Abstract
Brassica yellows virus (BrYV) mainly infects cruciferous crops and has been widely prevalent across China. To develop a rapid and highly sensitive method for detecting BrYV in oilseed rape, a reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay was established. Four specific primers were designed to target the conserved gene of BrYV, with total RNA extracted from BrYV-infected oilseed rape leaves used as the template for the RT-LAMP assay. The optimal reaction conditions were determined, including a primer concentration ratio of 1:8, MgSO4 concentration of 4 mM, reaction temperature of 64 °C, and a suitable reaction time of 60 min. Sensitivity analysis demonstrated that the RT-LAMP assay could detect total RNA at a concentration of 0.091 × 10−3 μg/μL, which was 100-fold more sensitive than conventional RT-PCR for BrYV detection. In addition to visualizing results by electrophoresis, the RT-LAMP assay could also be easily visualized using calcein-MnCl2. These results indicate the potential of the established RT-LAMP assay for rapid BrYV detection in oilseed rape plants, which can provide better technical support for field diagnosis, disease forecasting, and the implementation of effective control strategies against the virus.
1. Introduction
Brassica yellows virus (BrYV) is a member of the family Luteoviridae, genus Polerovirus, and mainly infects cruciferous crops, including cabbage, mustard, and oilseed rape. It is transmitted by aphids in a persistent, circulative non-propagative manner, infecting only the phloem and causing leaf yellowing and curling symptoms [1,2,3,4]. Oilseed rape plants infected with BrYV exhibit distinct red or purple leaves [3]. BrYV is a positive-sense, single-stranded RNA virus with three genotypes, BrYV-A, BrYV-B, and BrYV-C, which all occur in China, and its genome contains six open reading frames (ORFs) encoding six proteins [5,6]. BrYV is widely prevalent throughout China, with its presence detected in 21 provinces, such as Beijing, Inner Mongolia, Xinjiang, and Yunnan [7]. Therefore, a simple and rapid diagnostic method with a high sensitivity is required for detecting the virus.
Loop-mediated isothermal amplification (LAMP) was first described in 2000 by Notomi et al. [8] as a novel, isothermal technique for nucleic acid amplification. Reverse-transcription LAMP (RT-LAMP) is a one-step RNA amplification method conducted under isothermal conditions, achieved by adding reverse transcriptase to the original reaction [8,9]. The RT-LAMP specificity is extremely high because it uses four primers recognizing six distinct regions on the RNA template. Since the reaction occurs under isothermal conditions, there is no time loss associated with temperature changes. In our study, results can be observed by visually inspecting the color changes before and after the reaction. Adding a calcein-MnCl2 indicator before the reaction, and after the reaction ends, positive samples turn green, while negative samples remain orange. This technique has been widely used for detecting various pathogens [10,11,12,13,14,15,16,17,18]. Zhang et al. [19] established a multiplex RT-PCR assay system for BrYV detection. RT-PCR-based assays require specialized equipment and are more time-consuming than RT-LAMP. In this study, a rapid and accurate RT-LAMP method for detecting BrYV in oilseed rape was developed and optimized using total RNA extracted from oilseed rape. The relative sensitivity of RT-LAMP and RT-PCR for BrYV detection was evaluated. Our optimized and validated RT-LAMP method provides a simple, efficient, highly specific, and sensitive assay for the detection of BrYV. This method can offer improved technical support for field diagnosis, disease forecasting, and the implementation of effective control strategies against the virus.
2. Materials and Methods
2.1. Plant Material and Extraction of Total RNA
Five samples of oilseed rape leaves with BrYV-like infection symptoms were collected from Nanjing, Yancheng, and Yangzhou in Jiangsu province, China, during the 2020 oilseed rape growing season. Healthy oilseed rape plants were selected as a control. The collected samples were carefully wrapped in tin foil, rapidly frozen in liquid nitrogen, and stored at −80 °C.
Total RNA was extracted from 100 mg of symptomatic oilseed rape tissue using TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions, and the extracted RNA was stored at −80 °C. All samples were tested via RT-PCR to differentiate positive and negative samples, and the primers are shown in Table 1.

Table 1.
Sequences of primers.
2.2. Primer Design
Four specific target primers were designed using LAMP primer design software primer explorer V5, based on the conserved gene of BrYV (accession number OM469309). Additionally, two RT-PCR primers were designed using PCR primer design software Oligo 6.0, based on the BrYV CP sequence. The most optimal primer sequences were selected and are listed in Table 1.
2.3. RT-LAMP
The concentrations of primers, MgSO4, and the reaction temperature and time were individually optimized The 25 μL reaction mix contained the following components: 1 μL of 10 mM dNTP Mixture (Sangon biotech, Shanghai, China), 1 μL Bst3.0 DNA polymerase (NEB, Ipswich, MA, USA, 2.5 μL of 10 × Isothermal Amplification Buffer II (TOYOBO, Osaka, Japan), 0.2 μM F3 and B3, 1.6 μM FIP and BIP, 0.5 μL Rnase inhibitor (40 U/μL, Fermentas, Lafayette, CO, USA), 8 mM MgSO4, and 0.5 μL of total RNA from symptomatic oilseed rape plants as a template (total RNA from healthy oilseed rape as negative control). DEPC -treated water was added to adjust the volume to 25 μL. The reaction conditions were optimized for temperature and time, performed at 64 °C for 60 min, followed by termination at 80 °C for 5 min. Amplification products (5 μL) were visualized by agarose gel electrophoresis.
2.4. RT-PCR
The cDNA was synthesized via reverse transcription using 1 μL of total RNA as the template and HiScript III 1st Strand cDNA Synthesis Kit (+gDNA wiper) (Vazyme, Nanjing, China), according to the manufacturer’s instructions. For the PCR reaction, 1 μL of cDNA was used as the template, along with forward and reverse (F/R) primers. The PCR reaction mixture comprised 1 μL cDNA, 0.8 μL primer F, 0.8 μL primer R, and 10 μL 2 × GS Taq Master Mix. The PCR reaction protocol was as follows: initial denaturation at 95 °C for 3 min; followed by 34 cycles of denaturation at 95 °C for 30 s, annealing at 52 °C for 30 s, and extension at 72 °C for 30 s; with a final extension at 72 °C for 3 min. After the reaction, 5 μL of the PCR product was subjected to agarose gel electrophoresis for analysis.
2.5. RT-LAMP Sensitivity and Visualization Assay
The concentration of total RNA extracted from diseased oilseed rape plants was measured using a UV spectrophotometer (Thermo NanoDrop 2000C, Thermo Fisher Scientific, Waltham, MA, USA). After tenfold serial dilution with DEPC-treated water, 2 μL of the diluted RNA was used as a template for RT-LAMP or RT-PCR reactions independently. Total RNA of healthy oilseed rape was used as a negative control. The optimized RT-LAMP systems were employed for the reactions.
Calcein was employed as the fluorescence indicator. For its preparation, calcein was dissolved in DMSO to a final concentration of 5 mM and then diluted to 500 μM with double-distilled water (ddH2O). Before RT-LAMP reaction, 3 μL of this solution and 3 μL of 15 mM MnCl2 solution were added to the 25 μL reaction mixture.
3. Results
3.1. Oilseed Rape Plants with Leaf Yellowing and Curling Symptoms in Fields in Nanjinge
In 2020, oilseed rape plants showing symptoms of leaf yellowing and curling were collected from fields in Nanjing, Yancheng, and Yangzhou, Jiangsu Province. Red and purple leaf symptoms varied among the oilseed rape plants (Figure 1).

Figure 1.
BrYV symptoms on oilseed rape plants in field of Nanjing. (A): Visible symptoms of BrYV-infected oilseed rape plants in field. (B): Red and purple leaf symptoms in BrYV-infected oilseed rape plants.
RT-PCR results indicated that five oilseed rape samples were positive for BrYV. The sample utilized in this study was collected from Nanjing.
3.2. Optimization of RT-LAMP Reaction
Following an optimization test on primer concentration, amplification products were undetectable via electrophoresis when the external primer to internal primer concentration ratio was 1:2. However, ladder-like bands, which are characteristic of the LAMP reaction, were observed at concentration ratio of 1:4, 1:6, and 1:8. The optimal result was obtained with a concentration ratio of 1:8 (Figure 2A).
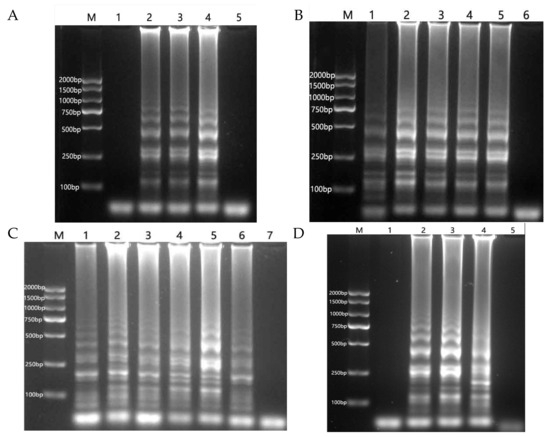
Figure 2.
Optimization of the RT-LAMP reaction system. (A): Primer concentration ratio. M: DNA marker. 1–4: 1:2, 1:4, 1:6, 1:8. 5: negative control. (B): MgSO4 concentration. M: DNA marker. 1–5: 2, 4, 6, 8, 10 mM. 6: negative control. (C): Reaction temperature. M: DNA marker. 1–6: 60–65 °C. 7: negative control. (D): Reaction time, (M): DNA marker. 1–4: 20, 40, 60, 80 min. 5: negative control.
The LAMP reaction exhibited ladder-like bands at final MgSO4 concentrations ranging from 2 to 10 mM, with the optimal result observed at 4 mM (Figure 2B).
Characteristic ladder-like bands of the LAMP reaction were observed at temperatures ranging from 60 °C to 65 °C, with the optimal results obtained at 64 °C (Figure 2C). The amplification product was not detected by electrophoresis at 20 min of constant temperature reaction. The amplification product was detectable at 40 min, but in small amounts; the amplification product increased after 60 min of reaction, and no significant change was observed when the reaction time was extended to 80 min. Therefore, a constant temperature reaction time of 60 min was determined to be most suitable (Figure 2D).
Following optimization, a 25 μL RT-LAMP reaction system was established as follows: 0.2 μM each of F3 and B3, 1.6 μM each of FIP and BIP, 1 μL of dNTP mixture, 10 mM solution, 1 μL of Bst 3.0 DNA polymerase, 2.5 μL of 10 × Isothermal Amplification Buffer II, 0.5 μL Rnase inhibitor, MgSO4 at a final concentration of 4 mM. The mixture was incubated at 64 °C for 60 min, followed by 5 min at 80 °C to terminate the reaction.
3.3. RT-LAMP Sensitivity and Visualization Assay
Total RNA was extracted from oilseed rape leaves, yielding a concentration of 0.091 μg/μL with an OD260/OD280 ratio of 2.18 measured by UV spectrophotometer (Thermo NanoDrop 2000C). The RT-LAMP method could detect total RNA at a concentration of 0.091 × 10−3 μg/μL, demonstrating a sensitivity 100 times higher than that of the RT-PCR method (Figure 3).
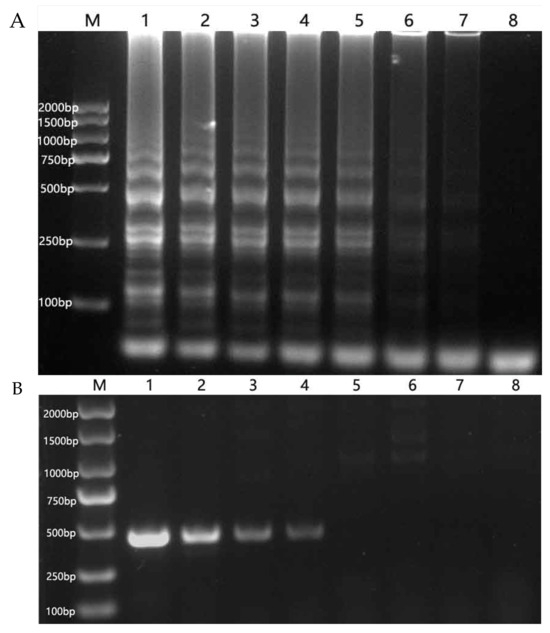
Figure 3.
Comparison of sensitivity of RT-LAMP (A) and RT-PCR (B). (A): M: DNA marker. 1–7: 9.1 × 10, 9.1, 9.1 × 10−1, 9.1 × 10−2, 9.1 × 10−3, 9.1 × 10−4, 9.1 × 10−5 μg/μL, 8: negative control. (B): M: DNA marker. 1–7: 9.1 × 10, 9.1, 9.1 × 10−1, 9.1 × 10−2, 9.1 × 10−3, 9.1 × 10−4, 9.1 × 10−5 μg/μL. 8: negative control.
Calcein was used as the fluorescence indicator. After the reaction ended, positive samples turned green while negative samples retained their original orange color. The detection results could be directly judged by observing these color changes (Figure 4).

Figure 4.
RT-LAMP results observed via visual inspection of color changes. 1: positive control, 2: negative control.
4. Discussion
China is not only the world’s largest consumer of cabbage, but also its largest producer [20]. Therefore, BrYV detection is crucial for the prevention and control of this disease. Traditional methods for detecting viruses, such as enzyme linked immunosorbent assay, RT-PCR, and quantitative PCR (qPCR), have limitations due to their complex production technology, lengthy processing cycles, and high production cost of monoclonal antibodies [21,22,23,24,25,26]. Multiplex RT-PCR assay and Taq Man real-time fluorescence quantitative RT-PCR have been established for the detection of BrYV [19,27]. However, RT-PCR and qPCR require the use of sophisticated instruments in the detection process, which imposes certain limitations in practical detection applications. This leads to numerous challenges for field diagnosis, disease forecasting, and disease prevention and control.
The development of RT-LAMP technology provides a new means for rapid detection of BrYV due to its advantages, including simple operation, isothermal conditions (60–65 °C), the requirement of only a one-step reaction, and visualization detection via the addition of a calcein-MnCl2 indicator [28,29]. However, because of its high sensitivity, rapid reaction, and large product amplification, RT-LAMP is prone to false positives during experiments [30]. Therefore, the operation process should strictly establish a test partition, avoiding aerosol contamination when the lid is opened, and ensure that the experimental environment, apparatus, and reagents are free from contamination by template RNA or amplification products, as such contamination can lead to false positive results and misjudgment.
The visualization method for LAMP involves adding a visualization indicator prior to the amplification reaction. This approach helps prevent potential aerosol contamination, as it does not require the reaction tube to be opened after amplification. Furthermore, aerosol generation can also be minimized by sealing the tube with paraffin oil during preparation. Visual LAMP has been widely applied in virus detection [31,32,33].
This rapid and specific RT-LAMP assay is suitable for the detection of BrYV in oilseed rape. The method developed in this study not only provides technical support for the rapid diagnosis and epidemiological study of the virus, but also holds significant implications for the prediction and control of Brassica yellowing disease in oilseed rape. However, whether the RT-LAMP can also detect BrYV in aphids remains to be verified.
5. Conclusions
In conclusion, an RT-LAMP assay was established for the rapid detection of BrYV in oilseed rape plants. In addition to observing results via electrophoresis, the detection results could be directly judged by observing color changes before and after the reaction. The established RT-LAMP assay can offer improved technical support for field diagnosis, disease forecasting, and the implementation of effective control strategies against the virus.
Author Contributions
Conceptualization, T.Z. and M.H.; methodology, L.D.; validation, T.Z. and Q.P.; formal analysis, L.D. and T.L.; investigation, L.D. and F.Z.; resources, Q.P. and X.Z.; data curation, L.D. and T.L.; writing—original draft preparation, L.D.; writing—review and editing, T.L. and J.K.K.; visualization, F.L.; supervision, T.Z. and F.Z.; project administration, T.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program, grant number 2023YFD1400300; and Guangxi Science and Technology Base and Talent Special Project, grant number guike AC22035090. J.K.K. is supported by the Ministry of Agriculture, Czech Republic, grant number MZE-RO0425.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Wang, Q.; Mao, J.; Xiang, Y.; Dong, H.; Sun, H.; Liu, S.; Liu, B. First Report of Brassica Yellows Virus on Tobacco in China. Plant Dis. 2015, 99, 1192. [Google Scholar] [CrossRef]
- Seungmo, L.; Hee, Y.; Davaajargal, I.; Fumei, Z.; Hyun, K.; Sun, M. Genome sequence of a recombinant brassica yellows virus infecting Chinese cabbage. Arch. Virol. 2015, 160, 597–600. [Google Scholar]
- Xiang, H.Y.; Dong, S.W.; Shang, Q.X.; Zhou, C.J.; Li, D.W.; Yu, J.L.; Han, C.G. Molecular characterization of two genotypes of a new polerovirus infecting brassicas in China. Arch. Virol. 2011, 156, 2251–2255. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Li, W.; Zhou, X.Y.; Sun, C.M.; Hou, Y.; Hu, M.L.; Fu, S.X.; Zhang, J.F.; Kundu, J.K.; Lei, L. Genetic Diversity Analysis of Brassica Yellows Virus Causing Aberrant Color Symptoms in Oilseed Rape. Plants 2023, 12, 1008. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Xiang, H.Y.; Zhou, C.J.; Li, D.W.; Yu, J.L.; Han, C.G. Complete genome sequence analysis identifies a new genotype of brassica yellows virus that infects cabbage and radish in China. Arch. Virol. 2014, 159, 2177–2180. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Dong, S.W.; Xiang, H.Y.; Chen, X.R.; Li, D.W.; Yu, J.L.; Han, C.G. Development of three full-length infectious cDNA clones of distinct brassica yellows virus genotypes for agrobacterium-mediated inoculation. Virus Res. 2015, 197, 13–16. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Peng, Y.M.; Xiang, H.Y.; Wang, Y.L.; Yu, D.W.; Han, C.G. Incidence and prevalence levels of three aphid-transmitted viruses in crucifer crops in China. J. Integr. Agric. 2022, 21, 774–780. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, 63. [Google Scholar] [CrossRef]
- Fukuta, S.; Iida, T.; Mizukami, Y.; Ishida, A.; Ueda, J.; Kanbe, M.; Ishimoto, Y. Detection of Japanese yam mosaic virus by RT-LAMP. Arch. Virol. 2003, 148, 1713–1720. [Google Scholar] [CrossRef]
- Gross, A.J.; Lopez, S., Jr.; Rogers, A.; Adkins, S.; Breitbart, M. A reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the rapid colorimetric detection of pepper mild mottle virus (PMMoV). J. Virol. Methods 2025, 338, 115225. [Google Scholar] [CrossRef]
- Yan, A.; Lei, Y.T.; Yu, Z.Y.; Wu, G.L.; Feng, W.Z.; Chen, X.R. Establishment of one-step reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the detection of potato aucuba mosaic virus. Crop Prot. 2025, 197, 107317. [Google Scholar] [CrossRef]
- Qin, Y.H.; Wang, F.L.; Wen, Y.; Zhao, Z.W.; Gao, S.X.; Zhang, D.S.; Li, S.J.; Liu, Y.K.; Liu, Y.X.; Lu, S.H.; et al. Establishment of RT-LAMP method for rapid detection of youcai mosaic virus. Acta Phytopathol. Sin. 2024, 54, 829–834. [Google Scholar]
- Pavon, R.D.N.; Rivera, W.L. Loop-Mediated Isothermal Amplification Assay for Visual Detection of Salmonella enterica Serovar Typhimurium in Food Animal Meat Products. Foods 2025, 14, 1731. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.M.; Yu, M.; Li, S.F.; Chen, Z.Y.; Chi, L.L.; Liu, J.; Yu, Z.H.; Zhang, M.Y.; Xu, S.Z. Establishment of a visual one step RT-LAMP for detection of bovine viral diarrhea virus. Chin. Vet. Sci. 2022, 52, 985–991. [Google Scholar]
- Wang, F.L.; Wang, F.; Lu, C.T.; Gao, S.X.; Liu, Y.X.; Wen, Y.; Yang, J.; Li, X.M.; Qi, W.P.; Liu, G.B.; et al. Establishment of RT-LAMP rapid detection method for yam latent virus. Acta Phytopathol. Sin. 2022, 53, 743–747. [Google Scholar] [CrossRef]
- Yang, Z.X.; Li, Z.; Li, Z.R.; Li, L.; Yang, H.; Niu, B.S.; Yao, P.F.; Zhu, J.B. Establishment and application of a visual reverse transcriptase loop mediated iso-thermal amplification method for detection of Tibetorbivirus. Chin. J. Vet. Sci. 2022, 42, 892–896+912. [Google Scholar]
- Liu, S.R.; Tao, F.F.; Xiong, Y.X.; Wen, L.; Guo, Y.L.; Kong, L.B. Establishment of the RT-LAMP Visual Detection Method for Novel Coronavirus. Biol. Disaster Sci. 2022, 45, 415–422. [Google Scholar]
- Xu, C.; Huang, H.; Luo, H.J.; Du, J.N.; Huang, X.Y.; Chen, Y.; Liang, W.Q.; Hu, S. Establishment of RT-LAMP-HNB Method for Detection of Cymbidium mosaic Virus and Odontoglossum Ringspot Virus. Guangdong Agric. Sci. 2022, 49, 95–101. [Google Scholar]
- Zhang, X.Y.; Peng, Y.M.; Wang, Y.; Zhang, Z.Y.; Li, D.W.; Yu, J.L.; Han, C.G. Simultaneous detection and differentiation of three genotypes of Brassica yellows virus by multiplex reverse transcription-polymerase chain reaction. Virol. J. 2016, 13, 189. [Google Scholar] [CrossRef]
- Wang, J.Y.; Hou, X.L.; Yang, X.D. Identification of conserved microRNAs and their targets in Chinese cabbage (Brassica rapa subsp. pekinensis). Genome 2011, 54, 1029–1040. [Google Scholar] [CrossRef]
- Wang, C. Study and Application of ELISA Detection Technology for Shallot Latent Virus in Tiller Onion. Master’s Thesis, Jilin Agricultural University, Changchun, China, 2022. [Google Scholar]
- Deng, K.H.; Chen, Z.F.; Yu, T.; Wang, Z.L.; Wang, X.W.; Guo, Z.X.; Li, S.B.; Wei, K.; He, D.S. Detection of Three Porcine Diarrhea Coronaviruses PEDV, TGEV and PDCoV by RT-LAMP Based on N gene. Chin. J. Anim. Infect. Dis. 2025, 33, 70–79. [Google Scholar]
- He, Q.; Yu, K.; Guo, M.; Li, Y.J.; Lu, S.; Liu, J.; Li, J.H.; Wei, Z.D. Establishment of an EvaGreen qPCR for the detection of bovine adenovirus type 3 in bovine serum. Prog. Microbiol. Immunol. 2022, 50, 15–20. [Google Scholar]
- Ren, C.M.; Yang, L.; Miao, Q.; Lu, F.; Ji, Y.H.; Chen, Z.B. Establishment and application of IC-RT-PCR for cucumber green mottle mosaic virus in seeds and seed lings. Plant Prot. 2022, 48, 212–219. [Google Scholar]
- Sun, X.; Zhang, J.; Yang, L.; Lu, Q.Y. Establishment and Application of SYBR Green I Based qPCR Assay for the Detection of Mulberry Crinkle Leaf Virus. Acta Sericologica Sin. 2020, 46, 140–145. [Google Scholar]
- Cui, X.H.; Chen, S.S.; Yu, C.; Yang, C.Y. Detection of Carnation ringspot virus by real-time fluorescent reverse transcription polymerase chain reaction. Acta Phytopathol. Sin. 2012, 42, 381–386. [Google Scholar]
- Zhao, X.L.; He, C.Y.; Xu, T.F.; Li, X.F.; Liu, J.J.; Li, S.F.; Wang, H.Q. Construction of Infectious cDNA Clone of Brassica Yellows Virus Isolated from Strawberry and Establishment of TaqMan RT-qPCR. Plants 2022, 11, 3380. [Google Scholar] [CrossRef]
- Tang, X.Z.; Chen, L.W.; Lu, R.B.; Zheng, X.T.; Huang, C.Q. Dectection of Porcine Epidemic Diarrhea Virus by Calcein-based Visual Loop-mediated Isothermal Amplification (LAMP) Assay. China Anim. Husb. Vet. Med. 2015, 42, 331–336. [Google Scholar]
- Zhu, H.P.; Zheng, X.; Wang, H.; Gao, J.; Si, S.H.; Du, T.F.; Mu, Y. Establishment and application of visual LAMP method for Actinobacillus pleuropneumoniae detection. Chin. Vet. Sci. 2022, 52, 837–845. [Google Scholar]
- He, L.; Zhou, Y.Q.; Oosthuizen, M.C.; Zhao, J.L. Loop-mediated isothermal amplification (LAMP) detection of Babesia orientalis in water buffalo (Bubalus babalis, Linnaeus, 1758) in China. Vet. Parasitol. 2009, 165, 36–40. [Google Scholar] [CrossRef]
- Li, F.X.; Xia, J.X.; Yu, Y.F.; Zhu, P.; Yang, S.B.; Zhao, W.H.; Gao, H.F. Development of visual loop-mediated isothermal amplification assay for field detection of Corynebacterium pseudotuberculosis. Chin. Vet. Sci. 2023, 53, 837–842. [Google Scholar] [CrossRef]
- Liu, N.N.; Yi, P.; Guo, A.Z.; Hu, C.M.; Chen, Y.Y. Establishment of visual LAMP method for detecting Mannheimia haemolytica in cattle. J. Huazhong Agric. Univ. 2023, 42, 32–37. [Google Scholar]
- Yin, X.Y.; Cao, J.J.; Li, X.; Piao, Y.Z. Rapid detection of cucumber green mottle mosaic virus based on visual RT-LAMP without nucleic acid extraction. Plant Prot. 2023, 49, 264–271. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).