Association Analysis and Identification of Candidate Genes for Sorghum Coleoptile Color
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Phenotypic Identification and Anthocyanin Content Analysis
2.3. Genome-Wide Association (GWAS) Analysis
2.4. Quantitative Real-Time PCR (qPCR) Analysis
2.5. Statistical Analysis
3. Results
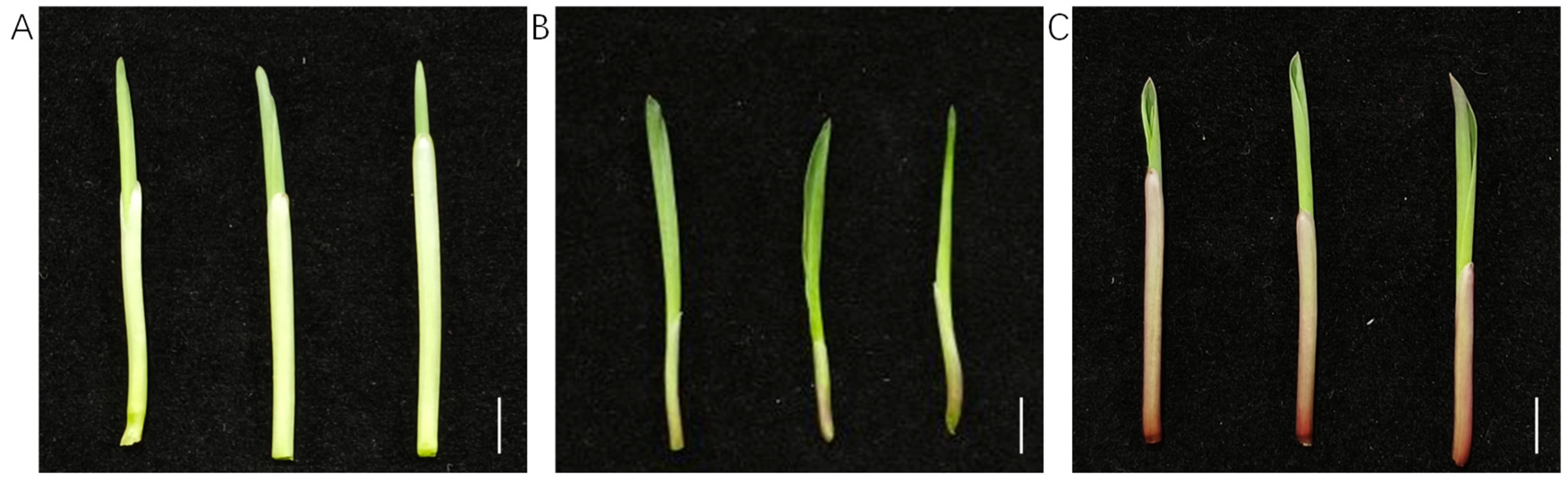
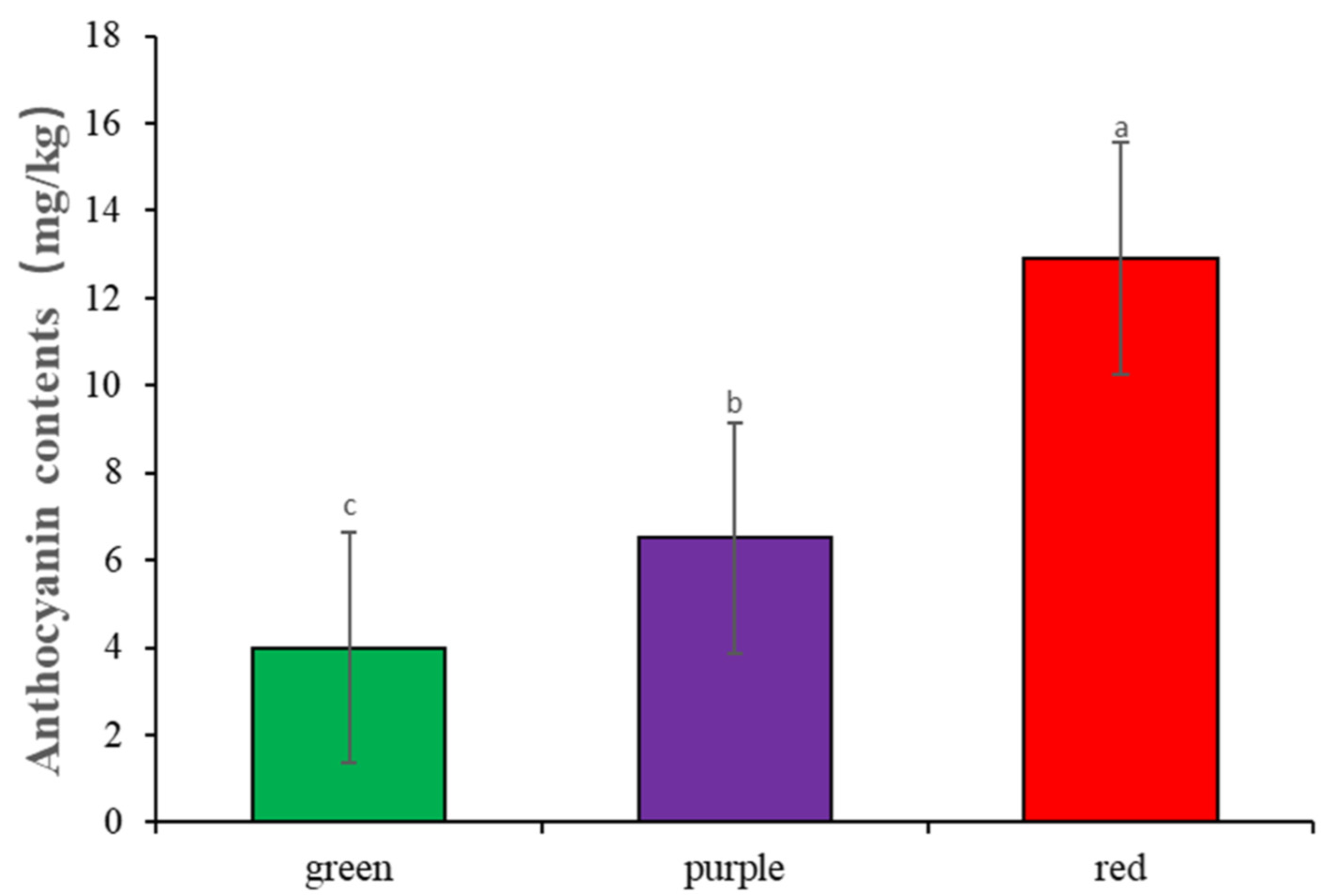
3.1. Phenotypic Analysis of Sorghum Coleoptile Color
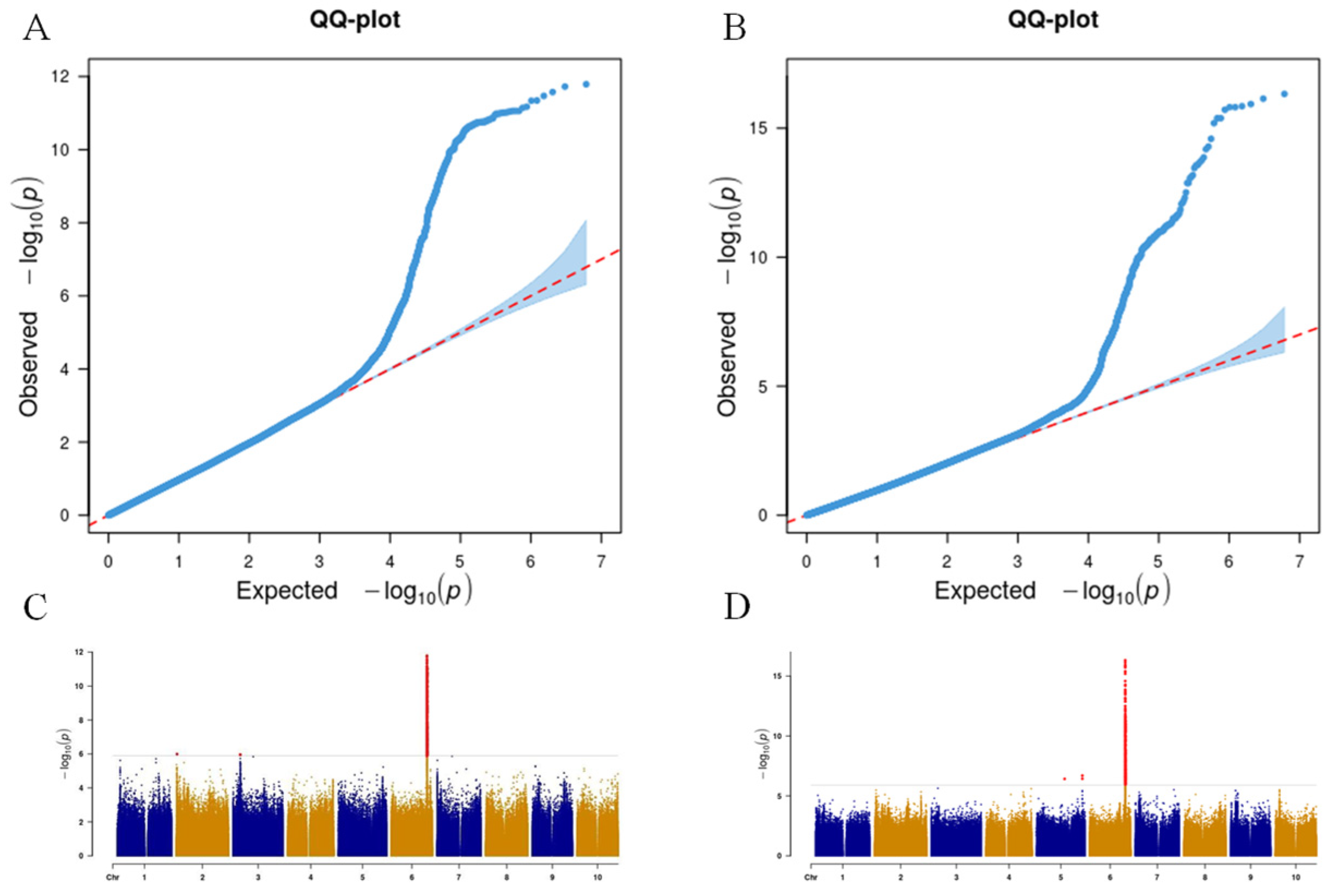
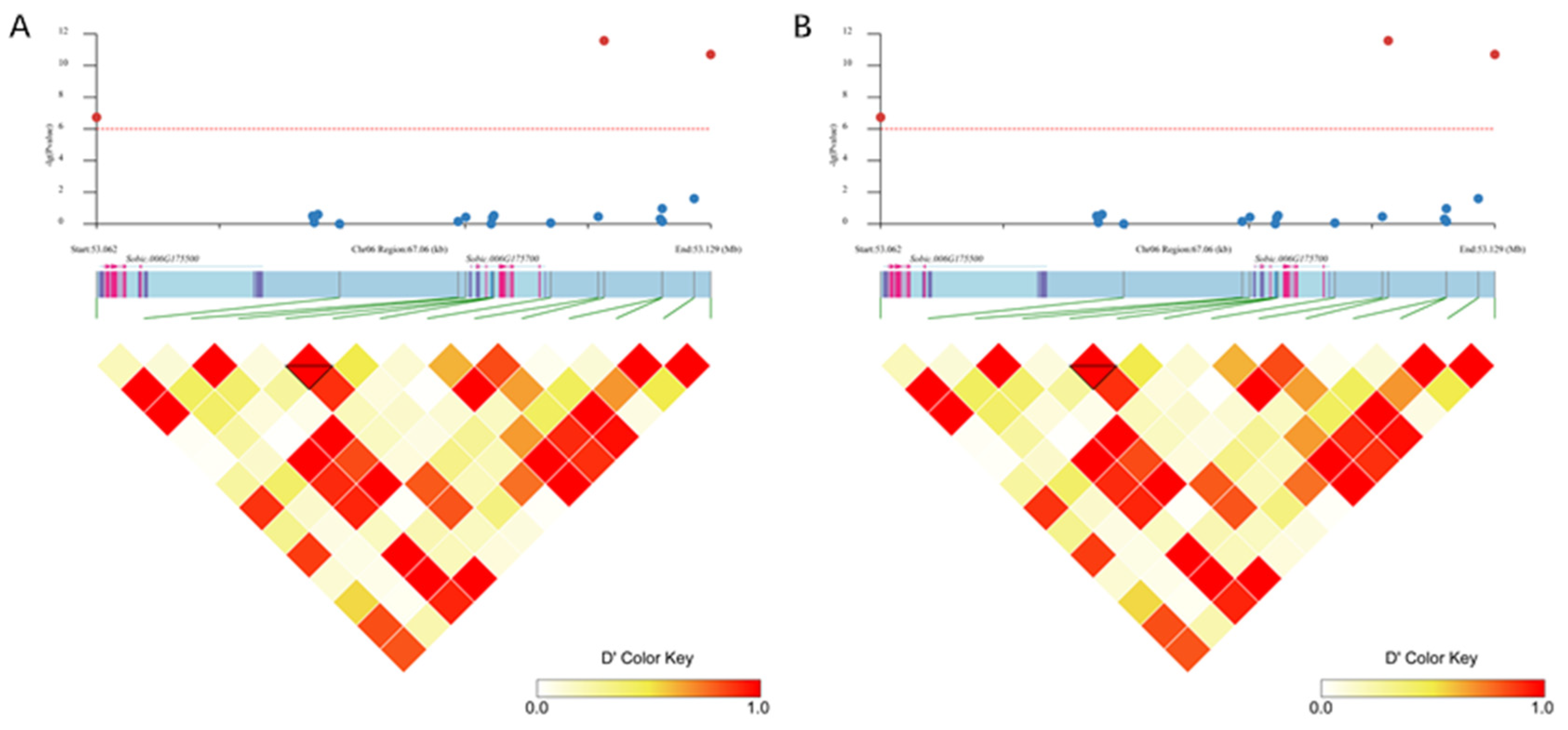
3.2. Genome-Wide Association Analysis of Sorghum Coleoptile Color
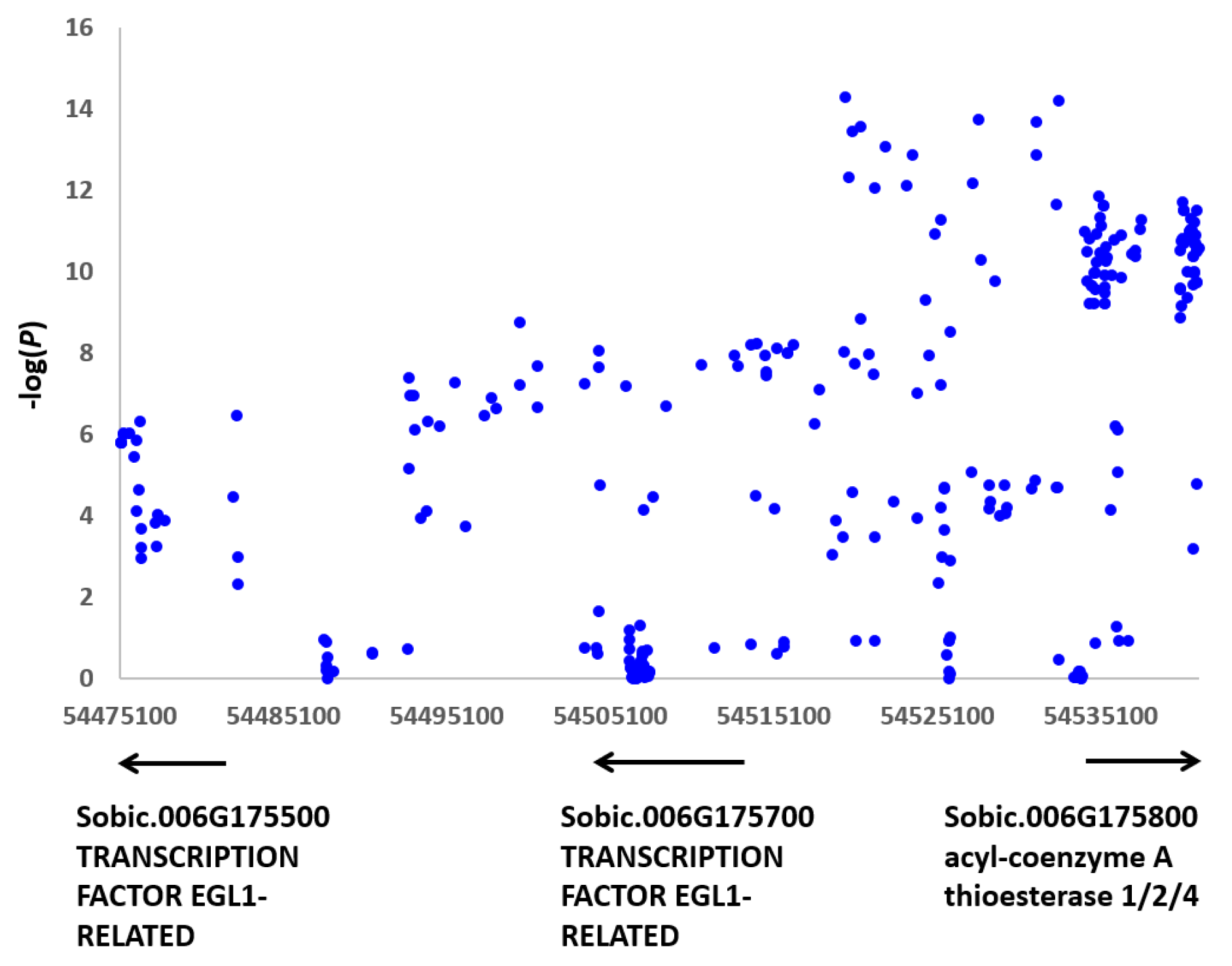
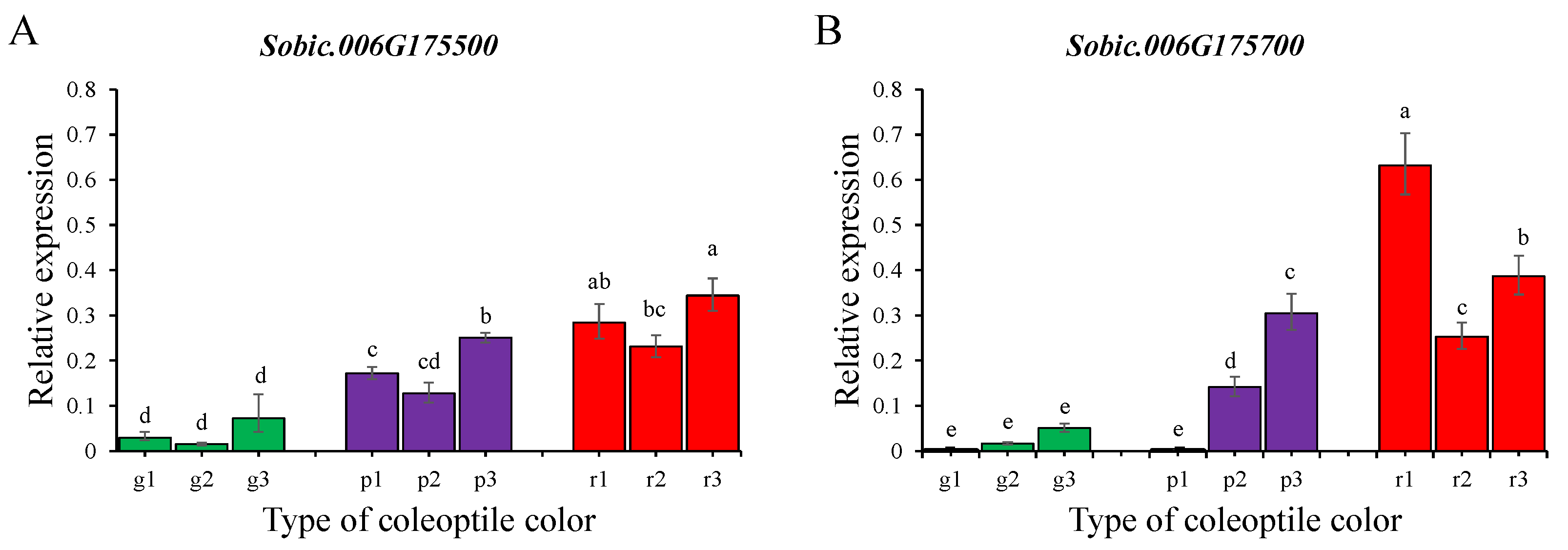
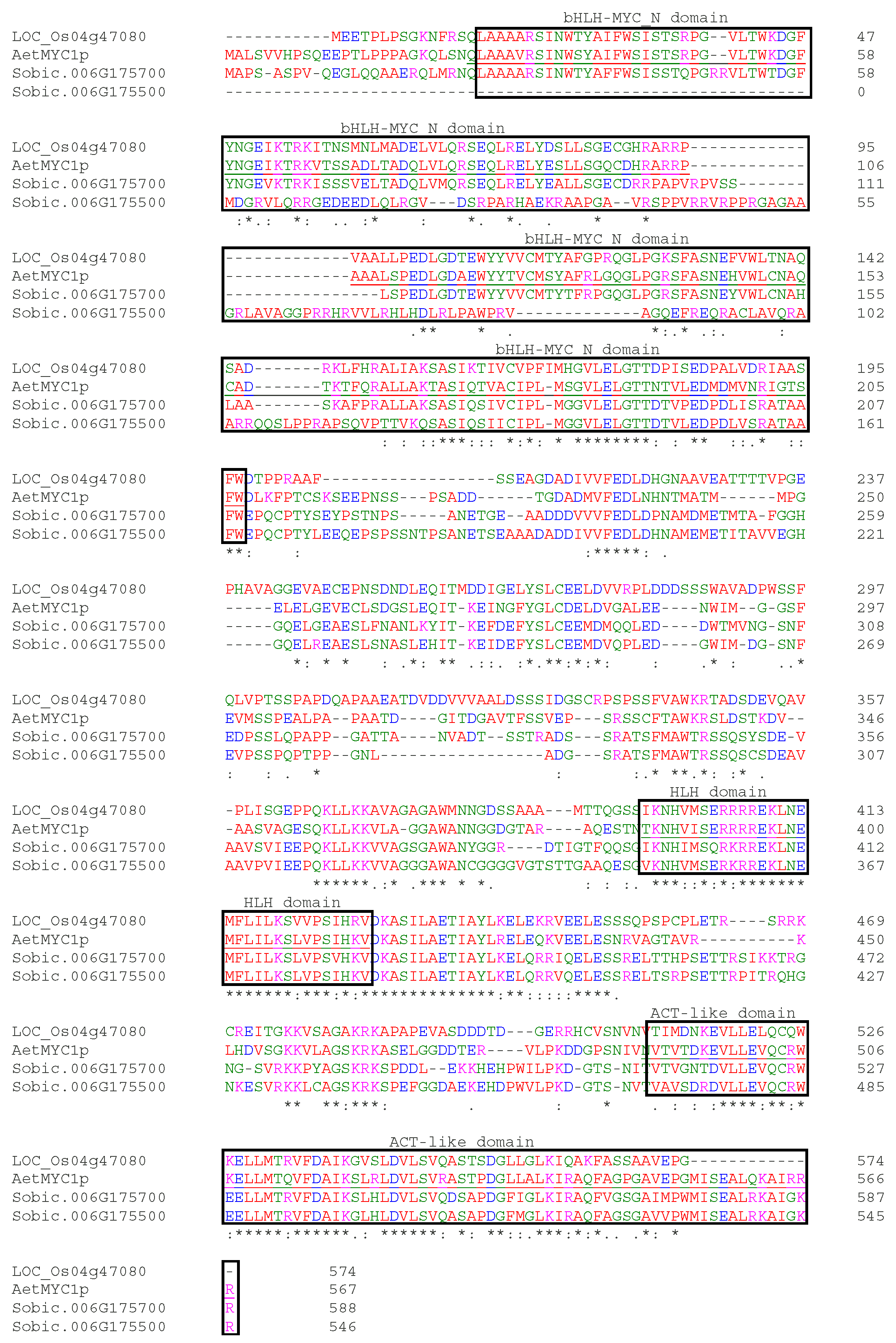
3.3. Candidate Genes and qPCR Analysis
3.4. Haplotype Analysis for the Candidate Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Proietti, I.; Frazzoli, C.; Mantovani, A. Exploiting Nutritional Value of Staple Foods in the World’s Semi-Arid Areas: Risks, Benefits, Challenges and Opportunities of Sorghum. Healthcare 2015, 3, 172–193. [Google Scholar] [CrossRef] [PubMed]
- Khoddami, A.; Messina, V.; Vadabalija Venkata, K.; Farahnaky, A.; Blanchard, C.L.; Roberts, T.H. Sorghum in foods: Functionality and potential in innovative products. Crit. Rev. Food Sci. Nutr. 2023, 63, 1170–1186. [Google Scholar] [CrossRef] [PubMed]
- National Research Council. Lost Crops of Africa: Volume I: Grains; The National Academies Press: Washington, DC, USA, 1996. [Google Scholar]
- Rhodes, D.H.; Hoffmann, L.; Rooney, W.L.; Herald, T.J.; Bean, S.; Boyles, R.; Kresovich, S.; Brenton, Z.W. Genetic architecture of kernel composition in global sorghum germplasm. BMC Genom. 2017, 18, 15. [Google Scholar] [CrossRef]
- Zhang, L.; Ding, Y.; Xu, J.; Gao, X.; Cao, N.; Li, K.; Feng, Z.; Cheng, B.; Zhou, L.; Ren, M.; et al. Selection Signatures in Chinese Sorghum Reveals Its Unique Liquor-Making Properties. Front. Plant Sci. 2022, 13, 923734. [Google Scholar] [CrossRef] [PubMed]
- Abreha, K.B.; Enyew, M.; Carlsson, A.S.; Vetukuri, R.R.; Feyissa, T.; Motlhaodi, T.; Ng’uni, D.; Geleta, M. Sorghum in dryland: Morphological, physiological, and molecular responses of sorghum under drought stress. Planta 2021, 255, 20. [Google Scholar] [CrossRef]
- Lemaire, G.; Charrier, X.; Hébert, Y. Nitrogen uptake capacities of maize and sorghum crops in different nitrogen and water supply conditions. Agronomie 1996, 16, 231–246. [Google Scholar] [CrossRef]
- Schittenhelm, S.; Schroetter, S. Comparison of drought tolerance of maize, sweet sorghum and sorghum-sudangrass hybrids. J. Agron. Crop Sci. 2013, 200, 46–53. [Google Scholar] [CrossRef]
- Rotundo, J.L.; Salinas, A.; Gomara, N.; Borras, L.; Messina, C. Maize outyielding sorghum under drought conditions helps explain land use changes in the US. Field Crops Res. 2024, 308, 109298. [Google Scholar] [CrossRef]
- Tillich, H.J. Seedling diversity and the homologies of seedling organs in the order Poales (Monocotyledons). Ann. Bot. 2007, 100, 1413–1429. [Google Scholar] [CrossRef]
- Srivastava, L.M. Plant Growth and Development: Hormones and Environment; Academic Press: Boston, MA, USA, 2002; pp. 341–379. [Google Scholar]
- Eiko, H.; Ahmed, N.; Kazuhiko, N. Colour genes (R and Rc) for grain and coleoptile upregulate flavonoid biosynthesis genes in wheat. Genome 2005, 48, 747–754. [Google Scholar]
- Shoeva, O.Y.; Gordeeva, E.I.; Arbuzova, V.S.; Khlestkina, E.K. Anthocyanins participate in protection of wheat seedlings from osmotic stress. Cereal Res. Commun. 2017, 45, 47–56. [Google Scholar] [CrossRef]
- Li, J.; Zhang, C.; Xu, X.; Su, Y.; Gao, Y.; Yang, J.; Xie, C.J.; Ma, J. A MYB family transcription factor TdRCA1 from wild emmer wheat regulates anthocyanin biosynthesis in coleoptile. Theor. Appl. Genet. 2024, 137, 208. [Google Scholar] [CrossRef] [PubMed]
- Harborne, J.B.; Williams, C.A. Anthocyanins and other flavonoids. Nat. Prod. Rep. 2001, 18, 310–333. [Google Scholar] [CrossRef]
- Tanaka, Y.; Brugliera, F.; Kalc, G.; Senior, M.; Dyson, B.; Nakamura, N.; Katsumoto, Y.; Chandler, S. Flower Color Modification by Engineering of the Flavonoid Biosynthetic Pathway: Practical perspectives. Biosci. Biotechnol. Biochem. 2010, 74, 1760–1769. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, L.; Zhang, D.; Su, D.; Li, W.; Wang, X.; Chen, Q.; Cai, W.; Xu, L.; Cao, F.; et al. Comprehensive analysis of metabolome and transcriptome reveals the mechanism of color formation in different leave of Loropetalum Chinense var. Rubrum. BMC Plant Biol. 2023, 23, 133. [Google Scholar] [CrossRef]
- Khoddami, A.; Mohammadrezaei, M.; Roberts, T.H. Effects of Sorghum Malting on Colour, Major Classes of Phenolics and Individual Anthocyanins. Molecules 2017, 22, 1713. [Google Scholar] [CrossRef]
- Liu, C.H.; Zhao, X.L.; Wang, Z.; Zheng, X.Z.; Xue, L.L.; Liu, C.; Chen, Z.Y.; Zhang, B.F. Evaluation of the antioxidant activity of blueberry ethanol extracts under microwave extraction. Int. J. Agric. Biol. Eng. 2022, 15, 205–213. [Google Scholar] [CrossRef]
- Lee, C.; Lee, J.Y.; Lee, J.W. Relationship of fruit color and anthocyanin content with related gene expression differ in strawberry cultivars during shelf life. Sci. Hortic. 2022, 301, 111109. [Google Scholar] [CrossRef]
- Wu, Y.Q.; Zhang, C.H.; Huang, Z.J.; Lyu, L.F.; Li, J.; Li, W.L.; Wu, W.L. The color difference of rubus fruits is closely related to the composition of flavonoids including anthocyanins. LWT 2021, 149, 111825. [Google Scholar] [CrossRef]
- Khlestkina, E.K.; Antonova, E.V.; Pershina, L.A.; Soloviev, A.A.; Badaeva, E.D.; Börner, A.; Salina, E.A. Variability of Rc (red coleoptile) alleles in wheat and wheat-alien genetic stock collections. Cereal Res. Commun. 2011, 39, 465–474. [Google Scholar] [CrossRef]
- Du, S.L.; Wang, Z.W.; Chen, Y.; Tan, Y.; Li, X.; Zhu, W.P.; He, G.H.; Lei, K.R.; Guo, L.B.; Zhang, Y. Coleoptile purple line regulated by A-P gene system is a valuable marker trait for seed purity identification in hybrid rice. Rice Sci. 2022, 29, 451–461. [Google Scholar] [CrossRef]
- Doggett, H. Sorghum, 2nd ed.; Longmans Scientific and Technical: Harlow, UK, 1988. [Google Scholar]
- Mace, E.S.; Jordan, D.R. Location of major effect genes in sorghum (Sorghum bicolor (L.) Moench). Theor. Appl. Genet. 2010, 121, 1339–1356. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.P.; Rhodes, D.H.; Brenton, Z.; Ramu, P.; Thayil, V.M.; Deshpande, S.; Hash, C.T.; Acharya, C.; Mitchell, S.E.; Buckler, E.S.; et al. Dissecting genome-wide association signals for loss-of-function phenotypes in sorghum flavonoid pigmentation traits. G3 2013, 3, 2085–2094. [Google Scholar] [CrossRef]
- Cone, K.C. Anthocyanin synthesis in maize aleurone tissue. Plant Cell Monogr. 2007, 8, 121–139. [Google Scholar]
- Li, Y.; Bernot, J.P.; Illingworth, C.; Lison, W.; Bernot, K.M.; Eggleston, W.B.; Fogle, K.J.; Dipaola, J.E.; Kermicle, J.; Alleman, M. Gene conversion within regulatory sequences generates maize r alleles with altered gene expression. Genetics 2001, 159, 1727–1740. [Google Scholar] [CrossRef]
- Walker, E.L.; Panavas, T. Structural features and methylation patterns associated with paramutation at the r1 locus of Zea mays. Genetics 2001, 159, 1201–1215. [Google Scholar] [CrossRef]
- Elena, K.K.; Elena, I.G.; Valentina, S.A. Molecular and functional characterization of wheat near-isogenic line ‘i:S29Ra’ having intensive anthocyanin pigmentation of the coleoptile, culm, leaves and auricles. Plant Breed. 2014, 133, 454–458. [Google Scholar]
- Riaz, B.; Chen, H.; Wang, J.; Du, L.; Wang, K.; Ye, X. Overexpression of Maize ZmC1 and ZmR Transcription Factors in Wheat Regulates Anthocyanin Biosynthesis in a Tissue-Specific Manner. Int. J. Mol. Sci. 2019, 20, 5806. [Google Scholar] [CrossRef]
- Qi, X.; Guo, S.; Zhong, Y.; Chen, B.; Liu, Z.; Yan, T.; Feng, B.; Ni, Z.; Chen, S.; Liu, C. Establishment of an efficient haploid identification system by engineering anthocyanin accumulation in the wheat embryo. Plant Commun. 2023, 4, 100568. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Hou, X.J.; Zhang, B.; Chen, W.J.; Liu, D.C.; Liu, B.L.; Zhang, H.G. Identification of a Candidate Gene for Rc-D1, a Locus Controlling Red Coleoptile Colour in Wheat. Cereal Res. Commun. 2016, 44, 35–46. [Google Scholar] [CrossRef]
- Himi, E.; Taketa, S. Isolation of candidate genes for the barley Ant1 and wheat Rc genes controlling anthocyanin pigmentation in different vegetative tissues. Mol. Genet. Genom. 2015, 290, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Khlestkina, E.K.; Pestsova, E.G.; Röder, M.S.; Börner, A. Molecular mapping, phenotypic expression and geographical distribution of genes determining anthocyanin pigmentation of coleoptiles in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2002, 104, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Chin, H.S.; Wu, Y.P.; Hour, A.L.; Hong, C.Y.; Lin, Y.R. Genetic and Evolutionary Analysis of Purple Leaf Sheath in Rice. Rice 2016, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Zhou, T.; Han, Z.; Tan, C.; Xing, Y. Dominant complementary interaction between OsC1 and two tightly linked genes, Rb1 and Rb2, controls the purple leaf sheath in rice. Theor. Appl. Genet. 2020, 133, 2555–2566. [Google Scholar] [CrossRef]
- Jiang, L.Q.; Lyu, S.W.; Yu, H.; Zhang, J.; Sun, B.G.; Liu, Q.; Mao, X.G.; Chen, P.G.; Pan, D.J.; Chen, W.F.; et al. Transcription factor encoding gene OsC1 regulates leaf sheath color through anthocyanidin metabolism in Oryza rufipogon and Oryza sativa. BMC Plant Biol. 2024, 24, 147. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Pundir, R.P.; Dwivedi, S.L.; Gowda, C.L.; Reddy, V.G.; Singh, S. Developing a mini core collection of sorghum for diversified utilization of germplasm. Crop Sci. 2009, 49, 1769–1780. [Google Scholar] [CrossRef]
- Yu, X. Analysis and Study of Anthocyanins in Plant Leaves. Mod. Instrum. 2000, 04, 37–38, (In Chinese with English Summary). [Google Scholar]
- Wang, L.; Upadhyaya, H.D.; Zheng, J.; Liu, Y.; Singh, S.K.; Gowda, C.L.; Kumar, R.; Zhu, Y.; Wang, Y.H.; Li, J. Genome-wide Association Mapping Identifies Novel Panicle Morphology Loci and Candidate Genes in Sorghum. Front. Plant Sci. 2021, 12, 743838. [Google Scholar] [CrossRef]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.Y.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Rosenberg, N.A.; Donnelly, P. Association mapping in structured populations. Am. J. Hum. Genet. 2000, 67, 170–181. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Wang, L.; Prakash, C.S.; Liu, Y.; Gao, L.; Meng, R.; Seetharam, K.; Gowda, C.L.L.; Ganesamurthy, K.; Singh, S.K.; et al. Genome-wide association mapping identifies an SNF4 ortholog that impacts biomass and sugar yield in sorghum and sugarcane. J. Exp. Bot. 2022, 73, 3584–3596. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.S.; He, W.M.; Ji, J.J.; Zhang, C.; Guo, Y.; Yang, T.L. LDBlockShow: A fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. Brief. Bioinform. 2021, 22, bbaa227. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Arvidsson, S.; Kwasniewski, M.; Riaño-Pachón, D.M.; Mueller-Roeber, B. QuantPrime—A flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinform. 2008, 9, 465. [Google Scholar] [CrossRef]
- Gao, S.; Peng, J.; Rong, M.; Liu, Y.; Xu, Y.; Wei, J. Screening and validation of reference genes in Dracaena cochinchinensis using quantitative real-time PCR. Sci. Rep. 2024, 14, 6165. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hu, J.; Anderson, B.; Wessler, S.R. Isolation and characterization of rice R genes: Evidence for distinct evolutionary paths in rice and maize. Genetics 1996, 142, 1021–1031. [Google Scholar] [CrossRef]
- Kalinger, R.S.; Pulsifer, I.P.; Hepworth, S.R.; Rowland, O. Fatty Acyl Synthetases and Thioesterases in Plant Lipid Metabolism: Diverse Functions and Biotechnological Applications. Lipids 2020, 55, 435–455. [Google Scholar] [CrossRef]
- Cao, D.; Ye, G.J.; Zong, Y.; Zhang, B.; Chen, W.J.; Liu, B.L.; Zhang, H.G. AetMYC1, the Candidate Gene Controlling the Red Coleoptile Trait in Aegilops tauschii Coss. Accession As77. Molecules 2017, 22, 2259. [Google Scholar] [CrossRef]
- Ahmed, N.; Maekawa, M.; Utsugi, S.; Rikiishia, K.; Ahmad, A.; Noda, K. The wheat Rc gene for red coleoptile colour codes for a transcriptional activator of late anthocyanin biosynthesis genes. J. Cereal Sci. 2006, 44, 54–58. [Google Scholar] [CrossRef]
- Zhang, Y. Map-Based Cloning and Application of Two Genes Related to Purple Line in Rice Coleoptile. Ph.D. Thesis, Southwest University, Chongqing, China, 2009. [Google Scholar]
- Gao, D.; He, B.; Zhou, Y.; Sun, L. Genetic and molecular analysis of a purple sheath somaclonal mutant in Japonica rice. Plant Cell Rep. 2011, 30, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Robbins, T.P.; Walker, E.L.; Kermicle, J.L.; Alleman, M.; Dellaporta, S.L. Meiotic instability of the R-r complex arising from displaced intragenic exchange and intrachromosomal rearrangement. Genetics 1991, 129, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Takehisa, H.; Kamatsuki, K.; Minami, H.; Namiki, N.; Ikawa, H.; Ohyanagi, H.; Sugimoto, K.; Antonio, B.A.; Nagamura, Y. RiceXPro Version 3.0: Expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2013, 41, D1206–D1213. [Google Scholar] [CrossRef]
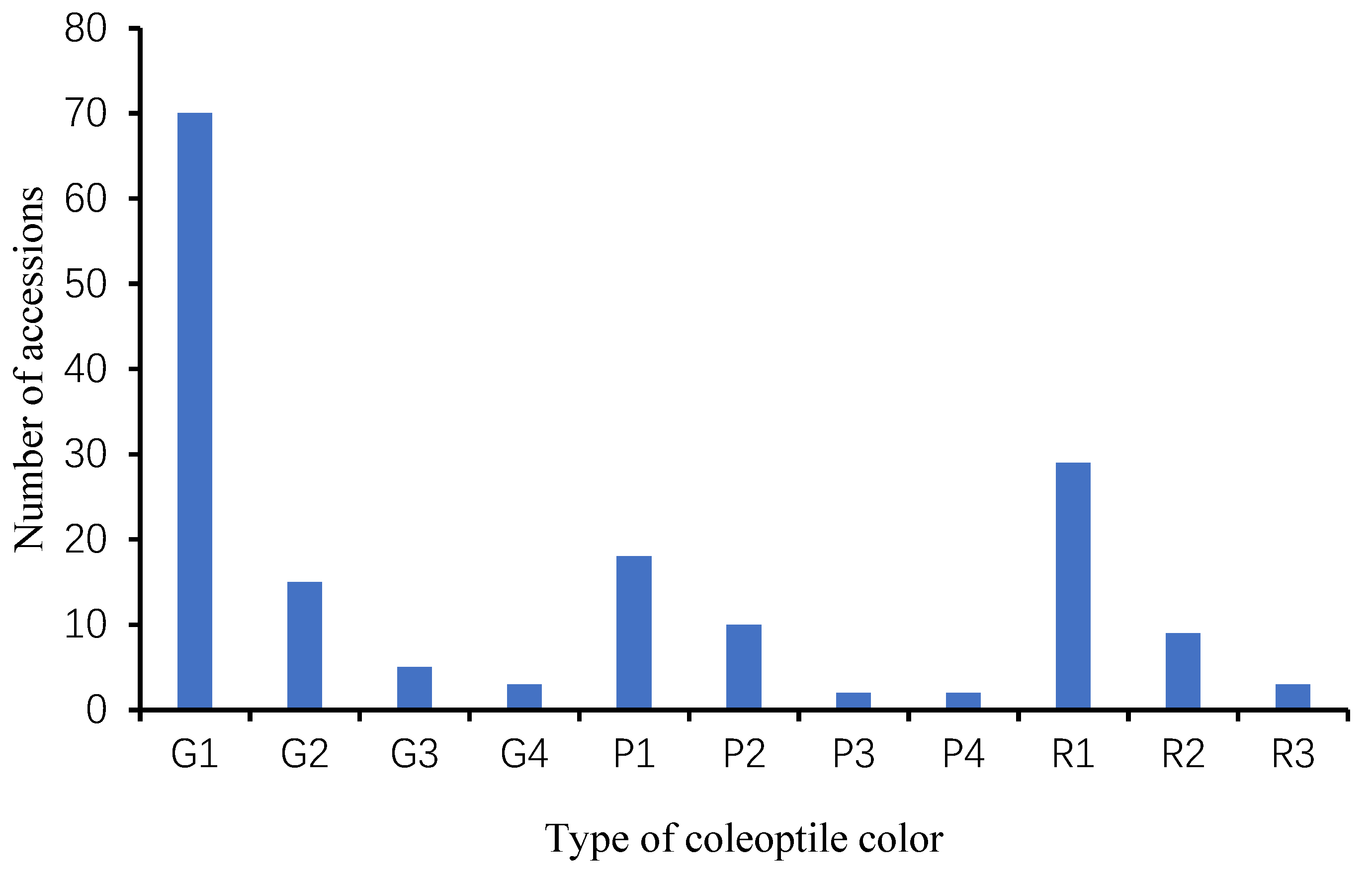
| Red | Purple | Green | Total | |
|---|---|---|---|---|
| 2022 | 52 | 61 | 122 | 235 |
| proportions | 22% | 26% | 52% | 100% |
| 2023 | 72 | 53 | 110 | 235 |
| proportions | 31% | 22% | 47% | 100% |
| Trait | Environment | QTL | CHR | POS (bp) | p-Value |
|---|---|---|---|---|---|
| Coleoptile color | 2022 | Chr02:1034741 | 2 | 1034741 | 1.021537 × 10−6 |
| Chr03:10703793 | 3 | 10703793 | 1.082520 × 10−6 | ||
| Chr06:53125544 | 6 | 53125544 | 1.627272 × 10−12 | ||
| 2023 | Chr05:67965506 | 5 | 67965506 | 2.025845 × 10−7 | |
| Chr05:41514284 | 5 | 41514284 | 3.798307 × 10−7 | ||
| Chr06:53032883 | 6 | 53117435 | 5.18 × 10−15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, K.; Wang, L.; Shen, Q.; Hu, L.; Xing, Z.; Wang, Y.; Li, J. Association Analysis and Identification of Candidate Genes for Sorghum Coleoptile Color. Agronomy 2025, 15, 688. https://doi.org/10.3390/agronomy15030688
Wang K, Wang L, Shen Q, Hu L, Xing Z, Wang Y, Li J. Association Analysis and Identification of Candidate Genes for Sorghum Coleoptile Color. Agronomy. 2025; 15(3):688. https://doi.org/10.3390/agronomy15030688
Chicago/Turabian StyleWang, Kai, Lihua Wang, Qi Shen, Lu Hu, Zhichao Xing, Yihong Wang, and Jieqin Li. 2025. "Association Analysis and Identification of Candidate Genes for Sorghum Coleoptile Color" Agronomy 15, no. 3: 688. https://doi.org/10.3390/agronomy15030688
APA StyleWang, K., Wang, L., Shen, Q., Hu, L., Xing, Z., Wang, Y., & Li, J. (2025). Association Analysis and Identification of Candidate Genes for Sorghum Coleoptile Color. Agronomy, 15(3), 688. https://doi.org/10.3390/agronomy15030688





