High-Throughput Sequence Analysis of Microbial Communities of Soybean in Northeast China
Abstract
1. Introduction
2. Materials and Methods
2.1. Soybean Cultivars
2.2. Traditional Detection of Seed Carrier Microbes
2.3. Identification of Fungi
2.4. Identification of Bacteria
2.5. Microbial DNA Extraction and Polymerase Chain Reaction
2.6. Bioinformatics Analysis
3. Results
3.1. Analysis of the Diversity of Microorganisms Carried by Soybean Seeds Using Traditional Detection Technique
3.1.1. Fungal Diversity on Soybean Seeds
3.1.2. Bacterial Diversity on Soybean Seeds
3.2. Microbial Diversity of Soybean Seeds Based on High-Throughput Sequencing Technology
3.2.1. Analysis of Fungi Using ITS Sequencing
3.2.2. Analysis of Bacteria Based on 16S rDNA Sequences
3.3. Alpha Diversity of Microorganisms Carried by Soybean Seeds
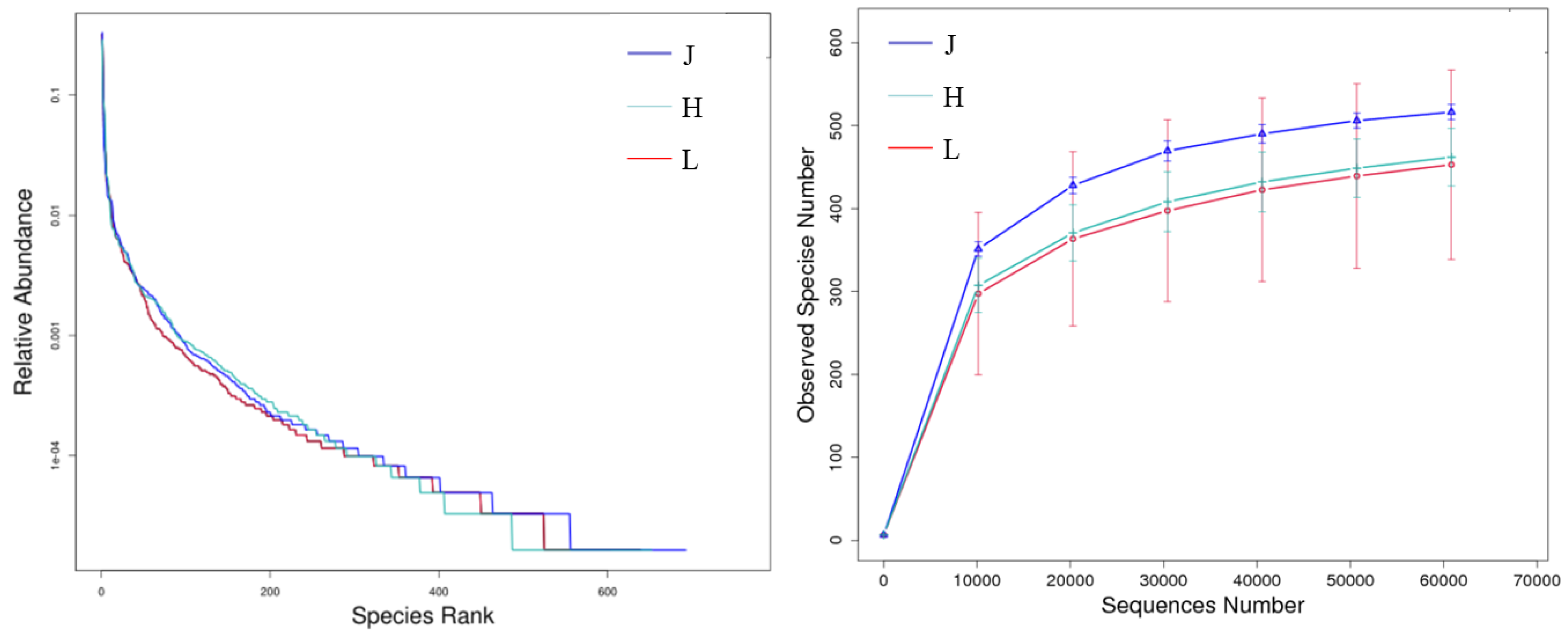
3.3.1. Alpha Diversity of Fungi
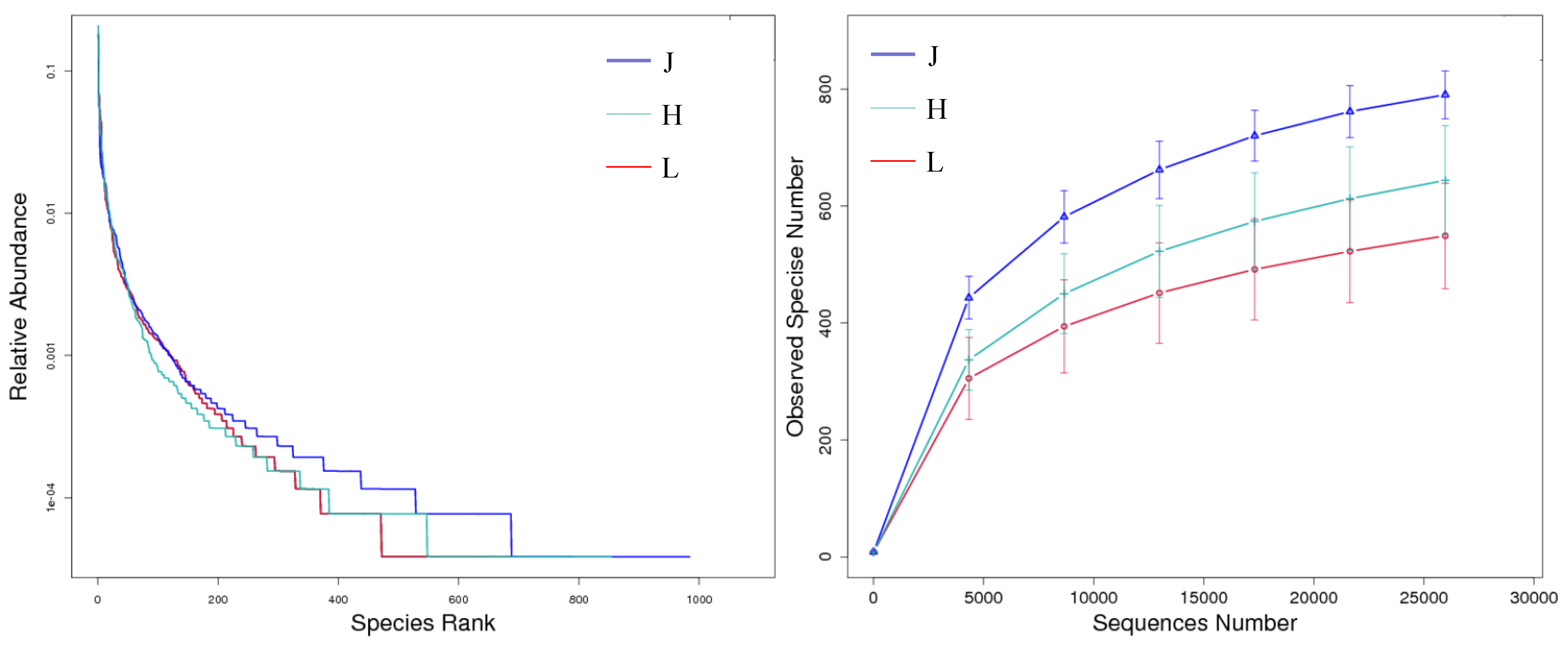
3.3.2. Alpha Diversity of Bacteria
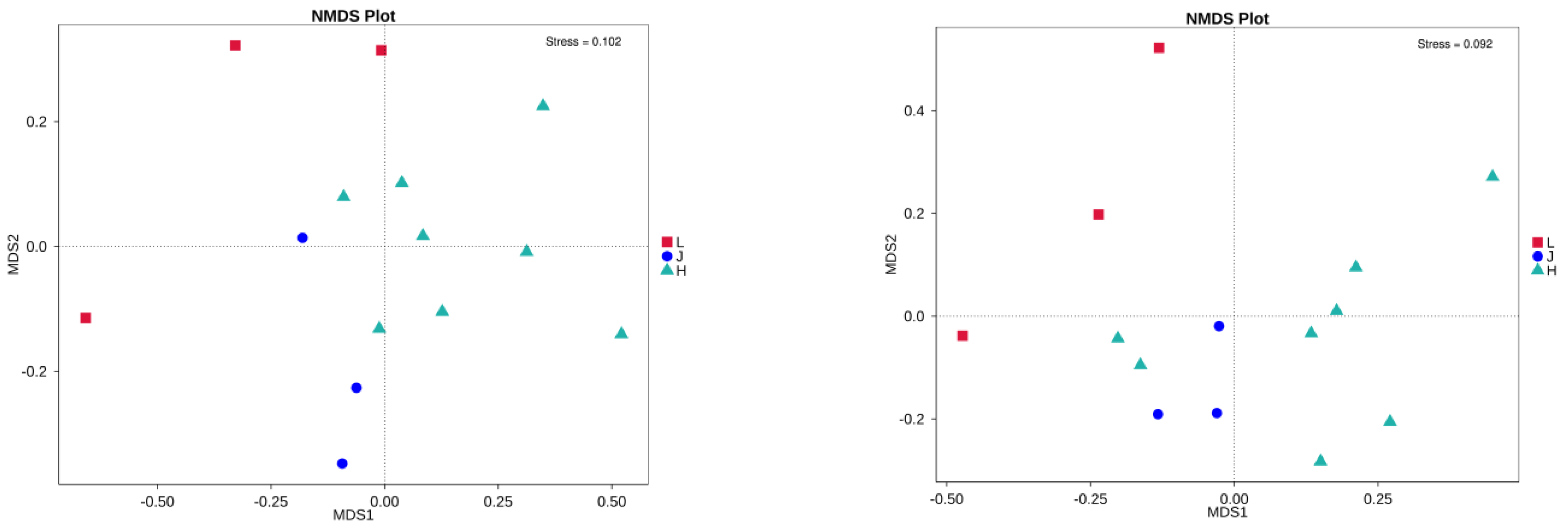
3.4. Characteristics of Microbial Communities Carried by Soybean Seeds
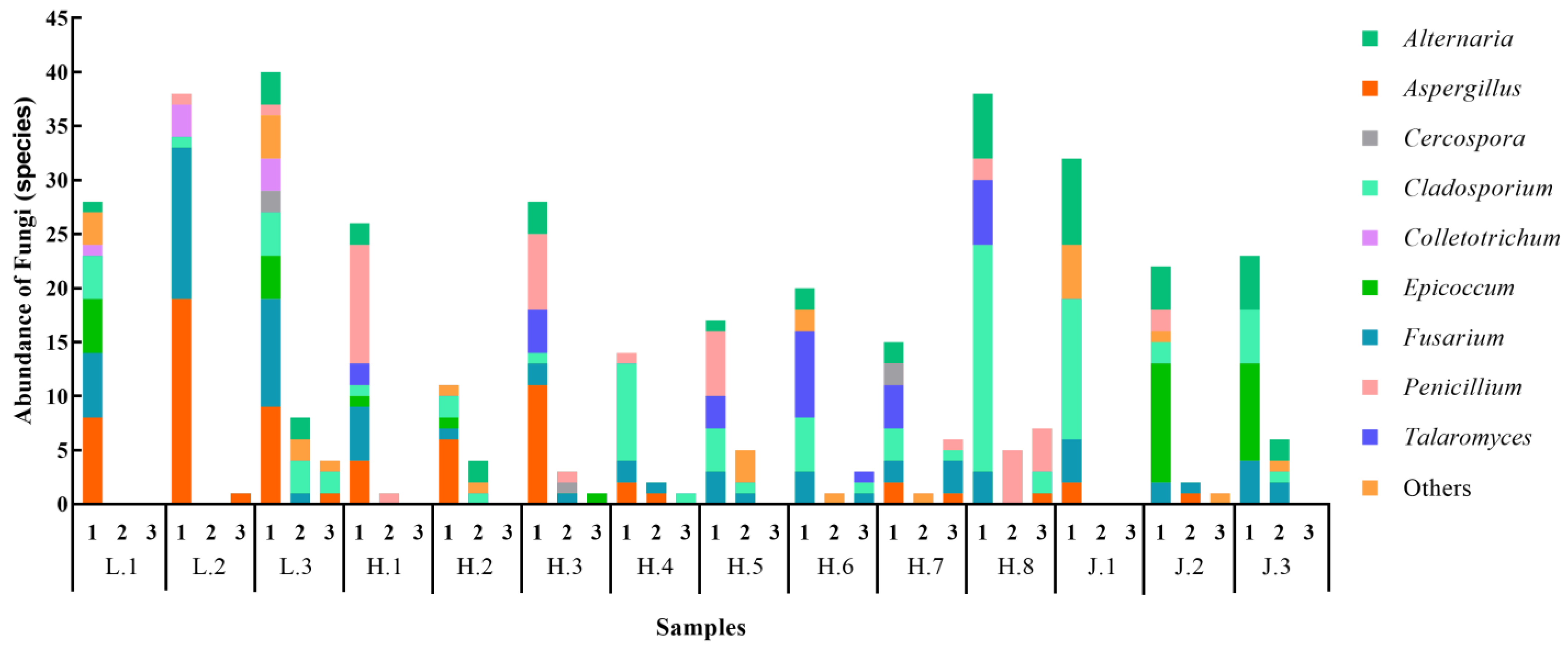
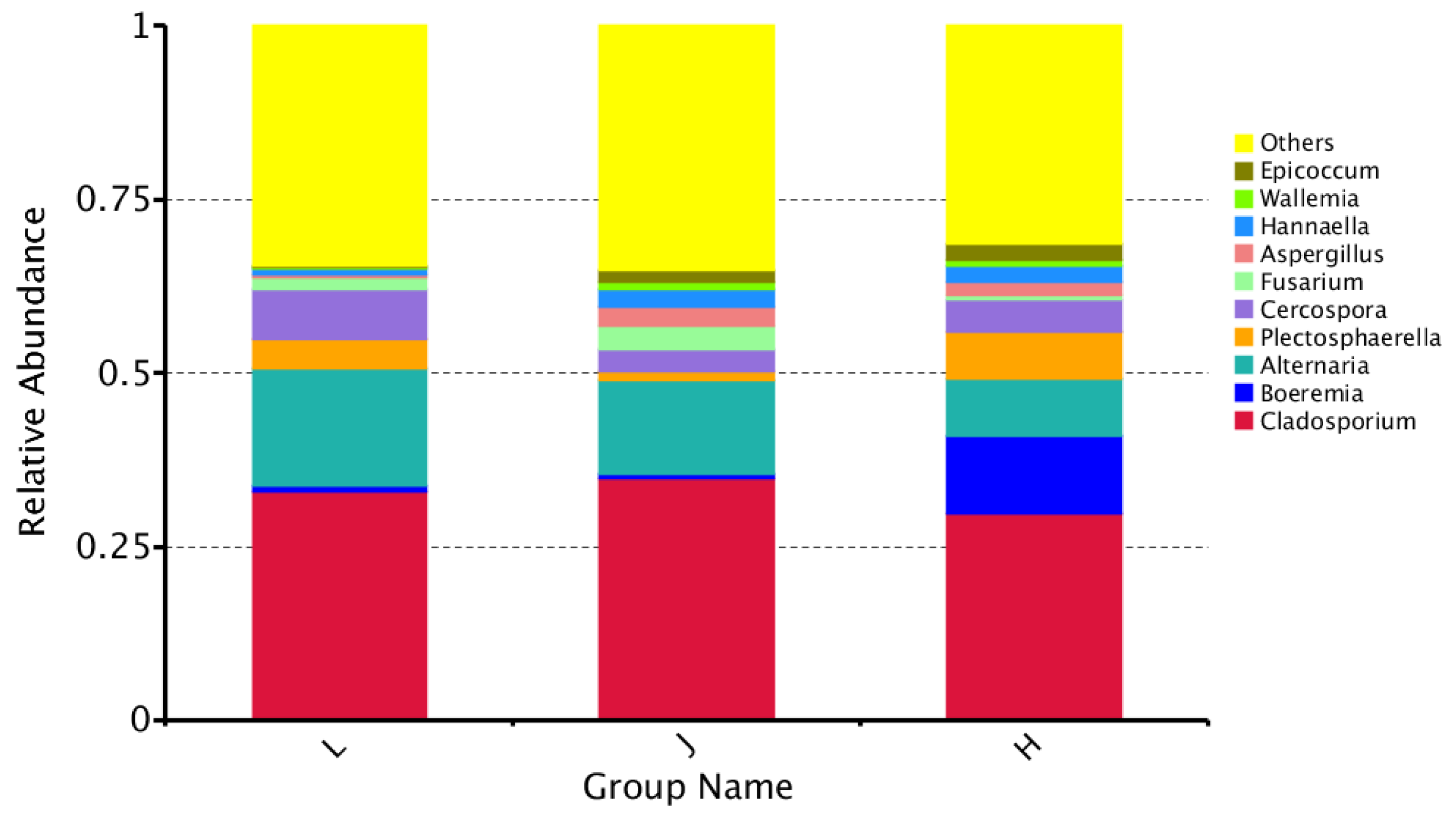
3.4.1. Characteristics of Fungal Communities
3.4.2. Characteristics of Bacterial Communities
3.5. Changes in Related Microbial Communities Carried by Soybean Seeds in Northeast China
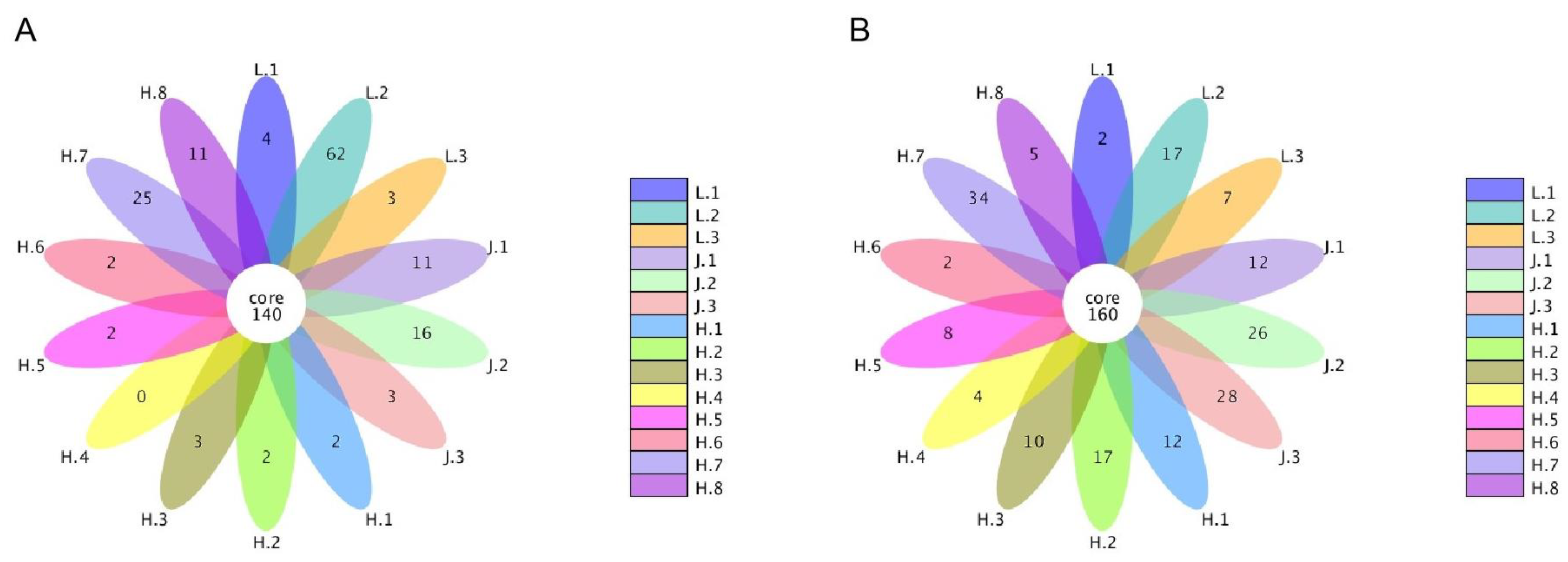
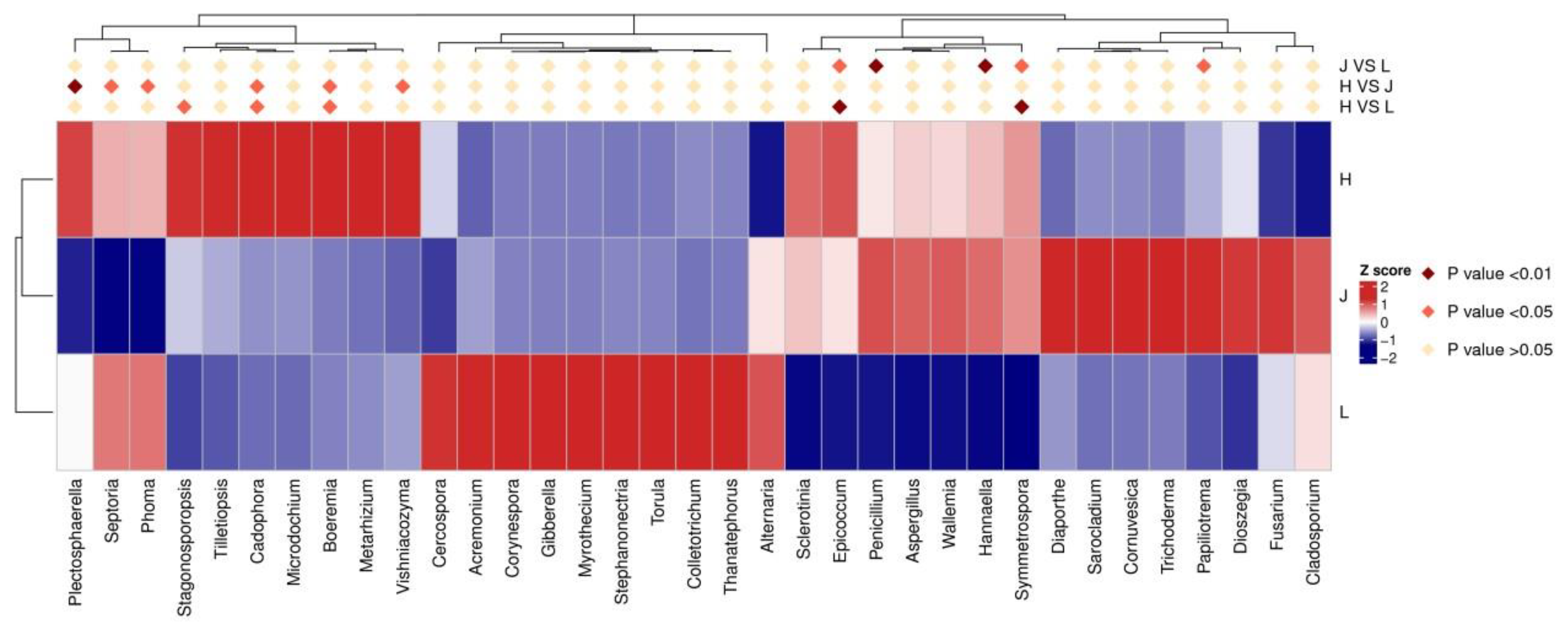
3.5.1. Changes in Fungal Communities
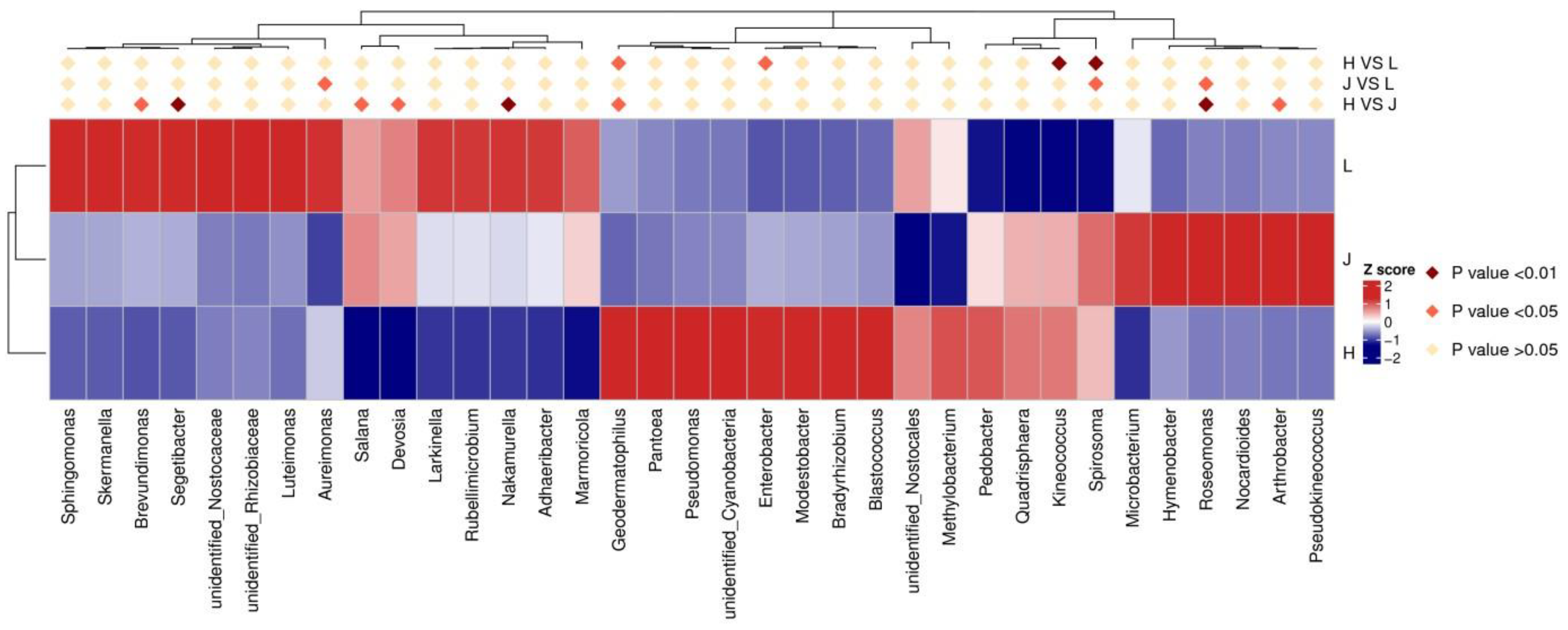
3.5.2. Changes in Related Bacterial Communities
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Colletti, A.; Attrovio, A.; Boffa, L.; Mantegna, S.; Cravotto, G. Valorisation of by-products from soybean (Glycine max (L.) Merr.) processing. Molecules 2020, 25, 2129. [Google Scholar] [CrossRef]
- Kockelmann, A.; Tilcher, R.; Fischer, U. Seed production and processing. SugarTech 2010, 12, 267–275. [Google Scholar] [CrossRef]
- Martín, I.; Gálvez, L.; Guasch, L.; Palmero, D. Fungal Pathogens and Seed Storage in the Dry State. Plants 2022, 11, 3167. [Google Scholar] [CrossRef] [PubMed]
- Barret, M.; Guimbaud, J.F.; Darrasse, A.; Jacques, M.A. Plant microbiota affects seed transmission of phytopathogenic microorganisms. Mol. Plant Pathol. 2016, 17, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Javaid, M.M.; Mahmood, A.; Alshaya, D.S.; AlKahtani, M.D.F.; Waheed, H.; Wasaya, A.; Khan, S.A.; Naqve, M.; Haider, I.; Shahid, M.A.; et al. Influence of environmental factors on seed germination and seedling characteristics of perennial ryegrass (Lolium perenne L.). Sci. Rep. 2022, 12, 9522. [Google Scholar] [CrossRef] [PubMed]
- Long, R.L.; Gorecki, M.J.; Renton, M.; Scott, J.K.; Colville, L.; Goggin, D.E.; Commander, L.E.; Westcott, D.A.; Cherry, H.; Finch-Savage, W.E. The ecophysiology of seed persistence: A mechanistic view of the journey to germination or demise. Biol. Rev. 2015, 90, 31–59. [Google Scholar] [CrossRef] [PubMed]
- Gitaitis, R.; Walcott, R. The epidemiology and management of seedborne bacterial diseases. Annu. Rev. Phytopathol. 2007, 45, 371–397. [Google Scholar] [CrossRef] [PubMed]
- Berg, T.; Tesoriero, L.; Hailstones, D. PCR-based detection of Xanthomonas campestris pathovars in Brassica seed. Plant Pathol. 2005, 54, 416–427. [Google Scholar] [CrossRef]
- Franco-Duarte, R.; Černáková, L.; Kadam, S.; Kaushik, K.S.; Salehi, B.; Bevilacqua, A.; Corbo, M.R.; Antolak, H.; Dybka-Stępień, K.; Leszczewicz, M.; et al. Advances in chemical and biological methods to identify microorganisms—From past to present. Microorganisms 2019, 7, 130. [Google Scholar] [CrossRef] [PubMed]
- Simonin, M.; Briand, M.; Chesneau, G.; Rochefort, A.; Marais, C.; Sarniguet, A.; Barret, M. Seed microbiota revealed by a large-scale meta-analysis including 50 plant species. New Phytol. 2022, 234, 1448–1463. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Ruan, Y.; Chen, X.-H.; Yu, H.-L.; Cheng, T.; Duan, X.-Y.; Liu, Y.-G.; Zhang, H.-Y.; Zhang, Q.-Y. Metabolites of pathogenic microorganisms database (MPMdb) and its seed metabolite applications. Microbiol. Spectr. 2024, 12, e02342-23. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y. Developmental Paths Differentiation and Genome Analysis of the Basidiomycota, Coprinopsis cinerea. ProQuest Dissertations & Theses, Hong Kong University of Science and Technology, Hong Kong, 2020. [Google Scholar]
- Olimi, E.; Kusstatscher, P.; Wicaksono, W.A.; Abdelfattah, A.; Cernava, T.; Berg, G. Insights into the microbiome assembly during different growth stages and storage of strawberry plants. Environ. Microbiome 2022, 17, 21. [Google Scholar] [CrossRef] [PubMed]
- Razzaq, A.; Saleem, F.; Kanwal, M.; Mustafa, G.; Yousaf, S.; Arshad, H.M.I.; Hameed, M.K.; Khan, M.S.; Joyia, F.A. Modern Trends in Plant Genome Editing: An Inclusive Review of the CRISPR/Cas9 Toolbox. Int. J. Mol. Sci. 2019, 20, 4045. [Google Scholar] [CrossRef]
- Hosseini, P.; Matthews, B.F. Regulatory interplay between soybean root and soybean cyst nematode during a resistant and susceptible reaction. BMC Plant Biol. 2014, 14, 300. [Google Scholar] [CrossRef] [PubMed]
- Roy, K.W.; Baird, R.E.; Abney, T.S. A review of soybean (Glycine max) seed, pod, and flower mycofloras in North America, with methods and a key for identification of selected fungi. Mycopathologia 2001, 150, 15–27. [Google Scholar] [CrossRef]
- Azerang, P.; Khalaj, V.; Kobarfard, F.; Owlia, P.; Sardari, S.; Shahidi, S. Molecular characterization of a fungus producing membrane active metabolite and analysis of the produced secondary metabolite. Iran. Biomed. J. 2019, 23, 121–128. [Google Scholar] [CrossRef]
- Manter, D.K.; Vivanco, J.M. Use of the ITS primers, ITS1F and ITS4, to characterize fungal abundance and diversity in mixed-template samples by qPCR and length heterogeneity analysis. J. Microbiol. Methods 2007, 71, 7–14. [Google Scholar] [CrossRef]
- Gerhardt, P.; Murray, R.G.E.; Costilow, R.; Nester, E.; Wood, W.; Krieg, N.; Phillips, G. Manual of Methods for General Bacteriology; American Society for Microbiology: Washington, DC, USA, 1981. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Zhao, J.; Liu, D.; Wang, Y.; Zhu, X.; Xuan, Y.; Liu, X.; Fan, H.; Chen, L.; Duan, Y. Biocontrol potential of Microbacterium maritypicum Sneb159 against Heterodera glycines. Pest Manag. Sci. 2019, 75, 3381–3391. [Google Scholar] [CrossRef] [PubMed]
- Edwards, U.; Rogall, T.; Blöcker, H.; Emde, M.; Böttger, E.C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989, 17, 7843–7853. [Google Scholar] [CrossRef]
- Lane, D. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Modern Microbiological Methods; John Wiley & Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Johnston Monje, D.; Raizada, M.N. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 2011, 6, e20396. [Google Scholar] [CrossRef] [PubMed]
- Lima, H.S.; Mancine, N.; Peruchi, G.B.; Francisco, C.S.; Wang, N.; de Souza, R.S.C.; Armanhi, J.S.L.; Della Coletta-Filho, H. Microbial community of cultivated and uncultivated citrus rhizosphere microbiota in Brazil. Sci. Data 2024, 11, 1294. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Bever, J.D.; Platt, T.G.; Morton, E.R. Microbial population and community dynamics on plant roots and their feedbacks on plant communities. Annu. Rev. Microbiol. 2012, 66, 265–283. [Google Scholar] [CrossRef] [PubMed]
- Carpentieri-Pipolo, V.; de Almeida Lopes, K.B.; Degrassi, G. Phenotypic and genotypic characterization of endophytic bacteria associated with transgenic and non-transgenic soybean plants. Arch. Microbiol. 2019, 201, 1029–1045. [Google Scholar] [CrossRef]
- Abdulmumeen, H.A.; Risikat, A.N.; Sururah, A.R. Food: Its preservatives, additives and applications. Int. J. Chem. Biochem. Sci. 2012, 1, 36–47. [Google Scholar]
- Barret, M.; Briand, M.; Bonneau, S.; Préveaux, A.; Valière, S.; Bouchez, O.; Hunault, G.; Simoneau, P.; Jacques, M.-A. Emergence shapes the structure of the seed microbiota. Appl. Environ. Microbiol. 2015, 81, 1257–1266. [Google Scholar] [CrossRef]
- Munkvold, G.P. Seed pathology progress in academia and industry. Annu. Rev. Phytopathol. 2009, 47, 285–311. [Google Scholar] [CrossRef] [PubMed]
- Mancini, V.; Murolo, S.; Romanazzi, G. Diagnostic methods for detecting fungal pathogens on vegetable seeds. Plant Pathol. 2016, 65, 691–703. [Google Scholar] [CrossRef]
- Sevik, M.A.; Arli-Sokmen, M. Estimation of the effect of Tomato spotted wilt virus (TSWV) infection on some yield components of tomato. Phytoparasitica 2012, 40, 87–93. [Google Scholar] [CrossRef]
- ElMasry, G.; ElGamal, R.; Mandour, N.; Gou, P.; Al-Rejaie, S.; Belin, E.; Rousseau, D. Emerging thermal imaging techniques for seed quality evaluation: Principles and applications. Food Res. Int. 2020, 131, 109025. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gu, A.; Zhang, X.; Xu, Y.; Luo, L.; Li, J. Comparative analysis of three kinds of testing methods for rice seed borne fungi. Food Res. Int. 2011, 11, 43–51. [Google Scholar]


| Location | Soybean Cultivars | Abbreviation |
|---|---|---|
| Liaoning | Tiefeng 31 | L.1 |
| Liaodou 21 | L.2 | |
| Liaodou 15 | L.3 | |
| Jilin | Huamidou 30 | J.1 |
| Jinong 38 | J.2 | |
| Dedou 10 | J.3 | |
| Heilongjiang | Nongqing Bean 20 | H.1 |
| Heinong 64 | H.2 | |
| Kangxian No. 9 | H.3 | |
| Beidou 17 | H.4 | |
| Dongsheng 9 | H.5 | |
| Heihe 43 | H.6 | |
| Qinong 10 | H.7 | |
| Longken 336 | H.8 |
| Sample Name | Raw Reads | Observed Species | Shannon | Simpson | Chao1 | ACE |
|---|---|---|---|---|---|---|
| L.1 | 81,918 | 421 | 4.464 | 0.852 | 458.558 | 459.703 |
| L.2 | 84,885 | 606 | 5.793 | 0.934 | 800.130 | 669.691 |
| L.3 | 82,460 | 332 | 3.047 | 0.732 | 367.646 | 370.855 |
| H.1 | 85,474 | 484 | 4.902 | 0.875 | 505.121 | 512.919 |
| H.2 | 84,255 | 493 | 5.339 | 0.888 | 522.647 | 537.702 |
| H.3 | 83,843 | 476 | 4.715 | 0.860 | 511.019 | 514.682 |
| H.4 | 85,262 | 492 | 4.863 | 0.882 | 538.250 | 543.746 |
| H.5 | 81,924 | 485 | 4.755 | 0.832 | 515.611 | 519.778 |
| H.6 | 84,103 | 390 | 4.125 | 0.838 | 447.468 | 444.756 |
| H.7 | 82,806 | 450 | 4.593 | 0.887 | 497.788 | 493.661 |
| H.8 | 87,901 | 426 | 4.917 | 0.913 | 462.167 | 468.367 |
| J.1 | 84,377 | 525 | 4.564 | 0.839 | 554.500 | 555.380 |
| J.2 | 85,514 | 520 | 5.390 | 0.905 | 535.769 | 537.281 |
| J.3 | 85,026 | 504 | 4.459 | 0.831 | 534.000 | 539.290 |
| Sample Name | Raw Reads | Observed Species | Shannon | Simpson | Chao1 | ACE |
| L.1 | 76,671 | 549 | 5.989 | 0.960 | 710.803 | 672.319 |
| L.2 | 86,246 | 660 | 6.310 | 0.953 | 811.076 | 795.754 |
| L.3 | 88,279 | 439 | 4.767 | 0.906 | 566.147 | 565.411 |
| H.1 | 87,645 | 751 | 5.934 | 0.941 | 918.632 | 918.984 |
| H.2 | 86,243 | 728 | 6.050 | 0.954 | 790.569 | 827.661 |
| H.3 | 81,837 | 672 | 5.336 | 0.911 | 861.000 | 842.736 |
| H.4 | 88,130 | 665 | 5.738 | 0.939 | 829.478 | 853.429 |
| H.5 | 87,417 | 629 | 5.969 | 0.960 | 853.438 | 814.259 |
| H.6 | 81,822 | 567 | 5.436 | 0.932 | 721.670 | 758.503 |
| H.7 | 81,953 | 699 | 6.458 | 0.965 | 814.636 | 832.801 |
| H.8 | 58,510 | 442 | 4.408 | 0.831 | 457.000 | 477.162 |
| J.1 | 88,793 | 742 | 6.245 | 0.955 | 919.300 | 919.734 |
| J.2 | 86,339 | 842 | 6.259 | 0.939 | 929.345 | 943.371 |
| J.3 | 86,450 | 788 | 6.803 | 0.965 | 833.200 | 854.305 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Bai, Q.; Meng, F.; Dong, W.; Fan, H.; Zhu, X.; Duan, Y.; Chen, L. High-Throughput Sequence Analysis of Microbial Communities of Soybean in Northeast China. Agronomy 2025, 15, 436. https://doi.org/10.3390/agronomy15020436
Wang Y, Bai Q, Meng F, Dong W, Fan H, Zhu X, Duan Y, Chen L. High-Throughput Sequence Analysis of Microbial Communities of Soybean in Northeast China. Agronomy. 2025; 15(2):436. https://doi.org/10.3390/agronomy15020436
Chicago/Turabian StyleWang, Yuanyuan, Qingyao Bai, Fanqi Meng, Wei Dong, Haiyan Fan, Xiaofeng Zhu, Yuxi Duan, and Lijie Chen. 2025. "High-Throughput Sequence Analysis of Microbial Communities of Soybean in Northeast China" Agronomy 15, no. 2: 436. https://doi.org/10.3390/agronomy15020436
APA StyleWang, Y., Bai, Q., Meng, F., Dong, W., Fan, H., Zhu, X., Duan, Y., & Chen, L. (2025). High-Throughput Sequence Analysis of Microbial Communities of Soybean in Northeast China. Agronomy, 15(2), 436. https://doi.org/10.3390/agronomy15020436






