Evaluation of Genetic Diversity in Wild Emmer Wheat (Triticum dicoccoides) and Durum Wheat (Triticum durum) Accessions Using CAAT and SCoT Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Extraction
2.3. CAAT-PCR and SCoT-PCR Amplification
2.4. Data Evaluation
3. Results and Discussion
3.1. Polymorphism Values for the Entire Set of Genotypes
3.2. Polymorphism Values Calculated Based on the Collected Region of Accessions
3.3. Parameters Obtained Using CAAT Molecular Markers
3.4. Parameters Obtained Using SCoT Molecular Markers
3.5. Mean Genetic Diversity Parameters for CAAT and SCoT Markers
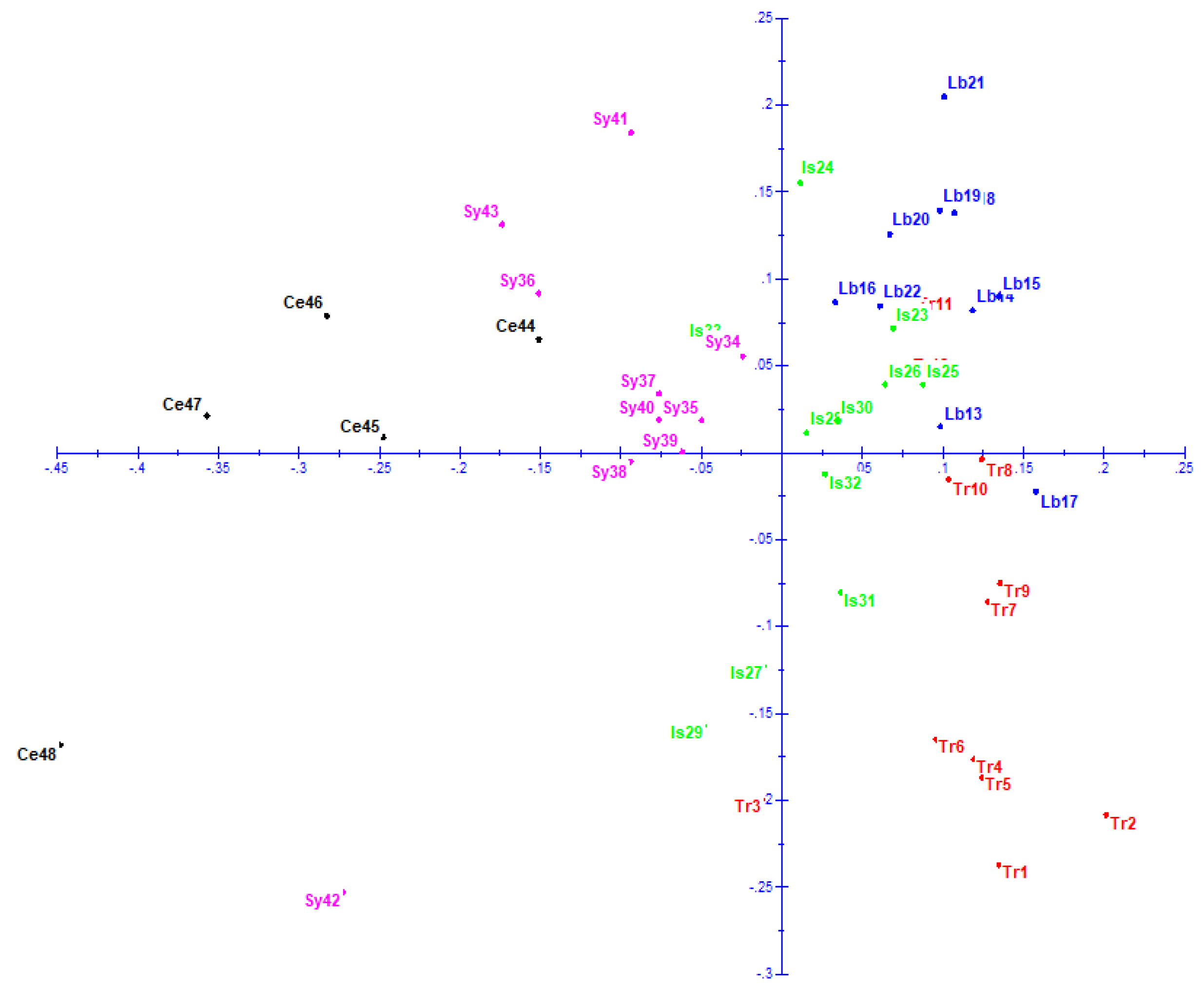
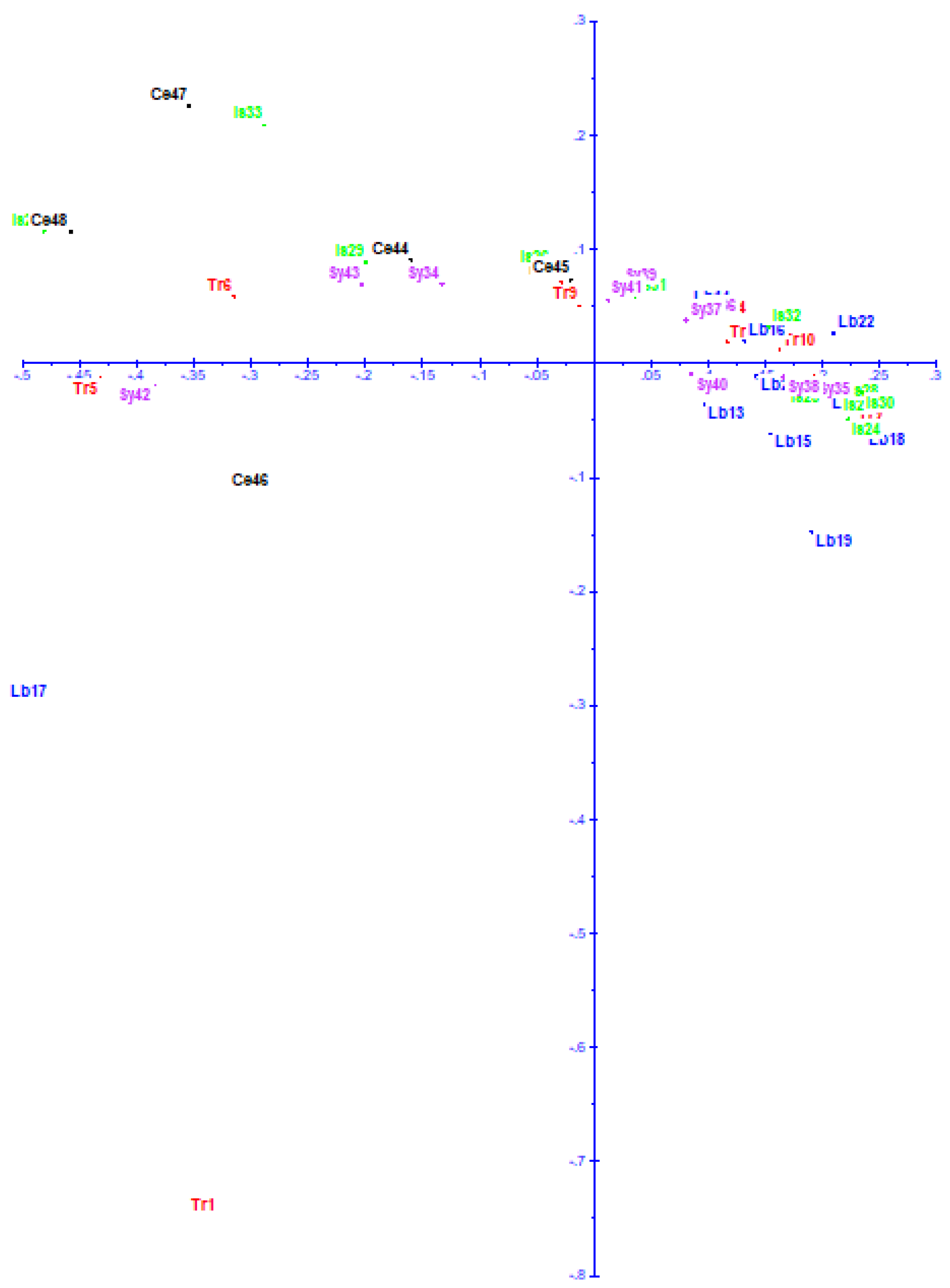
3.6. Principal Coordinate Analysis (PCoA)
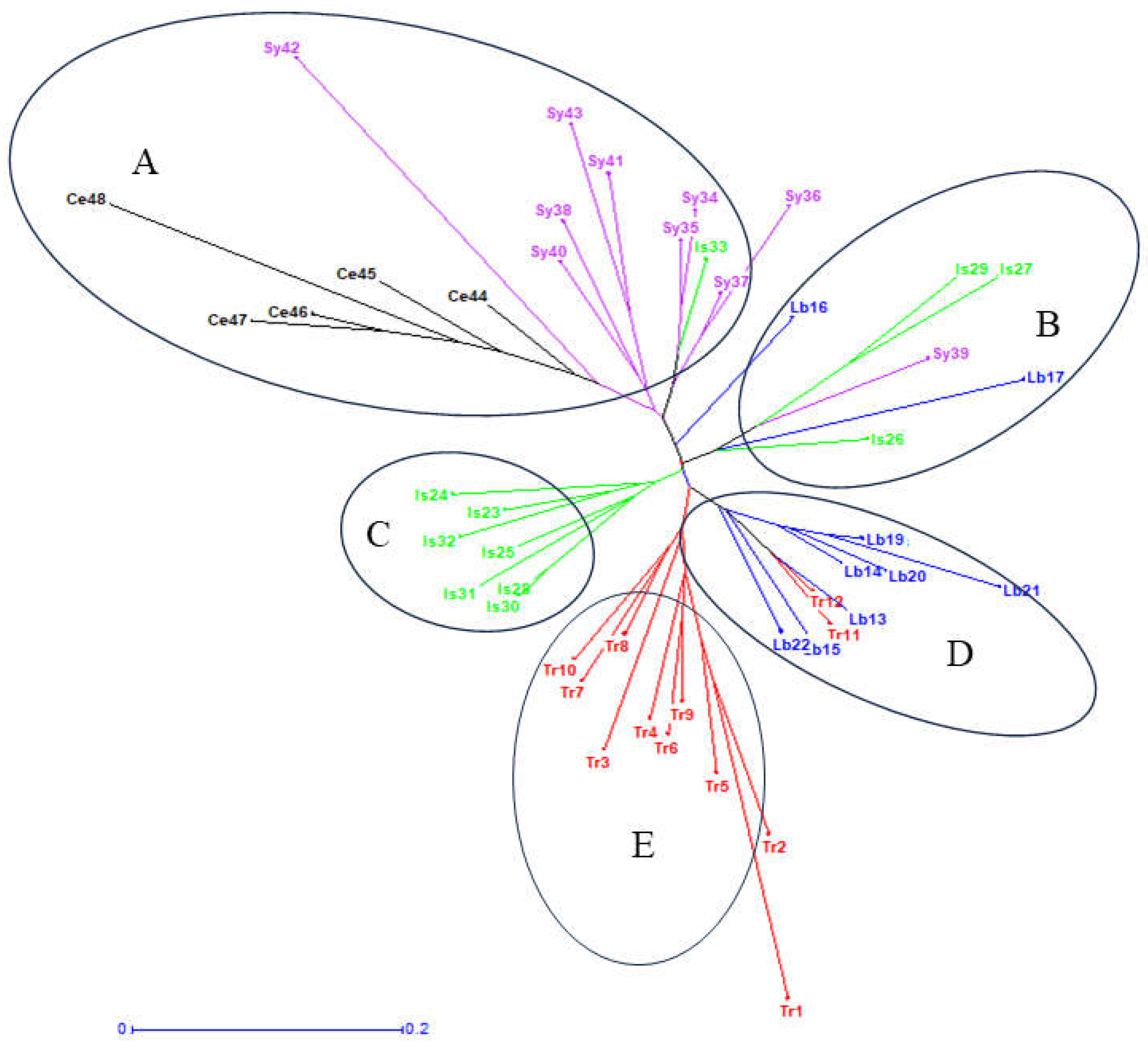
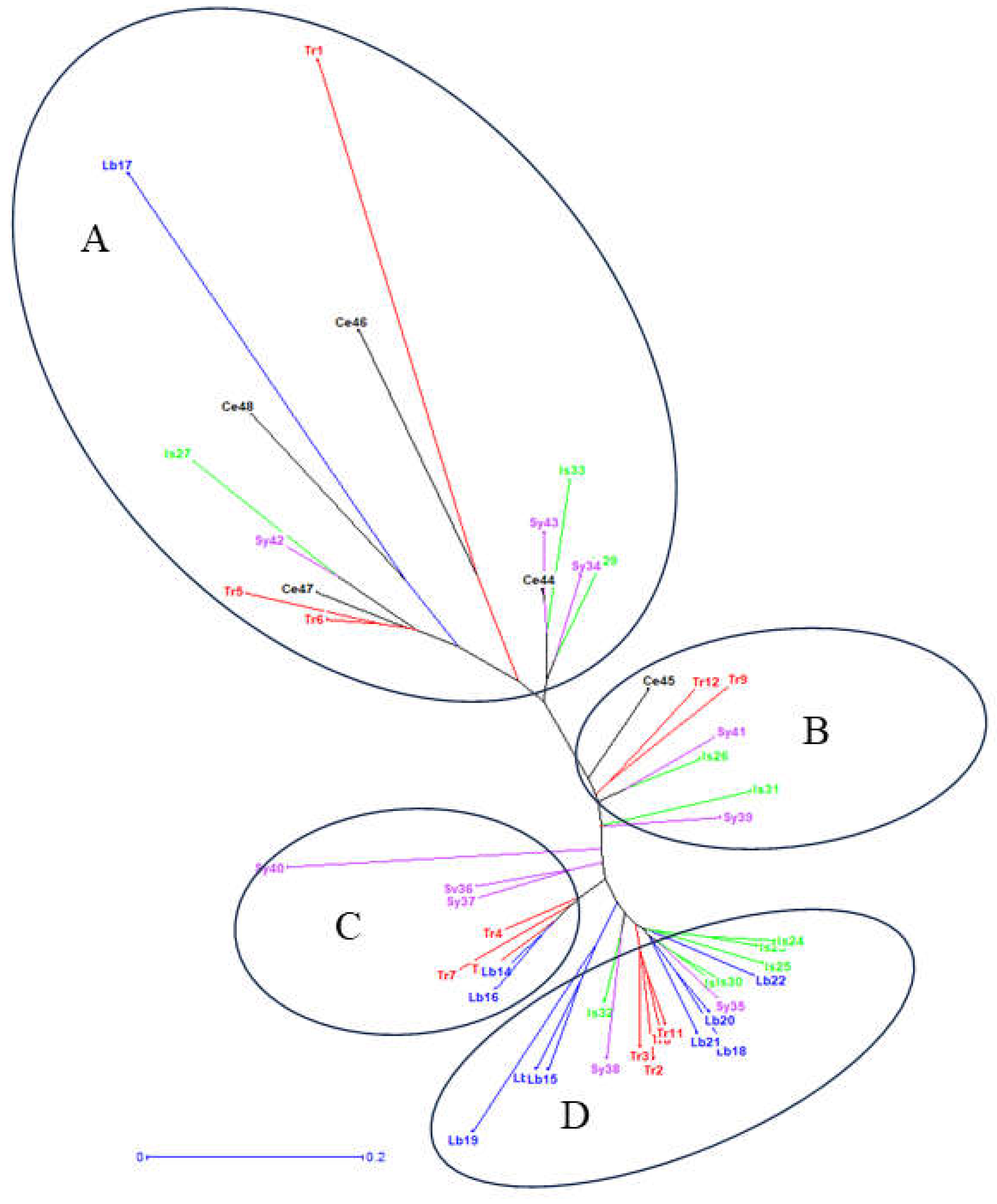
3.7. Neighbor-Joining Analyses
4. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poehlman, J.M. Breeding Field Crops; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2018, 32, 261–285. [Google Scholar] [CrossRef]
- El Haddad, N.; Kabbaj, H.; Zaïm, M.; El Hassouni, K.; Tidiane Sall, A.; Azouz, M.; Ortiz, R.; Baum, M.; Amri, A.; Gamba, F.; et al. Crop wild relatives in durum wheat breeding: Drift or thrift? Crop Sci. 2021, 61, 37–54. [Google Scholar] [CrossRef]
- El Haddad, N.; Sanchez-Garcia, M.; Visioni, A.; Jilal, A.; El Amil, R.; Sall, A.T.; Lagesse, W.; Kumar, S.; Bassi, F.M. Crop Wild Relatives Crosses: Multi-Location Assessment in Durum Wheat, Barley, and Lentil. Agronomy 2021, 11, 2283. [Google Scholar] [CrossRef]
- Dempewolf, H.; Baute, G.; Anderson, J.; Kilian, B.; Smith, C.; Guarino, L. Past and Future Use of Wild Relatives in Crop Breeding. Crop Sci. 2017, 57, 1070–1082. [Google Scholar] [CrossRef]
- Collard, B.C.; Mackill, D.J. Start codon targeted (SCoT) polymorphism: A simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol. Biol. Rep. 2009, 27, 86–93. [Google Scholar] [CrossRef]
- Xiong, F.; Zhong, R.; Han, Z.; Jiang, J.; He, L.; Zhuang, W.; Tang, R. Start codon targeted polymorphism for evaluation of functional genetic variation and relationships in cultivated peanut (Arachis hypogaea L.) genotypes. Mol. Biol. Rep. 2011, 38, 3487–3494. [Google Scholar] [CrossRef]
- Luo, C.; He, X.-H.; Chen, H.; Ou, S.-J.; Gao, M.-P. Analysis of diversity and relationships among mango cultivars using Start Codon Targeted (SCoT) markers. Biochem. Syst. Ecol. 2010, 38, 1176–1184. [Google Scholar] [CrossRef]
- Luo, C.; He, X.-H.; Chen, H.; Ou, S.-J.; Gao, M.-P.; Brown, J.S.; Tondo, C.T.; Schnell, R.J. Genetic diversity of mango cultivars estimated using SCoT and ISSR markers. Biochem. Syst. Ecol. 2011, 39, 676–684. [Google Scholar] [CrossRef]
- Luo, C.; He, X.-H.; Chen, H.; Hu, Y.; Ou, S.-J. Genetic relationship and diversity of Mangifera indica L.: Revealed through SCoT analysis. Genet. Resour. Crop Evol. 2012, 59, 1505–1515. [Google Scholar] [CrossRef]
- Gorji, A.M.; Poczai, P.; Polgar, Z.; Taller, J. Efficiency of arbitrarily amplified dominant markers (SCoT, ISSR and RAPD) for diagnostic fingerprinting in tetraploid potato. Am. J. Potato Res. 2011, 88, 226–237. [Google Scholar] [CrossRef]
- Guo, D.-L.; Zhang, J.-Y.; Liu, C.-H. Genetic diversity in some grape varieties revealed by SCoT analyses. Mol. Biol. Rep. 2012, 39, 5307–5313. [Google Scholar] [CrossRef] [PubMed]
- Joshi, C.P.; Zhou, H.; Huang, X.; Chiang, V.L. Context sequences of translation initiation codon in plants. Plant Mol. Biol. 1997, 35, 993–1001. [Google Scholar] [CrossRef] [PubMed]
- Sawant, S.V.; Singh, P.K.; Gupta, S.K.; Madnala, R.; Tuli, R. Conserved nucleotide sequences in highly expressed genes in plants. J. Genet. 1999, 78, 123–131. [Google Scholar] [CrossRef]
- Pakseresht, F.; Talebi, R.; Karami, E. Comparative assessment of ISSR, DAMD and SCoT markers for evaluation of genetic diversity and conservation of landrace chickpea (Cicer arietinum L.) genotypes collected from north-west of Iran. Physiol. Mol. Biol. Plants 2013, 19, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Daneshvar, Z.; Saeidi, A.; Zadeh, E.B. Comparison of the Performance of SCoT and RAPD Markers for Study of Genetic Diversity and Relationships of Anthurium (Anthurium andreanum) Cultivars. J. Crop Breed. 2018, 10, 101–109. [Google Scholar] [CrossRef]
- Abdein, M.; Abd El-Moneim, D.; Taha, S.S.; Al-Juhani, W.S.; Mohamed, S.E. Molecular characterization and genetic relationships among some tomato genotypes as revealed by ISSR and SCoT markers. Egypt. J. Genet. Cytol. 2018, 47, 139–159. [Google Scholar]
- Nouri, A.; Golabadi, M.; Etminan, A.; Rezaei, A.; Mehrabi, A.A. Comparative assessment of SCoT and ISSR markers for analysis of genetic diversity and population structure in some Aegilops tauschii Coss. accessions. Plant Genet. Resour. 2021, 19, 375–383. [Google Scholar] [CrossRef]
- Singh, A.K.; Rana, M.K.; Singh, S.; Kumar, S.; Kumar, R.; Singh, R. CAAT box- derived polymorphism (CBDP): A novel promoter -targeted molecular marker for plants. J. Plant Biochem. Biotechnol. 2014, 23, 175–183. [Google Scholar] [CrossRef]
- Aslan-Parviz, M.; Omidi, M.; Rashidi, V.; Etminan, A.; Ahmadzadeh, A. Evaluation of genetic diversity of durum wheat (Triticum durum desf.) genotypes using inter-simple sequence repeat (ISSR) and caat box-derived polymorphism (CBDP) markers. Genetika 2020, 52, 895–909. [Google Scholar] [CrossRef]
- Rai, M.K. Start codon targeted (SCoT) polymorphism marker in plant genome analysis: Current status and prospects. Planta 2023, 257, 34. [Google Scholar] [CrossRef]
- Andersen, J.R.; Lübberstedt, T. Functional markers in plants. Trends Plant Sci. 2003, 8, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Etminan, A.; Pour-Aboughadareh, A.; Mohammadi, R.; Noori, A.; Ahmadi-Rad, A. Applicability of CAAT box-derived polymorphism (CBDP) markers for analysis of genetic diversity in durum wheat. Cereal Res. Commun. 2018, 46, 1–9. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Kafkas, S.; Ozkan, H.; Ak, B.; Acar, I.; Atli, H.; Koyuncu, S. Detecting DNA polymorphism and genetic diversity in a wide pistachio germplasm: Comparison of AFLP, ISSR, and RAPD markers. J. Am. Soc. Hortic. Sci. 2006, 131, 522–529. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef]
- Gascuel, O. Concerning the NJ Algorithm and Its Unweighted Version UNJ in Mathematical Hierarchies and Biology; DIMACS Series in Discrete Mathematics and Theoretical Computer Science; Mirkin, B., McMorris, F.R., Roberts, F., Rzhetsky, A., Eds.; American Mathematical Society: Providence, RI, USA, 1997; pp. 149–171. [Google Scholar]
- Rasheed, A.; Hao, Y.; Xia, X.; Khan, A.; Xu, Y.; Varshney, R.K.; He, Z. Crop breeding chips and genotyping platforms: Progress, challenges, and perspectives. Mol. Plant 2017, 10, 1047–1064. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Ma, C.; Jia, Y.-H.; Wang, J.-Z.; Cao, S.-K.; Li, F.-F. The distribution and behavioral characteristics of plateau pikas (Ochotona curzoniae). ZooKeys 2021, 1059, 157. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, W.; Guo, X.; Chen, P.; Cheng, Y.; Mao, K.; Ma, F. Cation/Ca2+ exchanger 1 (MdCCX1), a plasma membrane-localized Na+ transporter, enhances plant salt tolerance by inhibiting excessive accumulation of Na+ and reactive oxygen species. Front. Plant Sci. 2021, 12, 746189. [Google Scholar] [CrossRef] [PubMed]
- Tene, M.; Adhikari, E.; Cobo, N.; Jordan, K.W.; Matny, O.; Del Blanco, I.A.; Roter, J.; Ezrati, S.; Govta, L.; Manisterski, J.; et al. GWAS for Stripe Rust Resistance in Wild Emmer Wheat (Triticum dicoccoides) Population: Obstacles and Solutions. Crops 2022, 2, 42–61. [Google Scholar] [CrossRef]
- Ren, J.; Chen, L.; Sun, D.; You, F.M.; Wang, J.; Peng, Y.; Nevo, E.; Beiles, A.; Sun, D.; Luo, M.-C. SNP-revealed genetic diversity in wild emmer wheat correlates with ecological factors. BMC Evol. Biol. 2013, 13, 169. [Google Scholar] [CrossRef] [PubMed]
- Shizuka, T.; Mori, N.; Ozkan, H.; Ohta, S. Chloroplast DNA haploty variation within two natural populations of wild emmer wheat (Triticum turgidum ssp. dicoccoides) in southern Turkey. Biotechnol. Biotechnol. Equip. 2015, 29, 423–430. [Google Scholar] [CrossRef]
- Abouseada, H.H.; Mohamed, A.-S.H.; Teleb, S.S.; Badr, A.; Tantawy, M.E.; Ibrahim, S.D.; Ellmouni, F.Y.; Ibrahim, M. Genetic diversity analysis in wheat cultivars using SCoT and ISSR markers, chloroplast DNA barcoding and grain SEM. BMC Plant Biol. 2023, 23, 193. [Google Scholar] [CrossRef] [PubMed]
- Hongtrakul, V.; Huestis, G.M.; Knapp, S.J. Amplified fragment length polymorphisms as a tool for DNA fingerprinting sunflower germplasm: Genetic diversity among oilseed inbred lines. Theor. Appl. Genet. 1997, 95, 400–407. [Google Scholar] [CrossRef]
- Garcia, A.A.; Benchimol, L.L.; Barbosa, A.M.; Geraldi, I.O.; Souza, C.L., Jr.; Souza, A.P.d. Comparison of RAPD, RFLP, AFLP and SSR markers for diversity studies in tropical maize inbred lines. Genet. Mol. Biol. 2004, 27, 579–588. [Google Scholar] [CrossRef]
- Sefc, K.; Lopes, M.; Lefort, F.; Botta, R.; Roubelakis-Angelakis, K.; Ibanez, J.; Pejić, I.; Wagner, H.; Glössl, J.; Steinkellner, H. Microsatellite variability in grapevine cultivars from different European regions and evaluation of assignment testing to assess the geographic origin of cultivars. Theor. Appl. Genet. 2000, 100, 498–505. [Google Scholar] [CrossRef]
- Chesnokov, Y.V.; Artemyeva, A.M. Evaluation of the measure of polymorphism information of genetic diversity. Сельскoхoзяйственная Биoлoгия 2015, 5, 571–578. [Google Scholar]
- Najaphy, A.; Parchin, R.A.; Farshadfar, E. Evaluation of genetic diversity in wheat cultivars and breeding lines using inter simple sequence repeat markers. Biotechnol. Biotechnol. Equip. 2011, 25, 2634–2638. [Google Scholar] [CrossRef]
- Gowayed, S.M.; Abd El-Moneim, D. Detection of genetic divergence among some wheat (Triticum aestivum L.) genotypes using molecular and biochemical indicators under salinity stress. PLoS ONE 2021, 16, e0248890. [Google Scholar] [CrossRef]
| Plant Species | Genbank Code | Country of Origin | Collsite | Lat (N) | Long (E) | Elevation |
|---|---|---|---|---|---|---|
| T. dicoccoides | PI 428089 | Tr1 | 37 km NE from kilis to Gaziantep | 37°20′19″ | 37°16′50″ | 830 |
| T. dicoccoides | PI 428023 | Tr2 | 36.2 km west of Diyarbakir in the Karacadag | 37°53′00″ | 39°52′00″ | 1200 |
| T. dicoccoides | PI 428046 | Tr3 | 12.9 km NW from Ovadag to Pirinclik | 37°47′31″ | 39°57′18″ | 1007 |
| T. dicoccoides | PI 428089 | Tr4 | 20.2 km east of Siverek | 37°43′00″ | 39°30′00″ | 1200 |
| T. dicoccoides | PI 656872 | Tr5 | 34 km ESE from Narli (WSW of Karadağ) | 37°20′12″ | 37°17′53″ | 780 (813) |
| T. dicoccoides | PI 428051 | Tr6 | 36.2 km west of Diyarbakir in the Karacadag | 37°53′00′’ | 39°52′00″ | 1200 |
| T. dicoccoides | ----------- | Tr7 | 37 km NE from Kilis to Gaziantep | 37°20′19″ | 37°16′50″ | 830 |
| T. dicoccoides | ----------- | Tr8 | Siverek Karakeçi Road Azemi Village | 37°36′51″ | 39°20′12″ | 733 |
| T. dicoccoides | PI 654321 | Tr9 | 4 km south of Siverek on Karakecili road | 37°43′05″ | 39°19′37″ | 720 |
| T. dicoccoides | PI 554581 | Tr10 | 25 km southwest of Diyarbakir | 37°45′00″ | 40°06′00″ | 1000 |
| T. dicoccoides | PI 538646 | Tr11 | 36.2 km west of Diyarbakir in the Karacadag | 37°53′00″ | 39°52′00″ | 1200 |
| T. dicoccoides | PI 554583 | Tr12 | 3 km southeast of the Junction of Karacadag Mt. road and Diyarbakir highway | 37°47′00″ | 39°47′00″ | 1350 |
| T. dicoccoides | PI 538713 | Lb13 | between Ain Harsch and Ain Ata | 33°26′00″ | 35°46′00″ | 1192 |
| T. dicoccoides | TRI 18478, PI 427998 | Lb14 | zwischen Kfarkouk und Aiha | 33°31′00″ | 35°52′00″ | 1216 |
| T. dicoccoides | TRI 18530, PI 538706 | Lb15 | zwischen Aiha und Kfarkouk, ca. 1 km von Aiha | 33°30′00″ | 35°52′00″ | 1216 |
| T. dicoccoides | PI 538703 | Lb16 | near Rashaya | 33°30′04″ | 35°50′22″ | 1000 |
| T. dicoccoides | PI 428133 | Lb17 | Aiha-Kfarkouk, above ‘sahlet’ | 33°31′00″ | 35°52′00″ | 1141 |
| T. dicoccoides | PI 538702 | Lb18 | near Rashaya | 33°30′04″ | 35°50′22″ | 1000 |
| T. dicoccoides | PI 538704 | Lb19 | near Rashaya | 33°30′04″ | 35°50′22″ | 1000 |
| T. dicoccoides | PI 428143 | Lb20 | between Rashaya and Aiha | 33°30′00″ | 35°50′00″ | 1000 |
| T. dicoccoides | CItr 17675 | Lb21 | outskirts of Rashaya | 33°30′04″ | 35°50′22″ | 1000 |
| T. dicoccoides | TRI 18499, PI 470979 | Lb22 | Mt. Hermon | 33°25′00″ | 35°52′00″ | 2655 |
| T. dicoccoides | PI 428105 | Is23 | 1 to 2 km south of Rosh Pinna toward Safad | 32°58′00″ | 35°32′00″ | 549 |
| T. dicoccoides | PI 538696 | Is24 | Between En haShofet and Daliyya | 32°35′00″ | 35°03′00″ | 122 |
| T. dicoccoides | PI 538670 | Is25 | Afula-Tiberias | 32°36′40″ | 35°17′30″ | 300 |
| T. dicoccoides | PI 538690 | Is26 | near Safad on the road to Rosh Pinna | 32°58′00″ | 35°29′40″ | 800 |
| T. dicoccoides | PI 538679 | Is27 | Afula-Tiberias | 32°36′40″ | 35°17′30″ | 300 |
| T. dicoccoides | PI 466972 | Is28 | Bat Shelomo | 32°35′48″ | 35°00′07″ | 105 |
| T. dicoccoides | PI 466969 | Is29 | Bat Shelomo | 32°35′48″ | 35°00′07″ | 105 |
| T. dicoccoides | PI 466974 | Is30 | Bat Shelomo | 32°35′48″ | 35°00′07″ | 105 |
| T. dicoccoides | PI 538696 | Is31 | Between En haShofet and Daliyya | 32°35′00″ | 35°03′00″ | 122 |
| T. dicoccoides | PI 428112 | Is32 | 1 to 2 km south of Rosh Pinna toward Safad | 32°58′00″ | 35°32′00″ | 549 |
| T. dicoccoides | PI 471041 | Is33 | Kokhav haShahar | 31°57′00″ | 35°20′00″ | 696 |
| T. dicoccoides | PI 466943 | Sy34 | Kazrin | 32°59′24″ | 35°41′24″ | 259 |
| T. dicoccoides | PI 470956 | Sy35 | Kazrin | 32°59′24″ | 35°41′24″ | 259 |
| T. dicoccoides | TRI 18506, PI487255 | Sy36 | Damaskus Provinz | 33°45′00″ | 36°05′00″ | 1240 |
| T. dicoccoides | TRI 18508, PI 487262 | Sy37 | Damaskus Provinz | 33°40′00″ | 36°02′00″ | 1300 |
| T. dicoccoides | PI 487260 | Sy38 | 32 km from Sweida between Sale and Malah | 32°38′52″ | 36°47′24″ | 1530 |
| T. dicoccoides | PI 487254 | Sy39 | Nawa | 32°52′10″ | 36°01′51″ | 551 |
| T. dicoccoides | PI 487264 | Sy40 | Aleppo-Abeen road after Aleppo-Afrin road, Aleppo Province | 36°30′00″ | 37°00′00″ | 350 |
| T. dicoccoides | PI 470945 | Sy41 | Kazrin | 32°59′24″ | 35°41′24″ | 259 |
| T. dicoccoides | TRI 18507, PI 487261 | Sy42 | Es Suweida (Soud) | 32°38′00″ | 36°46′00″ | 1450 |
| T. dicoccoides | PI 466947 | Sy43 | Kazrin | 32°59′24″ | 35°41′24″ | 259 |
| T. durum | Zardak | Ce44 | Iran | ----------- | ---------- | ----- |
| T. durum | Fırat-93 | Ce45 | TUR | ----------- | ---------- | ----- |
| T. durum | Zenit | Ce46 | ITALY | ----------- | ---------- | ----- |
| T. durum | Svevo | Ce47 | ITALY | ----------- | ---------- | ----- |
| T. durum | Timilia | Ce48 | ITALY | ----------- | ---------- | ----- |
| Primer Name | Sequencing (5′-3′) |
|---|---|
| CAAT PRIMERS | |
| CAAT10 | TGAGCACGATCCAATGTT |
| CAAT12 | TGAGCACGATCCAATATA |
| CAAT13 | TGAGCACGATCCAATGAG |
| CAAT14 | TGAGCACGATCCAATGCG |
| CAAT20 | CTGAGCACGATCCAATAT |
| CAAT21 | CTGAGCACGATCCAATCA |
| CAAT22 | CTGAGCACGATCCAATCG |
| SCoT PRIMERS | |
| SCOT7 | CAACAATGGCTACCACGG |
| SCOT8 | CAACAATGGCTACCACGT |
| SCOT9 | CAACAATGGCTACCAGCA |
| SCOT10 | CAACAATGGCTACCAGCC |
| SCOT11 | AAGCAATGGCTACCACCA |
| SCOT13 | ACGACATGGCGACCATCG |
| SCOT16 | CCATGGCTACCACCGGCC |
| SCOT17 | CATGGCTACCACCGGCCC |
| SCOT19 | GCAACAATGGCTACCACC |
| SCOT23 | ACCATGGCTACCACGGGC |
| Primer Name | Scored Bands | H | PIC | E | Hav | MI | D | R |
|---|---|---|---|---|---|---|---|---|
| CAAT Primers | ||||||||
| CAAT10 | 2 | 0.435 | 0.378 | 1.361 | 0.005 | 0.006 | 0.539 | 1.063 |
| CAAT12 | 7 | 0.328 | 0.419 | 1.446 | 0.001 | 0.001 | 0.958 | 1.617 |
| CAAT13 | 12 | 0.491 | 0.352 | 5.212 | 0.001 | 0.005 | 0.812 | 4.638 |
| CAAT14 | 8 | 0.499 | 0.348 | 3.872 | 0.001 | 0.005 | 0.766 | 3.829 |
| CAAT20 | 10 | 0.378 | 0.401 | 2.531 | 0.001 | 0.002 | 0.936 | 3.574 |
| CAAT21 | 12 | 0.498 | 0.349 | 6.361 | 0.001 | 0.006 | 0.719 | 7.531 |
| CAAT22 | 12 | 0.402 | 0.392 | 3.340 | 0.001 | 0.002 | 0.923 | 5.914 |
| Mean | 6.3 | 0.433 | 0.377 | 3.446 | 0.001 | 0.004 | 0.808 | 4.024 |
| SCoT Primers | ||||||||
| SCoT7 | 9 | 0.491 | 0.370 | 3.893 | 0.001 | 0.005 | 0.813 | 4.340 |
| SCoT8 | 10 | 0.487 | 0.371 | 4.191 | 0.001 | 0.004 | 0.825 | 5.489 |
| SCoT9 | 8 | 0.440 | 0.393 | 2.617 | 0.001 | 0.003 | 0.894 | 3.531 |
| SCoT10 | 7 | 0.498 | 0.366 | 3.255 | 0.002 | 0.005 | 0.784 | 2.936 |
| SCoT11 | 6 | 0.497 | 0.367 | 2.765 | 0.002 | 0.005 | 0.788 | 2.127 |
| SCoT13 | 7 | 0.447 | 0.390 | 2.361 | 0.001 | 0.003 | 0.887 | 3.319 |
| SCoT16 | 10 | 0.483 | 0.373 | 4.085 | 0.001 | 0.004 | 0.834 | 2.553 |
| SCoT17 | 7 | 0.499 | 0.365 | 3.361 | 0.002 | 0.005 | 0.770 | 2.382 |
| SCoT19 | 5 | 0.499 | 0.365 | 2.617 | 0.002 | 0.006 | 0.727 | 2.595 |
| SCoT23 | 7 | 0.500 | 0.365 | 3.468 | 0.002 | 0.005 | 0.755 | 4.723 |
| Mean | 7.6 | 0.484 | 0.373 | 3.261 | 0.001 | 0.005 | 0.808 | 3.400 |
| Countries | CAAT Primers | H | PIC | E | Hav | MI | D | R |
|---|---|---|---|---|---|---|---|---|
| caat10 | 0.497 | 0.350 | 1.083 | 0.021 | 0.022 | 0.717 | 1.166 | |
| caat12 | 0.278 | 0.435 | 1.166 | 0.003 | 0.004 | 0.974 | 2.000 | |
| caat13 | 0.481 | 0.358 | 4.833 | 0.003 | 0.016 | 0.839 | 4.333 | |
| caat14 | 0.500 | 0.349 | 4.083 | 0.005 | 0.021 | 0.742 | 3.500 | |
| Turkey | caat20 | 0.413 | 0.388 | 2.916 | 0.003 | 0.010 | 0.917 | 3.833 |
| caat21 | 0.472 | 0.362 | 7.416 | 0.003 | 0.024 | 0.620 | 6.500 | |
| caat22 | 0.353 | 0.411 | 2.750 | 0.002 | 0.007 | 0.949 | 5.166 | |
| mean | 0.428 | 0.379 | 3.464 | 0.006 | 0.015 | 0.823 | 3.785 | |
| caat10 | 0.397 | 0.411 | 1.454 | 0.018 | 0.026 | 0.481 | 1.090 | |
| caat12 | 0.329 | 0.435 | 1.454 | 0.004 | 0.006 | 0.959 | 1.818 | |
| caat13 | 0.500 | 0.364 | 5.909 | 0.004 | 0.022 | 0.759 | 3.090 | |
| caat14 | 0.499 | 0.365 | 3.818 | 0.006 | 0.022 | 0.775 | 3.090 | |
| Israel | caat20 | 0.388 | 0.414 | 2.636 | 0.004 | 0.009 | 0.932 | 3.636 |
| caat21 | 0.483 | 0.373 | 7.090 | 0.004 | 0.026 | 0.653 | 6.727 | |
| caat22 | 0.471 | 0.379 | 4.545 | 0.004 | 0.016 | 0.858 | 7.636 | |
| mean | 0.438 | 0.391 | 3.844 | 0.006 | 0.018 | 0.774 | 3.870 | |
| caat10 | 0.420 | 0.402 | 1.400 | 0.021 | 0.029 | 0.521 | 1.200 | |
| caat12 | 0.368 | 0.423 | 1.700 | 0.005 | 0.009 | 0.944 | 1.400 | |
| caat13 | 0.500 | 0.365 | 5.900 | 0.004 | 0.025 | 0.760 | 4.200 | |
| caat14 | 0.455 | 0.387 | 5.200 | 0.006 | 0.030 | 0.580 | 2.400 | |
| Lebanon | caat20 | 0.428 | 0.399 | 3.100 | 0.004 | 0.013 | 0.906 | 4.600 |
| caat21 | 0.500 | 0.365 | 5.900 | 0.004 | 0.025 | 0.760 | 8.200 | |
| caat22 | 0.439 | 0.394 | 3.900 | 0.004 | 0.014 | 0.896 | 5.400 | |
| mean | 0.444 | 0.391 | 3.315 | 0.007 | 0.021 | 0.767 | 3.914 | |
| caat10 | 0.320 | 0.403 | 1.600 | 0.016 | 0.026 | 0.368 | 0.800 | |
| caat12 | 0.368 | 0.387 | 1.700 | 0.005 | 0.009 | 0.944 | 0.600 | |
| caat13 | 0.477 | 0.341 | 4.700 | 0.004 | 0.019 | 0.849 | 3.800 | |
| caat14 | 0.480 | 0.339 | 3.200 | 0.006 | 0.019 | 0.843 | 3.600 | |
| Syria | caat20 | 0.320 | 0.403 | 2.200 | 0.003 | 0.006 | 0.962 | 2.800 |
| caat21 | 0.498 | 0.331 | 5.600 | 0.004 | 0.023 | 0.784 | 4.800 | |
| caat22 | 0.391 | 0.378 | 3.200 | 0.003 | 0.010 | 0.931 | 4.000 | |
| mean | 0.408 | 0.369 | 3.171 | 0.006 | 0.016 | 0.811 | 2.714 | |
| caat10 | 0.500 | 0.171 | 1.000 | 0.050 | 0.050 | 0.778 | 0.001 | |
| caat12 | 0.202 | 0.276 | 0.800 | 0.006 | 0.005 | 0.990 | 1.600 | |
| caat13 | 0.391 | 0.220 | 3.200 | 0.007 | 0.021 | 0.932 | 3.600 | |
| caat14 | 0.320 | 0.245 | 1.600 | 0.008 | 0.013 | 0.964 | 2.400 | |
| Durum Wheat | caat20 | 0.147 | 0.286 | 0.800 | 0.003 | 0.002 | 0.995 | 1.200 |
| caat21 | 0.406 | 0.214 | 3.400 | 0.007 | 0.023 | 0.923 | 2.000 | |
| caat22 | 0.095 | 0.292 | 0.600 | 0.002 | 0.001 | 0.998 | 1.200 | |
| mean | 0.295 | 0.243 | 1.628 | 0.012 | 0.016 | 0.940 | 1.714 |
| Countries | Primers | H | PIC | E | Hav | MI | D | R |
|---|---|---|---|---|---|---|---|---|
| Scot7 | 0.499 | 0.360 | 4.333 | 0.005 | 0.020 | 0.771 | 2.666 | |
| Scot8 | 0.486 | 0.370 | 4.166 | 0.004 | 0.017 | 0.828 | 5.333 | |
| Scot9 | 0.413 | 0.400 | 2.333 | 0.004 | 0.010 | 0.917 | 3.000 | |
| Scot10 | 0.472 | 0.380 | 2.666 | 0.006 | 0.015 | 0.858 | 1.333 | |
| Turkey | Scot11 | 0.486 | 0.370 | 2.500 | 0.007 | 0.017 | 0.830 | 2.333 |
| Scot13 | 0.436 | 0.390 | 2.250 | 0.005 | 0.012 | 0.899 | 2.166 | |
| Scot16 | 0.413 | 0.400 | 2.916 | 0.003 | 0.010 | 0.917 | 3.166 | |
| Scot17 | 0.486 | 0.370 | 4.083 | 0.006 | 0.024 | 0.663 | 1.833 | |
| Scot19 | 0.486 | 0.370 | 2.916 | 0.008 | 0.024 | 0.664 | 1.500 | |
| Scot23 | 0.490 | 0.370 | 4.000 | 0.006 | 0.023 | 0.676 | 2.333 | |
| Mean | 0.467 | 0.379 | 3.216 | 0.005 | 0.017 | 0.802 | 2.566 | |
| Scot7 | 0.485 | 0.382 | 3.727 | 0.005 | 0.018 | 0.831 | 4.000 | |
| Scot8 | 0.472 | 0.388 | 3.818 | 0.004 | 0.016 | 0.856 | 4.727 | |
| Scot9 | 0.469 | 0.390 | 3.000 | 0.005 | 0.016 | 0.862 | 2.545 | |
| Scot10 | 0.490 | 0.380 | 4.000 | 0.006 | 0.025 | 0.677 | 2.545 | |
| Israel | Scot11 | 0.498 | 0.376 | 2.818 | 0.008 | 0.021 | 0.783 | 1.636 |
| Scot13 | 0.499 | 0.375 | 3.636 | 0.006 | 0.024 | 0.733 | 2.545 | |
| Scot16 | 0.499 | 0.375 | 5.272 | 0.005 | 0.024 | 0.724 | 3.272 | |
| Scot17 | 0.499 | 0.375 | 3.363 | 0.006 | 0.022 | 0.772 | 2.727 | |
| Scot19 | 0.452 | 0.397 | 3.272 | 0.008 | 0.027 | 0.576 | 2.000 | |
| Scot23 | 0.481 | 0.384 | 4.181 | 0.006 | 0.026 | 0.646 | 2.727 | |
| Mean | 0.484 | 0.382 | 3.709 | 0.006 | 0.022 | 0.746 | 2.872 | |
| Scot7 | 0.500 | 0.375 | 4.600 | 0.006 | 0.026 | 0.742 | 3.200 | |
| Scot8 | 0.476 | 0.386 | 6.100 | 0.005 | 0.029 | 0.630 | 4.200 | |
| Scot9 | 0.480 | 0.384 | 3.200 | 0.006 | 0.019 | 0.843 | 3.200 | |
| Scot10 | 0.474 | 0.387 | 2.700 | 0.007 | 0.018 | 0.855 | 1.000 | |
| Scot11 | 0.499 | 0.375 | 2.900 | 0.008 | 0.024 | 0.771 | 1.400 | |
| Lebanon | Scot13 | 0.420 | 0.411 | 2.100 | 0.006 | 0.013 | 0.913 | 2.600 |
| Scot16 | 0.484 | 0.383 | 4.100 | 0.005 | 0.020 | 0.834 | 2.200 | |
| Scot17 | 0.474 | 0.387 | 4.300 | 0.007 | 0.029 | 0.626 | 1.000 | |
| Scot19 | 0.497 | 0.376 | 2.700 | 0.010 | 0.027 | 0.713 | 2.200 | |
| Scot23 | 0.474 | 0.387 | 4.300 | 0.007 | 0.029 | 0.626 | 2.200 | |
| Mean | 0.478 | 0.385 | 3.700 | 0.007 | 0.023 | 0.755 | 2.320 | |
| Scot7 | 0.470 | 0.356 | 3.400 | 0.005 | 0.018 | 0.860 | 2.400 | |
| Scot8 | 0.412 | 0.382 | 2.900 | 0.004 | 0.012 | 0.918 | 3.800 | |
| Scot9 | 0.439 | 0.370 | 2.600 | 0.005 | 0.014 | 0.897 | 3.200 | |
| Scot10 | 0.500 | 0.342 | 3.400 | 0.007 | 0.024 | 0.768 | 3.200 | |
| Syria | Scot11 | 0.498 | 0.343 | 3.200 | 0.008 | 0.027 | 0.720 | 2.400 |
| Scot13 | 0.420 | 0.379 | 2.100 | 0.006 | 0.013 | 0.913 | 2.200 | |
| Scot16 | 0.480 | 0.352 | 4.000 | 0.005 | 0.019 | 0.842 | 1.600 | |
| Scot17 | 0.485 | 0.349 | 2.900 | 0.007 | 0.020 | 0.832 | 1.000 | |
| Scot19 | 0.487 | 0.348 | 2.100 | 0.010 | 0.020 | 0.829 | 2.600 | |
| Scot23 | 0.353 | 0.405 | 1.600 | 0.005 | 0.008 | 0.950 | 2.400 | |
| Mean | 0.454 | 0.363 | 2.820 | 0.006 | 0.018 | 0.853 | 2.480 | |
| Scot7 | 0.411 | 0.316 | 2.600 | 0.009 | 0.024 | 0.921 | 2.800 | |
| Scot8 | 0.471 | 0.289 | 3.800 | 0.009 | 0.036 | 0.860 | 2.000 | |
| Scot9 | 0.180 | 0.384 | 0.800 | 0.005 | 0.004 | 0.992 | 1.600 | |
| Scot10 | 0.496 | 0.277 | 3.800 | 0.014 | 0.054 | 0.713 | 0.400 | |
| Durum Wheat | Scot11 | 0.391 | 0.323 | 1.600 | 0.013 | 0.021 | 0.936 | 1.200 |
| Scot13 | 0.202 | 0.379 | 0.800 | 0.006 | 0.005 | 0.990 | 0.400 | |
| Scot16 | 0.461 | 0.294 | 3.600 | 0.009 | 0.033 | 0.875 | 2.000 | |
| Scot17 | 0.202 | 0.379 | 0.800 | 0.006 | 0.005 | 0.990 | 1.600 | |
| Scot19 | 0.365 | 0.333 | 1.200 | 0.015 | 0.018 | 0.950 | 1.200 | |
| Scot23 | 0.408 | 0.317 | 2.000 | 0.012 | 0.023 | 0.924 | 2.000 | |
| Mean | 0.359 | 0.329 | 1.860 | 0.010 | 0.022 | 0.915 | 1.440 |
| H | PIC | E | Hav | MI | D | R | ||
|---|---|---|---|---|---|---|---|---|
| CAAT PRIMERS | ||||||||
| Turkey | 0.428 | 0.379 | 3.464 | 0.006 | 0.015 | 0.823 | 3.785 | |
| Israel | 0.438 | 0.391 | 3.844 | 0.006 | 0.018 | 0.774 | 3.870 | |
| Lebanon | 0.444 | 0.391 | 3.315 | 0.007 | 0.021 | 0.767 | 3.914 | |
| Syria | 0.408 | 0.369 | 3.171 | 0.006 | 0.016 | 0.811 | 2.714 | |
| Durum Wheat | 0.295 | 0.243 | 1.628 | 0.012 | 0.016 | 0.940 | 1.714 | |
| SCoT PRIMERS | ||||||||
| Turkey | 0.467 | 0.379 | 3.216 | 0.005 | 0.017 | 0.802 | 2.566 | |
| Israel | 0.484 | 0.382 | 3.709 | 0.006 | 0.022 | 0.746 | 2.872 | |
| Lebanon | 0.478 | 0.385 | 3.700 | 0.007 | 0.023 | 0.755 | 2.320 | |
| Syria | 0.454 | 0.363 | 2.820 | 0.006 | 0.018 | 0.853 | 2.480 | |
| Durum Wheat | 0.359 | 0.329 | 2.100 | 0.010 | 0.022 | 0.915 | 1.440 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Çakır, E. Evaluation of Genetic Diversity in Wild Emmer Wheat (Triticum dicoccoides) and Durum Wheat (Triticum durum) Accessions Using CAAT and SCoT Markers. Agronomy 2025, 15, 284. https://doi.org/10.3390/agronomy15020284
Çakır E. Evaluation of Genetic Diversity in Wild Emmer Wheat (Triticum dicoccoides) and Durum Wheat (Triticum durum) Accessions Using CAAT and SCoT Markers. Agronomy. 2025; 15(2):284. https://doi.org/10.3390/agronomy15020284
Chicago/Turabian StyleÇakır, Esra. 2025. "Evaluation of Genetic Diversity in Wild Emmer Wheat (Triticum dicoccoides) and Durum Wheat (Triticum durum) Accessions Using CAAT and SCoT Markers" Agronomy 15, no. 2: 284. https://doi.org/10.3390/agronomy15020284
APA StyleÇakır, E. (2025). Evaluation of Genetic Diversity in Wild Emmer Wheat (Triticum dicoccoides) and Durum Wheat (Triticum durum) Accessions Using CAAT and SCoT Markers. Agronomy, 15(2), 284. https://doi.org/10.3390/agronomy15020284






