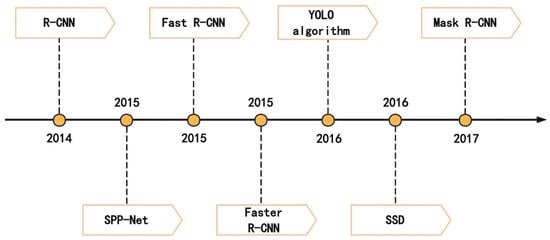
Figure 4.
Research progress of CNN-based target identification algorithm.
In a recent study, Sangha H S et al. [38] defined an agricultural scene as a complex background with low contrast. The effect of the model size and photometric image enhancement methods on single-stage (RetinaNet) and two-stage (Faster-RCNN) models was investigated in low-contrast complex backgrounds. With a single stage, smaller models performed better, while with two stages, larger models improved the performance; all enhancement methods, except the random contrast image enhancement method, significantly improved the model performance.
On complex backgrounds with low contrast, crop pests and diseases display extremely diverse morphological features, including shape, size, texture, colour, background, arrangement and light conditions at different times of the day. The above factors make the identification of pests and diseases extremely complex and challenging, which seriously restricts the accuracy improvement of pest and disease identification algorithms based on traditional machine learning. With the development of CNN-based network models, researchers have proposed many CNN-based solutions for pest and disease identification.
Xu Cong et al. [39] used CNNs for the identification of rice pests and diseases in the field, taking advantage of the feature of CNNs to automatically learn features during the training process, avoiding the errors generated by manual selection. However, due to the pooling operation in this method, the model can only give rough location information. It is not effective at recognising complex or blurred images of pests and diseases and there are limitations in the generalisation ability of the model.
Thakur P S et al. [40], after comparing various types of state-of-the-art models in 2022, found that the VGG-16 model had a deeper network structure; could extract richer image features; and the identification effect was significantly improved compared with the basic CNN, with an accuracy as high as 99.53%, and the model’s generalisation ability was also improved. However, the model had a complex network structure, longer training time, and greater demand for computational resources and storage space, and its performance in object detection still needs to be improved.
In the task of object detection, R-CNN has made breakthrough progress. R-CNN belongs to the two-stage object detection algorithm, which first generates a series of sample candidate frames by the algorithm and then classifies them by a convolutional neural network. The method suffers from the problems of complex training, inefficiency and slow computation speed. To solve the problem of a slow R-CNN, Fast R-CNN and Faster R-CNN were derived. Table 2 provides a systematic comparison of these three models in terms of speed, accuracy and resource usage.

Table 2.
Comparison of three models R-CNN, Fast R-CNN and Faster R-CNN.
In pest and disease detection applications, Gong X et al. [41] introduced Faster R-CNN for apple leaf disease identification with an improvement: advanced Res2Net and feature pyramid network architectures were introduced for extracting reliable multidimensional features. In addition, RoIAlign replaced RoIPool in order to generate accurate candidate regions to localise object positions. Compared with VGG-16, this algorithm not only adopts an integrated process in pest and disease identification but can also achieve high-precision pest and disease region localisation, but it is less computationally efficient and requires a large amount of computational resources and storage space, which is suitable for scenarios that do not require a high level of real-time application. Lei Du et al. [42] proposed an end-to-end model based on the Faster R-CNN for a Pest R-CNN end model for maize leaf pest and disease identification. Based on the original model, an FPN algorithm, deformable convolution and multi-scale strategy were adopted, which improved the object detection accuracies by 12% and 19% compared with Faster R-CNN and YOLOv5, respectively. However, the method still has some problems in real time.
In pest and disease identification, the algorithm complexity and detection accuracy are a pair of conflicting problems. Complex models improve the accuracy of model detection but consume computational resources, affecting the real-time performance; lightweight architectures facilitate real-time diagnosis but reduce the accuracy of object detection at the expense of fine-grained feature extraction. Therefore, according to different research fields and needs, different scholars have optimised CNN models from the above two perspectives.
In terms of the detection accuracy, Rong M et al. [43] proposed an improved model based on Mask R-CNN. The authors used an improved FPN algorithm, which effectively solved the multi-scale problem, improved the weight coefficients, and utilised semantic and localization information to achieve more accurate identification and localisation, and improved the detection accuracy and resolution robustness. Liu S et al. [44] constructed a GA-Mask R-CNN identification model for rice stem borer and rice leafroller and combined it with field detection equipment. The basic idea was to introduce the GAN into the Mask R-CNN network to improve the sensitivity of the model to the pest information and the identification accuracy. The specific process is, first, a single pest sample is extracted based on the biological habit of rice pests; then, the GAN is used to generate the pest samples, generate images of the real environment and engage in deep learning by multiplexed enhancement; and then, the channel attention ECA module is added into the Mask R-CNN, which improves the connection of the residual blocks in the backbone network ResNet101, reduces the training volume of the model and improves the accuracy of the model. In addition, the experimental results proved that the average precision, recall and balanced score F1 were improved by 7.07%, 7.65% and 8.83%, respectively, compared with the original Mask R-CNN.
Lee M G et al. [45] proposed the SegNet algorithm based on semantic segmentation, which is capable of recognising objects one by one and maintaining the integrity of high-frequency details, which is important for the precise identification and counting of pests and diseases. However, since the algorithm requires pixel-level segmentation, the computational complexity is relatively high, the training time is long, and the hardware requirements are also high.
In terms of real-time detection, KR Li et al. [46] proposed a lightweight, location-aware LLA-RCNN model for pest detection and real-time monitoring. The model uses MobileNet V3 to replace the backbone of the original full-size backbone network, which reduces the computational complexity and compresses the size of the model to speed up the pest detection. A Coordinate Attention (CA) block is used to enhance the location information, and a GloU loss function is introduced, which is more accurate than the regression loss function smoothL1 used by the original R-CNN. Experiments demonstrated that the model not only significantly reduced the number of parameters and floating-point computations (FLOPs) but also achieved better performance than some popular pest detection methods and greatly improved the detection speed while ensuring a high accuracy.
To solve the problems of real-time and computational resource requirements of two-stage object detection algorithms, the YOLO (You Only Look Once) algorithm transforms the object detection problem into a regression problem, predicting the bounding box coordinates and object categories directly from the image pixels. Unlike two-stage algorithms, such as R-CNN, the YOLO algorithm completes the analysis of the image at one time and does not require multiple scans of the image, which provides a significant advantage in terms of speed and real-time performance and is suitable for application scenarios that require high speed and low resource consumption. However, the YOLO algorithm needs to be improved regarding the detection of small targets, the detection of complex backgrounds and the detection ability under extreme lighting conditions. Table 3 shows the comparison of the characteristics of the YOLO algorithm and typical object detection algorithms. Overall, two-stage algorithms, such as Faster R-CNN and Mask R-CNN, are better in terms of the detection accuracy; single-stage algorithms, such as YOLO and SSD, are superior in terms of the detection speed. In addition, the RetinaNet algorithm, which uses focal loss (the mathematical expression is shown in Equation (5)) can be used when the categories are unbalanced; and lightweight design algorithms, such as EfficientDet and CenterNet, can be used when the resources are limited.

Table 3.
Comparison of object detection algorithms.
pt is the adjusted predictive probability, reflecting the model’s predicted value for the correct category. αt is the category balancing factor, usually taken as α (positive category weight) or 1 − α (negative category weight), where α ∈ [0,1]. γ is the focusing parameter (focusing parameter), where usually γ ≥ 0.
The comparison of the algorithms in Table 3 not only provides clear criteria for algorithm selection in pest and disease identification (accuracy, speed, resources, data balance) but also reveals that the application of deep learning in this field needs to be closely adapted to the actual agricultural scenarios. Future research will pay more attention to the adaptability of algorithms (e.g., cross-crop generalisation ability) and deployment feasibility (compatibility of edge devices) so as to promote the innovation of the whole chain from laboratory research to field implementation, and to realise the dynamic balance of ‘speed-accuracy-robustness’ under limited resources.
In the field of agriculture, the YOLO algorithm has been rapidly used in the detection and identification of crop pests and diseases and the real-time analysis of images taken by agricultural drones or ground robots.
Since the YOLO algorithm improves the detection speed and reduces the demand for computational resources while guaranteeing the detection accuracy, it has quickly become one of the main models in the current pest and disease identification methods and has made many research advances.
In 2023, Dong Q et al. [47] introduced the YOLO algorithm into pest and disease identification, and its identification accuracy could reach a 92.3% accuracy at a detection speed of 112.56 FPS; meanwhile, the advantages achieved in accuracy and real-time made it an important choice for agricultural pest and disease identification.
Jun Liu et al. [48] improved YOLOv3 to achieve multi-scale feature detection, which can quickly identify the location and category of tomato pests and diseases. MJA Soeb et al. [49] trained YOLOv7 on a self-built dataset, and the experimental results show that the YOLOv7 algorithm outperformed CNN, D-CNN, DNN, the improved DCNN, YOLOv5 and multi-objective image segmentation (Table 4).

Table 4.
YOLO (v3–v7) improvement comparison.
To address the low accuracy of the YOLO algorithm in small object detection, Wang L et al. [50] proposed an improved SSD model, which better solves the problem of small object detection by extracting feature maps at different scales. In addition, it achieves a better balance between detection speed and accuracy.
However, for pest and disease detection with complex backgrounds, CNN models, such as AlexNet, VGG16, MobileNet-V2, Faster R-CNN and YOLO, are all limited by the local perceptual property of the convolutional kernel, which makes it difficult to deal with long-range dependency such that the models have a weak generalisation ability in complex backgrounds and the identification effect is not good [48,49].
As seen above, CNN-based object detection models have their own advantages and disadvantages in the pest and disease identification task. In practical applications, the appropriate model should be selected according to the specific needs and scenarios. For example, in scenes with high real-time requirements, models with faster detection speeds, such as YOLO or SSD, can be selected; in scenes requiring accurate segmentation and counting, pixel semantic segmentation networks, such as SegNet, can be selected. At the same time, it is also possible to consider using multiple models in combination to make full use of their respective advantages and compensate for each other’s shortcomings. In addition, transfer learning approaches are also useful. M Turkoglu et al. [51] proposed a pre-trained convolutional neural network (MLP-CNN) based on multi-model LSTM as an integrated majority voting classifier for detecting plant pests and diseases. The authors considered real-time images obtained from different conditions (e.g., light, background variations) for experiments and constructed a hybrid model for apple pest and disease detection, which showed that the detection rate of up to 15 FPS could satisfy real-time applications and performed better in comparison with the pre-trained deep architecture.
3.2.2. Vision Transformer-Based Models
In comparison with conventional computer vision (CV) methodologies that depend on hand-designed features with restricted generalisation capabilities, CNN, which automatically extracts features through end-to-end learning, has achieved considerable success in recent years and has become the prevailing technology in the CV domain. In the domain of natural language processing (NLP), the Transformer architecture eschews the cyclic structure of the Recurrent Neural Network (RNN) and adopts a self-attentive mechanism to process sequential data. This mechanism has been demonstrated to effectively capture long-range dependencies and achieve global information acquisition [52]. Moreover, the Transformer architecture boasts powerful parallel computing capability, a factor that has contributed to its rapid emergence as the prevailing architecture in the field of NLP.
The self-attention mechanism in a Transformer, akin to a CNN, is also concerned with the extraction of features. Alternatively, an image can be regarded as comprising multiple patches, and by assigning positional encoding to each patch, the image can be converted into a sequence. This approach enables finding effective solutions of image processing problems through the utilisation of Transformer technology. Building on this, Alexey Dosovitskiy et al. [53] proposed the Vision Transformer (ViT) algorithm in 2020, which marked the first real direct application of the Transformer architecture to CV. As shown in Table 5, the main advantage of the Transformer architecture over CNNs is its ability to capture global features and long-range dependencies with good robustness under occlusion noise, making it more suitable for complex tasks requiring global semantic understanding. However, it is important to note its limitations, which include low sensitivity to local details and the inability to capture local fine features (e.g., lines, shapes). Moreover, challenges such as computational efficiency and data dependency emerge as significant obstacles in the practical implementation of a Transformer.

Table 5.
Comparison of Transformer and CNN.
The Transformer’s capabilities for global semantic understanding have been demonstrated to be more accurate than CNN in the identification of crop pests and diseases in complex contexts. Reference [54] utilised the ViT algorithm for the classification of plant leaf diseases, attaining an accuracy of 97.98%. This further substantiates the efficacy of the Transformer architecture in handling CV tasks, such as the detection of pests and diseases. In a related study, Chen Dongmei et al. [55] proposed a target detection model based on the Transformer architecture. This model introduced a multi-scale feature fusion mechanism and utilised multi-scale datasets to enhance the Transformer’s performance in capturing small targets and complex backgrounds. This, in turn, improved the detection accuracy and robustness for multi-scale and dense complex pest detection scenarios. The construction of a small pest dataset (MTC-PAWPD) containing rice flies, aphids and wheat spiders was undertaken, and the design effectiveness of the DINO-based [56] pest detection model was verified. The experimental findings demonstrate that the proposed model attained a mean average precision (mAP@50) of 70.0% on the MTC-PAWPD test set, which signified a 2.3 percentage point enhancement in comparison with other prevalent models, such as AF-RCNN [57]. In related research, Yinshuo Zhang et al. [58] developed a Multimodal Fine-Grained Transformer (MMFGT) architecture for pest identification tasks, incorporating advanced feature extraction mechanisms across multiple sensory modalities. The experimental findings demonstrate that the MMFGT model exhibited a substantial enhancement in performance when compared with the baseline methods, such as ViT and SwinT, particularly on small sample datasets. Furthermore, MMFGT enhanced the accuracy by 17.43% over ResNet101 on a 29-class IDADP pest dataset and 7.74% on a dataset comprising eight tomato pests. In a recent study, Wu et al. [54] proposed a novel segmentation method, termed DS-DETR, for the classification of tomato pests. This approach utilises the DETR network as a foundation, incorporating pre-training to expedite the convergence process of the model. The efficacy of this method was demonstrated by its ability to enhance the accuracy of the model. The DS-DETR model attained an APmask score of 0.6823, which represented 12.87%, 8.25%, 3.67%, 1.95%, 10.27% and 9.52% enhancements over the state-of-the-art Mask RCNN, BlendMask, CondInst, SOLOv2, ISTR and DETR models, respectively. The model’s segmentation results demonstrated an accuracy of 0.9640 in disease classification. QH Cap et al. [59] proposed a LeafGAN algorithm, which integrates CycleGAN and a Transformer. The LeafGAN algorithm augmented the sample dataset using CycleGAN, addressed the challenge of Transformer training reliance on extensive datasets and substantially enhanced the accuracy of cucumber leaf disease diagnosis.
3.2.3. CNN–Transformer Hybrid Models
Notwithstanding the Transformer-based models’ advantageous properties, including its capacity for robust generalisation and the effective handling of complex tasks in the domains of pests and diseases detection, its deployment on devices with limited resources in agriculture is constrained by its substantial computational and spatial complexity. Additionally, Transformer model training necessitates substantial data volumes, which curtails the model’s generalisation capabilities due to the limited size and uneven distribution of categories across all datasets in the agricultural domain. Consequently, the combination of the advantages of CNN’s local feature extraction and Transformer’s advantages in global information modelling to construct a CNN–Transformer hybrid model has become a current research hotspot.
In the CNN–Transformer hybrid model, feature fusion is the key link, and the following are several common feature fusion methods.
(1) Sequential fusion: This mode connects a CNN and Transformer sequentially, as shown in Figure 5. Specifically, it can be divided into two categories. One class uses a CNN to extract local features first, which are then input into a Transformer for global modelling, such as DETR and CoAtNet. This type of model first uses a CNN to extract local features (such as edges and texture) of the image and then spreads the feature map into a sequence and inputs it into a Transformer for global modelling so as to achieve information fusion and processing from local to global. The other category is to process the input sequence with the Transformer first and then refine the local structure by a CNN. For example, using TransGAN [60] in an image generation task, the input sequence is first processed with a Transformer to generate global contextual features, and then the local structure is refined by a GAN to generate finer image details.
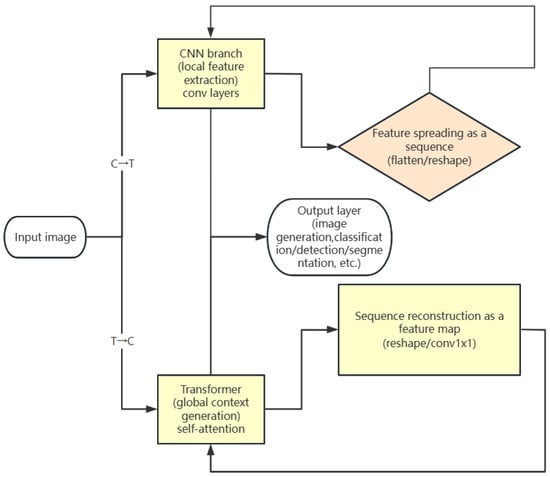
Figure 5.
Serial fusion mode diagram.
(2) Parallel fusion (PF) mode, as shown in Figure 6: Common practices include feature map splicing, weighted sum or attention fusion, such as the CMT model [61], which first processes the inputs by a CNN and Transformer separately, and then combines the results by feature map splicing, weighted sum or attention fusion so that the features extracted by the two models complement each other and enhance the expressive ability of the models. Another common practice is to use the cross-modal attention mechanism to achieve feature dynamic interaction using the CoTr model [62]. It adopts the cross-modal attention mechanism to achieve feature dynamic interaction between the parallel branches of a CNN and Transformer so that the two models can work better together and jointly extract more valuable feature information.
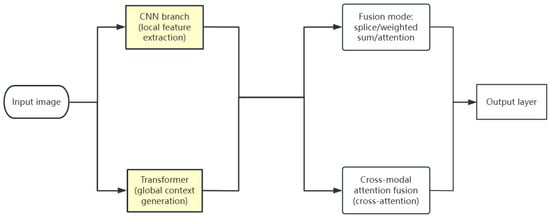
Figure 6.
Parallel fusion model diagram.
(3) Cross-Stage Fusion (CSF) model: The common practice of this model is to borrow the common structure of a CNN to achieve efficient feature extraction, and there are also models that stack CNNs and Transformers alternately to achieve deeper fusion layer by layer. For example, Swin Transformer [63], FPN-Transformer [64] and UNeXt [65] refer to the multi-scale pyramid structure, Resnet structure and UNet structure in CNNs, respectively; these models show better scalability and robustness with complex backgrounds on different tasks and datasets and can efficiently deal with multi-scaled image information in complex backgrounds. The CoAtNet [66] model deepens feature fusion layer by layer by stacking CNNs and Transformers alternately so that the model can make full use of the advantages of the two models at different levels to extract richer and more representative features (Figure 7).
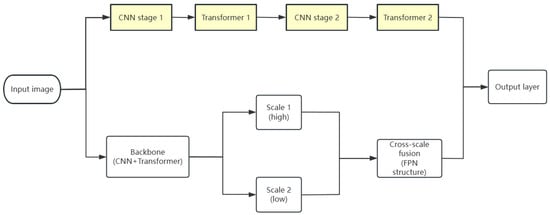
Figure 7.
Cross-phase integration model diagram.
(4) Hybrid model: Aiming at the data characteristics, model deployment environment and other special needs, multiple fusion methods are used in the fusion mode. For example, Yanghao Li et al. [67] proposed the MViTv2 model for the problem of large differences in target size in the target detection task. This model improves the performance in multi-scale vision tasks by introducing an Improved Pooling Attention mechanism, combining window attention and pooling attention, and using multi-scale feature extraction while reducing the computational and memory overheads, making the model more advantageous when dealing with different-sized targets. MobileViTv3 [68] improves the accuracy of the model while optimising the latency and throughput by splicing features from local and global feature blocks and introducing residual concatenation, making it more suitable for deployments on mobile devices and meeting the needs of applications in resource-constrained environments.
In the pest and disease identification task, Meng Zhang et al. [69] addressed the problem of low identification accuracy of traditional pest identification methods in the face of diverse pest species, different sizes, varied morphologies and complex field backgrounds, and based on the ResNet50 architecture, introduced the Relation-aware Global Attention (RGA) module, Multi-scale Feature Fusion (MSFF) module and Generalised Mean Pooling (GeMP) module to design an AM-MSFF network. The experimental results show that AM-MSFF achieved an accuracy of 72.64% on the IP102 dataset and 99.05% on the D0 dataset, which was better than most of the existing methods. Ruotong Yang et al. [70], in response to the problem of insufficient accuracy in cucumber leaf spot segmentation under natural conditions, proposed an ECA-SegFormer-based cucumber leaf spot semantic segmentation method based on ECA-SegFormer. The method introduces the ECA module into the decoder of SegFormer, which enhances the model’s ability to capture important information by extracting attention in the channel dimension. Meanwhile, the multi-scale feature fusion using the FPN module improves the scale robustness of feature representation. The experimental results show that the model outperformed DeepLabV3+, U-Net, PSPNet, HRNet, SETR and SegFormer in both the mIoU and MPA metrics on the self-built dataset. Xiaopeng Li et al. [71], in order to solve the problems of the complex background of apple leaf disease and the low contrast between the disease region and the background, proposed a lightweight apple disease identification model based on the alternating fusion of CNNs and Transformers called ConvViT, which effectively reduces the number of parameters and computational complexity of the model by improving the patch embedding method and adopting depth-separable convolution. The experimental results show that on the self-built dataset, its identification accuracy was comparable with that of the Swin-Tiny model, but the number of parameters and FLOPs were only 32.7% and 21.7% of those of Swin-Tiny, which makes it more feasible to be deployed in resource-constrained edge devices. Atul Kumar Dixit et al. [72], for the insufficient accuracy of feature extraction in rice images, proposed a DST model, where D stands for DCNN, S stands for SVM and T stands for transfer learning, which effectively improves the efficiency of feature extraction and classification accuracy, and the experimental results show that the model achieved a 95% training accuracy and 85% validation accuracy in multiple datasets, which significantly improved the accuracy of detection and classification of rice plant diseases.
As can be seen from the above, the current deep learning-based pest and disease identification methods have significantly surpassed the traditional machine learning methods in terms of feature expression ability and identification accuracy. Among them, CNN-based models, as the mainstream methods, can efficiently extract local features, such as the texture and spots of pests and diseases, by virtue of the local sensory field characteristics of the convolutional kernel and reach the practical level in tasks such as leaf spot segmentation [73] and pest counting [74]. ViT et al.’s model based on a Transformer, which relies on its global feature extraction advantage, has an outstanding performance in the identification of targets under complex backgrounds, and is especially applicable to large scales. The performance is outstanding, and it is especially suitable for long-distance dependent modelling of pests and diseases in large-scale field images, such as the identification of disease propagation patterns between plants. However, Transformer-based models require high computational resources and memory, and large-scale data samples are needed for training. CNN–Transformer hybrid models combine CNN and Transformer features through certain fusion strategies so that the model enhances the global semantic understanding in complex backgrounds while maintaining the sensitivity of the local details, such as CoATNeT [66]. Hybrid models are able to better distinguish between similar diseases and natural aging spots. However, the research on hybrid models is still in its infancy, and there are still many problems that need to be solved.
4. Discussion
The crop pest and disease identification task is unique and complex in the CV domain, and this section discusses the key challenges and possible solutions for crop pest and disease identification methods based on deep learning in terms of the following aspects.
4.1. Small Sample Size of Data
The training of models based on deep learning, especially those based on the Transformer architecture, is highly dependent on the size and quality of the training data. However, in the field of crop pest and disease identification, there is a significant gap between available datasets and model requirements. First, the sizes of publicly available datasets are limited. Currently, there are relatively few public datasets and limited sample sizes for agricultural pest and disease identification. The Plant Village dataset and IP102 dataset, for example, have 54,305 plant leaf images (including disease and health) and 75,222 pest images, which are orders of magnitude different from the common datasets in the CV domain, and are only 0.4% and 0.5% of the ImageNet dataset (14 million annotated images) and much lower than the amount of data required for regular pre-training of Transformer models. Taking the ViT model as an example, its demand for data volume in the pre-training process shows an obvious scale effect. The performance is not as good as ResNet on the ImageNet-1K (1.3M sheets) small dataset, which is comparable with ResNet on the ImageNet-21K (14M) medium- and large-scale datasets and significantly outperforms ResNet on the JFT-300M (303M) mega dataset. Second, the self-built dataset faces the difficulty of data collection. Data annotation requires the participation of plant protection experts, and the annotation cost is high. In addition, the dataset is not balanced in terms of categories, and there are fewer data samples of certain pests and diseases, which also leads to the lack of identification ability of the model for a few categories during the training process.
To address the above problems, on the one hand, there is a need to expand data collection in collaboration with agricultural research institutions and agricultural practitioners. By sharing data resources, a larger open dataset can be constructed. At the same time, technologies, such as drones and IoT devices, are utilised to collect data automatically and improve the efficiency and quality of the data collection. On the other hand, with the existing dataset size, the following methods can be used to alleviate the problem of insufficient data. For example, a GAN is used to synthesise realistic and high-quality images of pests and diseases in order to enrich the diversity of data and expand the dataset. In addition, transfer learning is also a commonly used and effective solution to solve the problem of insufficient data samples. It utilises models pre-trained on large-scale datasets and fine-tuned with a small amount of new data to migrate them to a specific crop pest and disease identification task to achieve better performance on a limited dataset. Finally, federated learning can also be introduced to achieve collaborative modelling based on data illiquidity, which can effectively solve the problem of data silos while meeting privacy protection needs.
4.2. Diversity of Pest and Disease Patterns
The wide variety of pests and diseases, their diverse forms, and the fact that the forms change in different geographical areas and at different times of the year pose great challenges to the task of identifying and classifying pests and diseases. This is specifically manifested in small-object identification. Some pests and diseases account for a small proportion of the image, the characteristics are not significant enough, and some pests and diseases account for only about 1% of the image, which greatly increases the identification difficulty; regarding multi-scale feature extraction, this is mainly reflected in the pest and disease identification and classification task of the size and shape of the individual large differences. In terms of fine-grained identification, there exists a high degree of similarity between the classes and a high degree of intra-class variability in the external morphology. For example, different species of aphids may be very similar morphologically, but their resistance to specific pesticides is different; the aphid species can be accurately judged through fine-grained identification, and then appropriate pesticides can be selected for control. Taking aphids and leafhoppers as examples, these two pests have many morphological similarities, which makes the existing identification models misjudge up to 35% in practical applications (Figure 8 and Figure 9).
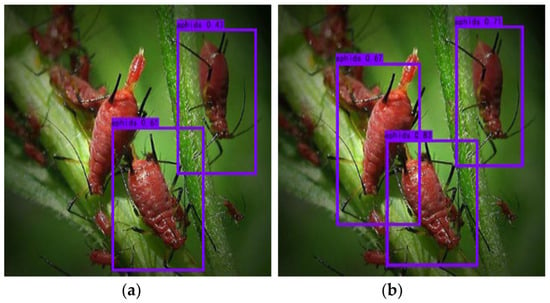
Figure 8.
Missed detections in multi-scale feature extraction [75].

Figure 9.
Morphological similarities in fine-grained recognition: (a) aphids and (b) leafhoppers.
In addition, there exists variability in pest and disease symptoms exhibited across regions and time periods. To address the above problems, the following solutions can be adopted:
(1) Enhanced local texture feature enhancement: Taking aphids and leafhoppers as examples, the method can focus on extracting the fine texture of their body’s surface, such as the tiny bumps on the body surface. Then, the local texture features are dynamically and accurately extracted and quantified with the help of algorithms, such as texture analysis. On this basis, the cross-layer non-local module fuses multi-scale spatial features through Reinforcement Learning (RL) to improve the ability to capture the edges and textures of the spots and enhance the identification accuracy of the model regarding the fine features.
(2) Adoption of multimodal data fusion: Multi-spectral, thermal imaging and other data commonly used in the agricultural field and various types of sensor data are fused. In the data-preprocessing stage, the multimodal data are standardised and normalised to eliminate the differences in scale and noise interference between different modal data. In the feature extraction stage, corresponding feature extraction methods are adopted for the characteristics of different modal data. Finally, the fusion method based on the attention mechanism can be used to dynamically assign the weights of different modal features through self-attention and cross-attention modules, highlighting important features and suppressing irrelevant features. In addition, a fusion method based on a graph neural network can be adopted to construct the multimodal data into a graph structure and perform feature fusion using a graph convolution network (GCN) or graph attention network (GAT) to deal with the complex relationship and topology between multimodal data.
4.3. Adaptation Challenges in Complex Backgrounds
There are still significant limitations in the effectiveness of the current crop pest and disease identification models in real production scenarios. The specific performance is as follows: first, the scene generalisation ability is insufficient. In the existing training dataset, the laboratory environment samples account for a large proportion of the lighting conditions and shooting angles, and the background complexity is significantly different from the real farmland environment, especially regarding rainy weather, plant shading and other complex conditions; the laboratory samples cannot fully simulate the diversity of real farmland. Second, the problem of cross-domain feature drift. For example, there are geographic domain differences (the difference in the disease spot area between rice blast in the Yangtze River Valley and Northeast China is 40–60 mm2), temporal domain differences (the coefficients of variation of phenotypic features of the same disease are different at different fertility stages of the crop), varietal domain differences (there is a shift in the key features in disease characterisation between disease-resistant varieties and susceptible varieties), equipment domain differences (there are differences in the features captured by multispectral cameras and visible light cameras) and spatial differences (Figure 10).

Figure 10.
Problems with cross-domain feature drift in plant diseases. The top and bottom images in column (A) are from the laboratory environment and the field environment (geographic area differences). The images in column (B) are apple leaves with rust (temporal domain differences). In column (C), the top and bottom images are potato and tomato leaves with late blight, respectively (varietal domain differences) [76].
To address the above problems, on the one hand, data enhancement techniques, such as a GAN, can be used to achieve the migration of laboratory data to field backgrounds (e.g., light, cloudy and rainy) to increase the diversity of the data. By generating realistic field environment images, the training dataset is enriched to improve the model’s adaptability to different scenes. On the other hand, multi-domain joint training is used to introduce Domain Adaptation (DA) algorithms, such as Adversarial Domain Alignment (DANN), to reduce the distributional differences by means of feature space alignment. In addition, a meta-learning framework can also be introduced to design feature extractors that are shared across crops, combined with MAML (Model-Agnostic Meta-Learning) to achieve rapid adaptation for pest and disease identification in new scenarios. The meta-learning framework enables the model to quickly adapt to new scenarios with a small number of samples and improves the generalisation ability and adaptability of the model. In addition, multimodal data fusion is also an effective method to solve the problem.
4.4. Model Demands for Lightweight and Real-Time Performance
Model lightweighting and real-time optimisation are the core aspects of crop pest and disease identification systems. On the one hand, models are usually deployed on mobile terminals (e.g., drones and mobile phones) or edge computing devices, which require lightweight processing due to the limitations of power consumption, storage and computing resources of the terminal devices. The main contradiction faced by lightweight processing is the trade-off between accuracy, generalisation ability and efficiency. On the other hand, the dynamics and complexity of the farmland environment require the model to complete real-time reasoning with a low latency. For example, in the process of precision pest and disease control, plant protection machinery is required to respond quickly to adjust the amount of medicine applied, while the traditional model often fails to meet the real-time requirements due to the high computational complexity in processing high-resolution image input.
To address the above problems, efficient and lightweight network architectures, such as MobileNet, ShuffleNet or EfficientNet, can be developed, and the models can be customised for specific hardware platforms to balance the algorithmic power and accuracy, making them suitable for resource-constrained equipment. Model compression methods, such as distillation, pruning or quantisation, can also be used to reduce the number of model parameters and computational complexity; in addition, edge–cloud collaborative inference mechanisms can be added to perform preliminary screening of lightweight models on mobile or edge devices and upload complex samples to the cloud for high-precision analysis in order to balance the needs of real-time and accuracy.
5. Conclusions
Compared with traditional machine learning, pest and disease identification models based on deep learning have the ability to automatically learn features and process more complex data, significantly improving the model identification accuracy. In this paper, the infrastructure of the model and the pest and disease identification methods based on deep learning are divided into three categories: CNN, Transformer and CNN–Transformer hybrid models, and the characteristics, advantages and shortcomings of the common models in these three categories, as well as the related application research in pest and disease identification, are deeply analysed. Specifically, identification methods based on CNN are relatively mature and have been widely adopted in practical applications; the Transformer, based on its advantage of global semantic understanding, shows a higher accuracy than CNN in dealing with crop pest and disease detection tasks with complex backgrounds. However, the Transformer also has the problems of large computational and spatial complexity, and relies on large-scale training samples, which limits its application in the field of agriculture to a certain extent; the approach based on the hybrid model of CNN–Transformer integrates the advantages of a CNN and Transformer, and shows greater application potential in the field of crop pest and disease identification potential. However, this method started relatively late and still needs further in-depth research in terms of a feature fusion strategy and new model architecture.
This paper concludes with a detailed discussion of the four main challenges faced by crop pest and disease identification techniques in practical applications, i.e., small data samples, diverse pest and disease morphologies, the influence of complex backgrounds, and the need for model lightweighting and real-time performance, and proposes corresponding solutions or future research directions.
Overall, future research should focus on the following key directions: (1) Construct high-quality large-scale datasets to provide richer and more representative data resources for model training in order to improve the generalisation ability and identification accuracy of the model. (2) Further improve the model’s identification accuracy and generalisation ability in complex scenarios, such as small-target identification, fine-grained identification, multi-scale identification and complex backgrounds, while taking a real-time model into consideration. (3) Consider the lightweight deployment of the model on resource-constrained agricultural equipment, optimise the model structure and algorithms, reduce the dependence of the model on hardware resources, and improve the deployability and application efficiency of the model in the actual agricultural production environment. (4) Study the fusion methods of remote sensing, multi-spectral, sensors, and other multi-sources of data, and explore the relationship between agronomic laws and AI models. The deep fusion of agronomic laws and AI models, as well as the edge–cloud collaborative inference mechanism, can be used to promote the application of pest and disease identification methods in the field of intelligent agriculture through multidisciplinary crossover and technological innovation, and to provide more accurate and efficient pest and disease monitoring and control solutions for agricultural production.
Funding
This research was funded by the [The Agricultural Machinery Purchase Subsidy Business Management Project of the Ministry of Agriculture and Rural Affairs], grant number [29012308]. And the APC was funded by the [China Agricultural University].
Acknowledgments
This project was supported by the Agricultural Machinery Purchase Subsidy Business Management Project of the Ministry of Agriculture and Rural Affairs (No. 29012308), Yantai Municipal Education Bureau 2023 campus integration project (No. 2023XDRHXMPT12) and Yantai Science and Technology Plan Project (2022XDRH025).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zhang, X.; Yin, H.; Zhuang, C.; Ren, G.; Li, C.; Yang, Q.; Zhou, Y.; Feng, B. Current Situation and Analysis of the Standardization of Crop Pests, Diseases and Weeds. Stand. Sci. 2024, (Suppl. S2), 132–139. [Google Scholar]
- Li, Z.; Li, B.; Li, Z.; Zhan, Y.; Wang, L.; Gong, Q. Research progress in crop disease and pest identification based on deep learning. Hubei Agric. Sci. 2023, 62, 165–169. [Google Scholar]
- Wang, B.; Yang, M.; Cao, P.; Liu, Y. A novel embedded cross framework for high-resolution salient object detection. Appl. Intell. 2025, 55, 277. [Google Scholar] [CrossRef]
- Cai, H.; Wang, Y.; Luo, Y.; Mao, K. A Dual-Channel Collaborative Transformer for continual learning. Appl. Soft Comput. 2025, 171, 112792. [Google Scholar] [CrossRef]
- Saro, J.; Kavit, A. Review: Study on simple K-mean and modified K-mean clustering technique. Int. J. Comput. Sci. Eng. Technol. 2016, 6, 279–281. [Google Scholar]
- Suykens, J.A.K.; Vandewalle, J. Least squares support vector machine classifiers. Neural Process. Lett. 1999, 9, 293–300. [Google Scholar] [CrossRef]
- Jin, Y. Image Recognition of Four Kinds of Fruit Tree Diseases. Master’s Thesis, Liaoning Normal University, Dalian, China, 2021. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 2, 273–297. [Google Scholar] [CrossRef]
- Pearson, K. On lines and planes of closest fit to systems of points in space. Philos. Mag. 1901, 2, 559–572. [Google Scholar] [CrossRef]
- Pantazi, X.E.; Moshou, D.; Tamouridou, A.A. Automated leaf disease detection in different crop species through image features analysis and One Class Classifiers. Comput. Electron. Agric. 2019, 156, 96–104. [Google Scholar] [CrossRef]
- Li, Y. Research on Image Segmentation Algorithm Based on Superpixel and Graph Theory. Master’s Thesis, Chongqing University, Chongqing, China, 2020. [Google Scholar]
- Liu, H.; Zhu, S.; Shen, Y.; Tang, J. Fast Segmentation Algorithm of Tree Trunks Based on Multi-feature Fusion. Trans. Chin. Soc. Agric. Mach. 2020, 51, 221–229. [Google Scholar]
- Zhang, N. Color Image Segmentation Based on Contrast and GrabCut. Master’s Thesis, Hebei University, Baoding, China, 2018. [Google Scholar]
- Li, K.; Feng, Q.; Zhang, J. Co-Segmentation Algorithm for Complex Background Image of Cotton Seedling Leaves. J. Comput.-Aided Des. Comput. Graph. 2017, 29, 1871–1880. [Google Scholar]
- Lei, Y.; Han, D.; Zeng, Q.; He, D. Grading Method of Disease Severity of Wheat Stripe Rust Based on Hyperspectral Imaging Technology. Trans. Chin. Soc. Agric. Mach. 2018, 49, 226–232. [Google Scholar]
- Pan, Y.; Zhang, H.; Yan, J.; Zhang, H. Source Identification of Radix Glycyrrhizae(Licorice) Based on the Fusion of Hyperspectral and Texture Feature. J. Instrum. Anal. 2024, 43, 1745–1753. [Google Scholar]
- Dong, C.; Yang, T.; Chen, Q.; Liu, L.; Xiao, X.; Wei, Z.; Shi, C.; Shao, Y.; Gao, D. Application of hyperspectral imaging technology in non-destructive detection of apple quality. J. Fruit Sci. 2024, 41, 2582–2594. [Google Scholar]
- Mei, X.; Hu, Y.; Zhang, H.; Cai, Y.; Luo, K.; Meng, Y.; Song, Y.; Shan, W. Evaluation of drought status of potato leavesbased on hyperspectral imaging. Agric. Res. Arid. Areas 2024, 42, 246–254. [Google Scholar]
- Liu, Y.; Yin, Y.; Liu, J.; Yin, Y.; Chu, T.; Jiang, Y. Characterization and identification of weeds in the field using hyperspectral imaging. J. Tarim Univ. 2024, 36, 89–97. [Google Scholar]
- Singh, V. Sunflower leaf diseases detection using image segmentation based on particle swarm optimization. Artif. Intell. Agric. 2019, 3, 62–68. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, M.; Shen, Z.; Chen, C.; Fang, S.; Du, K.; Yang, L.; Deng, T. Research on the Number of Crack in Wood Based on Acoustic Emission. For. Eng. 2025, 41, 59–66. [Google Scholar]
- Qiao, X.; Pan, X.; Wang, X.; Peng, J.; Zhao, X. Image segmentation of potato pests and diseases based on g-r component and k-means. J. Inn. Mong. Agric. Univ. (Nat. Sci. Ed.) 2021, 42, 84–87. [Google Scholar]
- Zhou, R.; Xi, J.; Ding, Y.; Duan, J.; Qi, B.; Yao, T.; Dong, S.; Liu, Y.; Ding, C.; Yang, G.; et al. Research on Curing Tobacco Image Segmentation Based on K-means Clustering Algorithm. J. Anhui Agric. Sci. 2024, 52, 232–237. [Google Scholar]
- Li, H.; Liu, J.; Wu, K. Image segmentation based on K-means algorithm. Mod. Comput. 2024, 30, 49–51+91. [Google Scholar]
- Lyu, S.; Yang, H.; Fan, X. Image recognition technology for potato diseases and pests based on FT and superpixel fuzzy C-means clustering. Digit. Microgr. Imaging 2023, 3–5. [Google Scholar]
- Yuan, Q.; Deng, H.; Wang, X. Citrus disease and insect pest area segmentation based on superpixel fast fuzzy C-means clustering and support vector machine. J. Comput. Appl. 2021, 41, 563–570. [Google Scholar]
- Liu, J.; Wang, X. Plant diseases and pests detection based on deep learning: A review. Plant Methods 2021, 17, 22. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, Z.; Liu, X.; Zeng, N.; Liu, Y.; Alsaadi, F.E. A survey of deep neural network architectures and their applications. Neurocomputing 2017, 234, 11–26. [Google Scholar] [CrossRef]
- Zhang, Y.; Wa, S.; Zhang, L.; Lv, C. Automatic plant disease detection based on tranvolution detection network with GAN modules using leaf images. Front. Plant Sci. 2022, 13, 875693. [Google Scholar] [CrossRef]
- Thenmozhi, K.; Reddy, U.S. Crop pest classification based on deep convolutional neural network and transfer learning. Comput. Electron. Agric. 2019, 164, 104906. [Google Scholar] [CrossRef]
- Abbas, A.; Jain, S.; Gour, M.; Vankudothu, S. Tomato plant disease detection using transfer learning with C-GAN synthetic images. Comput. Electron. Agric. 2021, 187, 106279. [Google Scholar] [CrossRef]
- Hu, K.; Liu, Y.M.; Nie, J.; Zheng, X.; Zhang, W.; Liu, Y.; Xie, T. Rice pest identification based on multi-scale double-branch GAN-ResNet. Front. Plant Sci. 2023, 14, 1167121. [Google Scholar] [CrossRef]
- Bates, E.; Popović, M.; Marsh, C.; Clark, R.; Kovac, M.; Kocer, B.B. Leaf level Ash Dieback Disease Detection and Online Severity Estimation with UAVs. IEEE Access 2025, 13, 55499–55511. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, J.; Song, Y.; LI, X.; Xu, W. Fusing visual quantified features for heterogeneous traffic flow prediction. Promet-Traffic Transp. 2024, 36, 1068–1077. [Google Scholar] [CrossRef]
- Chen, J.; Ye, H.; Ying, Z.; Sun, Y.; Xu, W. Dynamic Trend Fusion Module for Traffic Flow Prediction. arXiv 2025, arXiv:2501.10796. [Google Scholar] [CrossRef]
- Chen, J.; Pan, S.; Peng, W.; Xu, W. Bilinear Spatiotemporal Fusion Network: An efficient approach for traffic flow prediction. Neural Netw. 2025, 187, 107382. [Google Scholar] [CrossRef] [PubMed]
- Rahman, C.R.; Arko, P.S.; Ali, M.E.; Khan, M.A.I.; Apon, S.H.; Nowrin, F.; Wasif, A. Identification and recognition of rice diseases and pests using convolutional neural networks. Biosyst. Eng. 2020, 194, 112–120. [Google Scholar] [CrossRef]
- Sangha, H.S.; Darr, M.J. Influence of Model Size and Image Augmentations on Object Detection in Low-Contrast Complex Background Scenes. AI 2025, 6, 52. [Google Scholar] [CrossRef]
- Xu, C.; Yu, C.; Zhang, S.; Wang, X. Multi-scale convolution-capsule network for crop insect pest recognition. Electronics 2022, 11, 1630. [Google Scholar] [CrossRef]
- Thakur, P.S.; Sheorey, T.; Ojha, A. VGG-ICNN: A Lightweight CNN model for crop disease identification. Multimed. Tools Appl. 2023, 82, 497–520. [Google Scholar] [CrossRef]
- Gong, X.; Zhang, S. A high-precision detection method of apple leaf diseases using improved faster R-CNN. Agriculture 2023, 13, 240. [Google Scholar] [CrossRef]
- Du, L.; Sun, Y.; Chen, S.; Feng, J.; Zhao, Y.; Yan, Z.; Zhang, X.; Bian, Y. A novel object detection model based on faster R-CNN for spodoptera frugiperda according to feeding trace of corn leaves. Agriculture 2022, 12, 248. [Google Scholar] [CrossRef]
- Rong, M.; Wang, Z.; Ban, B.; Guo, X. Pest Identification and Counting of Yellow Plate in Field Based on Improved Mask R-CNN. Discret. Dyn. Nat. Soc. 2022, 2022, 1913577. [Google Scholar] [CrossRef]
- Liu, S.; Fu, S.; Hu, A.; Ma, P.; Hu, X.; Tian, X.; Zhang, H.; Liu, S. Research on Insect Pest Identification in Rice Canopy Based on GA-Mask R-CNN. Agronomy 2023, 13, 2155. [Google Scholar] [CrossRef]
- Lee, M.G.; Cho, H.B.; Youm, S.K.; Kim, S.W. Detection of pine wilt disease using time series UAV imagery and deep learning semantic segmentation. Forests 2023, 14, 1576. [Google Scholar] [CrossRef]
- Li, K.R.; Duan, L.J.; Deng, Y.J.; Liu, J.L.; Long, C.F.; Zhu, X.H. Pest Detection Based on Lightweight Locality-Aware Faster R-CNN. Agronomy 2024, 14, 2303. [Google Scholar] [CrossRef]
- Dong, Q.; Sun, L.; Han, T.; Cai, M.; Gao, C. PestLite: A novel YOLO-based deep learning technique for crop pest detection. Agriculture 2024, 14, 228. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X. Tomato diseases and pests detection based on improved Yolo V3 convolutional neural network. Front. Plant Sci. 2020, 11, 898. [Google Scholar] [CrossRef]
- Soeb, M.J.A.; Jubayer, M.F.; Tarin, T.A.; AI Mamun, M.R.; Ruhad, F.M.; Parven, A.; Mubarak, N.M.; Karri, S.L.; Meftaul, I.M. Tea leaf disease detection and identification based on YOLOv7 (YOLO-T). Sci. Rep. 2023, 13, 6078. [Google Scholar] [CrossRef]
- Wang, L.; Shi, W.; Tang, Y.; Liu, Z.; He, X.; Xiao, H.; Yang, Y. Transfer Learning-Based Lightweight SSD Model for Detection of Pests in Citrus. Agronomy 2023, 13, 1710. [Google Scholar] [CrossRef]
- Türkoğlu, M.; Hanbay, D. Plant disease and pest detection using deep learning-based features. Turk. J. Electr. Eng. Comput. Sci. 2019, 27, 1636–1651. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 6000–6010. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Wu, J.; Wen, C.; Chen, H.; Ma, Z.; Zhang, T.; Su, H.; Yang, C. DS-DETR: A model for tomato leaf disease segmentation and damage evaluation. Agronomy 2022, 12, 2023. [Google Scholar] [CrossRef]
- Chen, D.; Lin, J.; Wang, H.; Wu, K.; Lu, Y.; Zhou, X.; Zhang, J. Pest detection model based on multi-scale dataset. Trans. Chin. Soc. Agric. Eng. 2024, 40, 196–206. [Google Scholar]
- Zhang, H.; Li, F.; Liu, S.; Zhang, L.; Su, H.; Zhu, J.; Ni, L.M.; Shum, H.Y. Dino: Detr with improved denoising anchor boxes for end-to-end object detection. arXiv 2022, arXiv:2203.03605. [Google Scholar]
- Jiao, L.; Dong, S.; Zhang, S.; Xie, C.; Wang, H. AF-RCNN: An anchor-free convolutional neural network for multi-categories agricultural pest detection. Comput. Electron. Agric. 2020, 174, 105522. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, L.; Yuan, Y. Multimodal fine-grained transformer model for pest recognition. Electronics 2023, 12, 2620. [Google Scholar] [CrossRef]
- Cap, Q.H.; Uga, H.; Kagiwada, S.; Iyatomi, H. Leafgan: An effective data augmentation method for practical plant disease diagnosis. IEEE Trans. Autom. Sci. Eng. 2020, 19, 1258–1267. [Google Scholar] [CrossRef]
- Jiang, Y.; Chang, S.; Wang, Z. Transgan: Two pure transformers can make one strong gan, and that can scale up. Adv. Neural Inf. Process. Syst. 2021, 34, 14745–14758. [Google Scholar]
- Guo, J.; Han, K.; Wu, H.; Tang, Y.; Chen, X.; Wang, Y.; Xu, C. Cmt: Convolutional neural networks meet vision transformers. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 12175–12185. [Google Scholar]
- Shen, Z.; Fu, R.; Lin, C.; Zheng, S. COTR: Convolution in transformer network for end to end polyp detection. In Proceedings of the 2021 7th International Conference on Computer and Communications (ICCC), Chengdu, China, 10–13 December 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 1757–1761. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Nashville, TN, USA, 20–25 June 2021; pp. 10012–10022. [Google Scholar]
- Zhang, T.; Li, K.; Chen, X.; Zhong, C.; Luo, B.; Grijalva, I.; McCornack, B.; Flippo, D.; Sharda, A.; Wang, G.; et al. Aphid cluster recognition and detection in the wild using deep learning models. Sci. Rep. 2023, 13, 13410. [Google Scholar] [CrossRef] [PubMed]
- Chang, Z.; Xu, M.; Wei, Y.; Lian, J.; Zhang, C.; Li, C. UNeXt: An Efficient Network for the Semantic Segmentation of High-Resolution Remote Sensing Images. Sensors 2024, 24, 6655. [Google Scholar] [CrossRef]
- Dai, Z.; Liu, H.; Le, Q.V.; Tan, M. Coatnet: Marrying convolution and attention for all data sizes. Adv. Neural Inf. Process. Syst. 2021, 34, 3965–3977. [Google Scholar]
- Li, Y.; Wu, C.Y.; Fan, H.; Mangalam, K.; Xiong, B.; Malik, J.; Feichtenhofer, C. Mvitv2: Improved multiscale vision transformers for classification and detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–22 June 2022; pp. 4804–4814. [Google Scholar]
- Mehta, S.; Rastegari, M. Mobilevit: Light-weight, general-purpose, and mobile-friendly vision transformer. arXiv 2021, arXiv:2110.02178. [Google Scholar]
- Zhang, M.; Yang, W.; Chen, D.; Fu, C.; Wei, F. AM-MSFF: A Pest Recognition Network Based on Attention Mechanism and Multi-Scale Feature Fusion. Entropy 2024, 26, 431. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Guo, Y.; Hu, Z.; Gao, R.; Yang, H. Semantic segmentation of cucumber leaf disease spots based on ECA-SegFormer. Agriculture 2023, 13, 1513. [Google Scholar] [CrossRef]
- Li, X.; Li, S. Transformer help CNN see better: A lightweight hybrid apple disease identification model based on transformers. Agriculture 2022, 12, 884. [Google Scholar] [CrossRef]
- Dixit, A.K.; Verma, R. Advanced hybrid model for multi paddy diseases detection using deep learning. EAI Endorsed Trans. Pervasive Health Technol. 2023, 9. [Google Scholar] [CrossRef]
- Yu, H.; Song, J.; Chen, C.; Heidari, A.A.; Liu, J.; Chen, H.; Zaguia, A.; Mafarj, M. Image segmentation of Leaf Spot Diseases on Maize using multi-stage Cauchy-enabled grey wolf algorithm. Eng. Appl. Artif. Intell. 2022, 109, 104653. [Google Scholar] [CrossRef]
- Hong, S.J.; Nam, I.; Kim, S.Y.; Kim, E.; Lee, C.H.; Ahn, S.; Park, I.K.; Kim, G. Automatic pest counting from pheromone trap images using deep learning object detectors for matsucoccus thunbergianae monitoring. Insects 2021, 12, 342. [Google Scholar] [CrossRef]
- Kang, J.; Zhang, W.; Xia, Y.; Liu, W. A Study on Maize Leaf Pest and Disease Detection Model Based on Attention and Multi-Scale Features. Appl. Sci. 2023, 13, 10441. [Google Scholar] [CrossRef]
- Wu, X.; Fan, X.; Luo, P.; Choudhury, S.D.; Tjahjadi, T.; Hu, C. From laboratory to field: Unsupervised domain adaptation for plant disease recognition in the wild. Plant Phenomics 2023, 5, 0038. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).