The GRF9-6Ab Allele Compensates for the Pleiotropic Deficits of the Ddw1 Dwarfing Gene in Triticale
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Field Experiments and Phenotyping
2.3. DNA Extraction, PCR and Sequencing
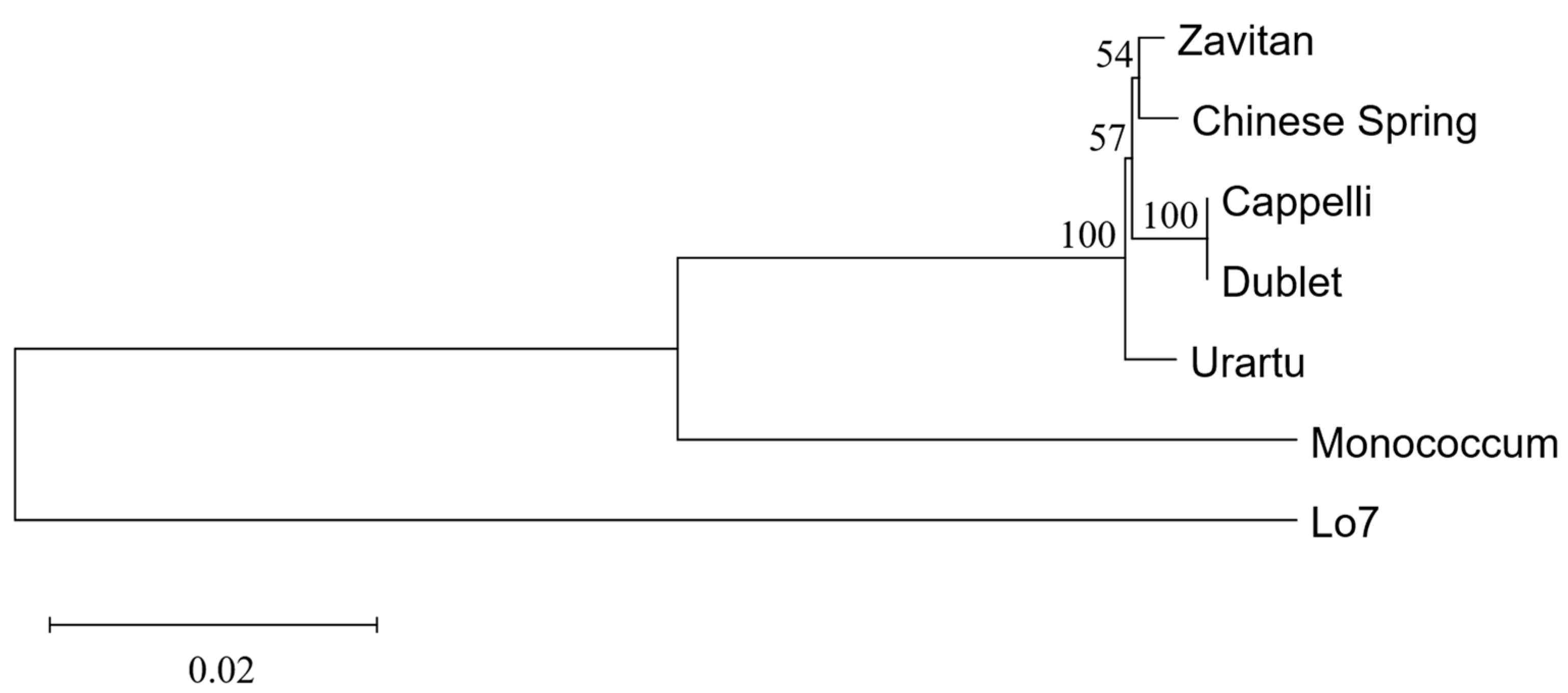
2.4. Evolutionary Analysis
2.5. Visualization
2.6. Genotyping
2.7. Statistical Analysis
3. Results
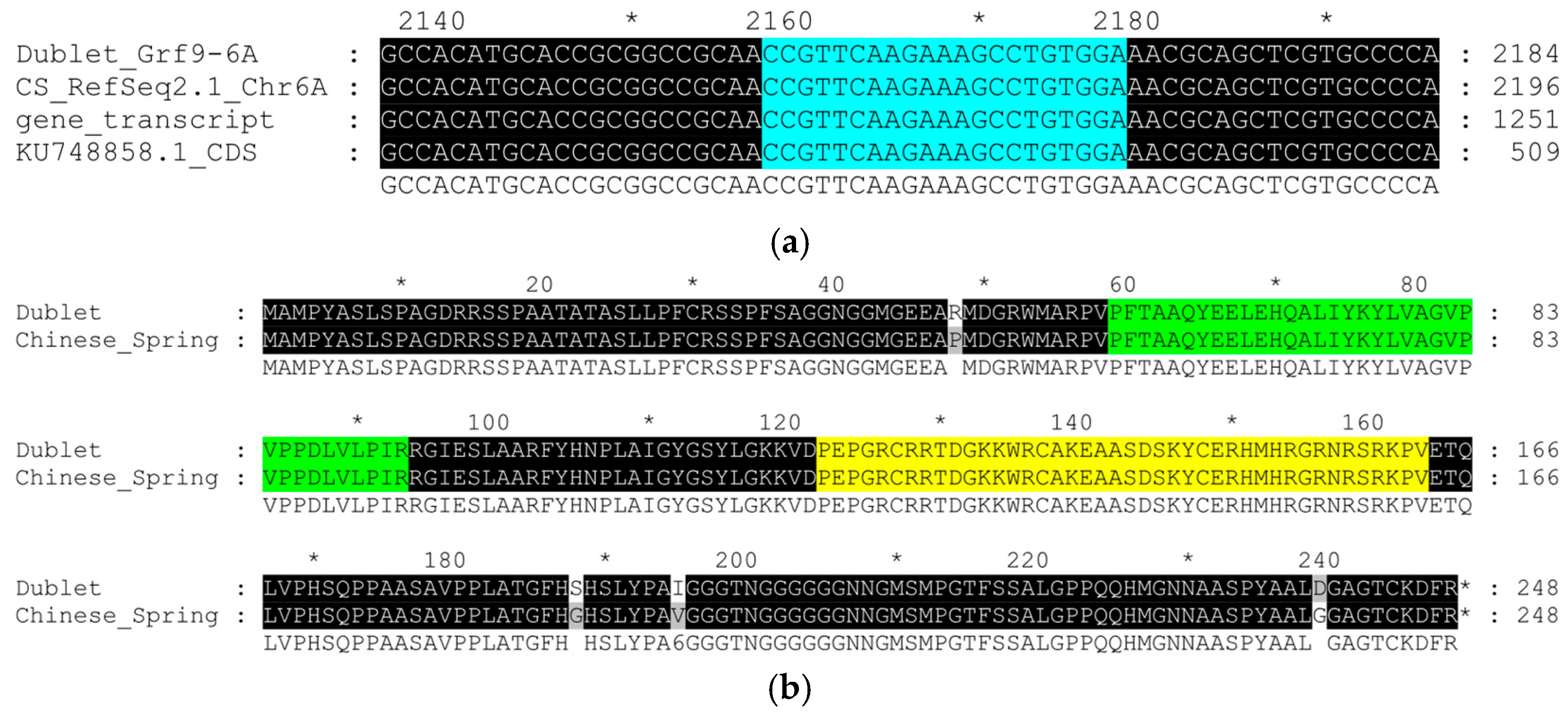
3.1. Sequencing of GRF9-6A in Triticale ‘Dublet’
3.2. Genotyping of the RIL Population for Ddw1 and GRF9-6A Loci
3.3. Phenotypic Effects of the Ddw1 and GRF9-6A Genes and Their Interaction
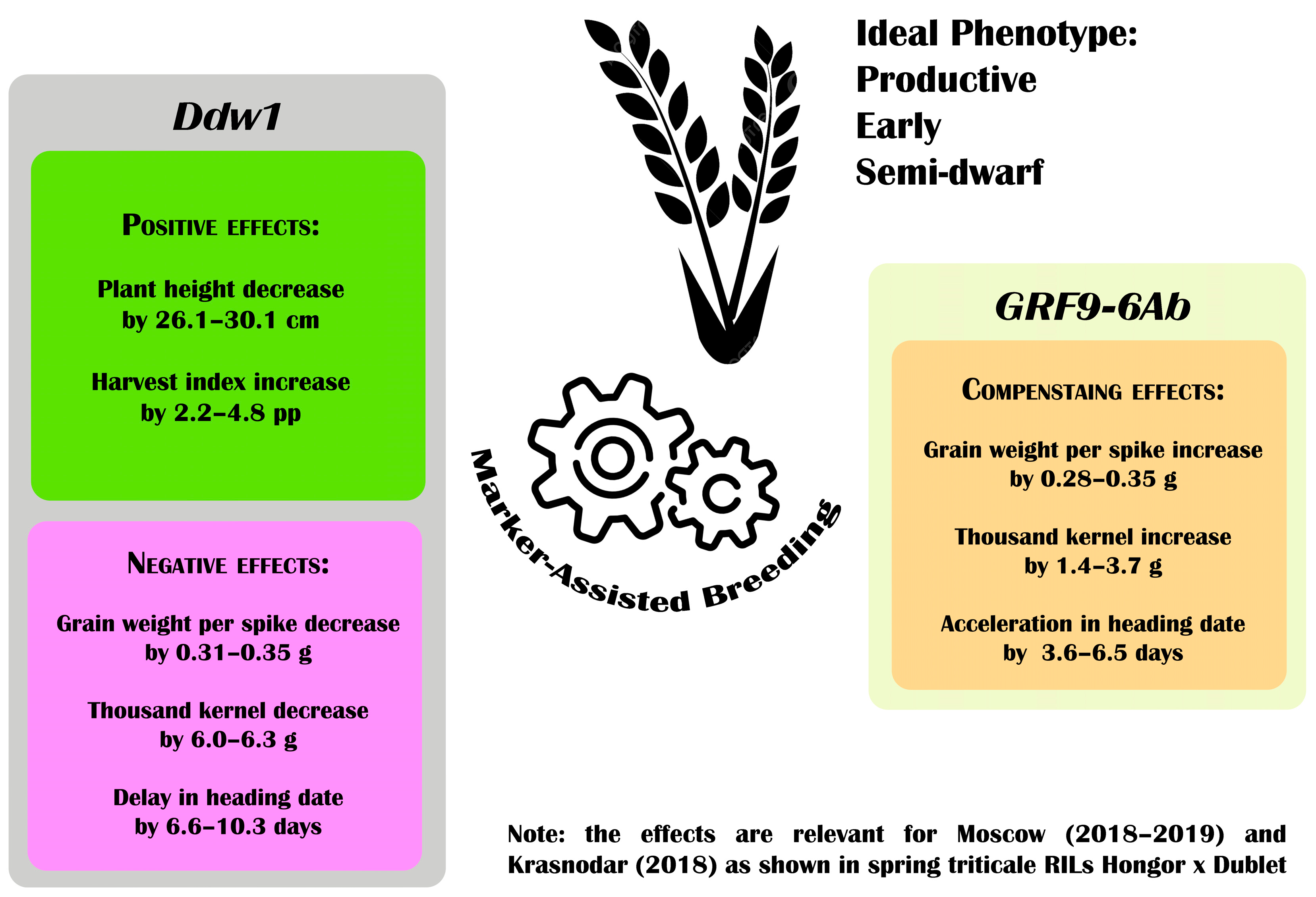
3.3.1. Effects of the Ddw1 Gene
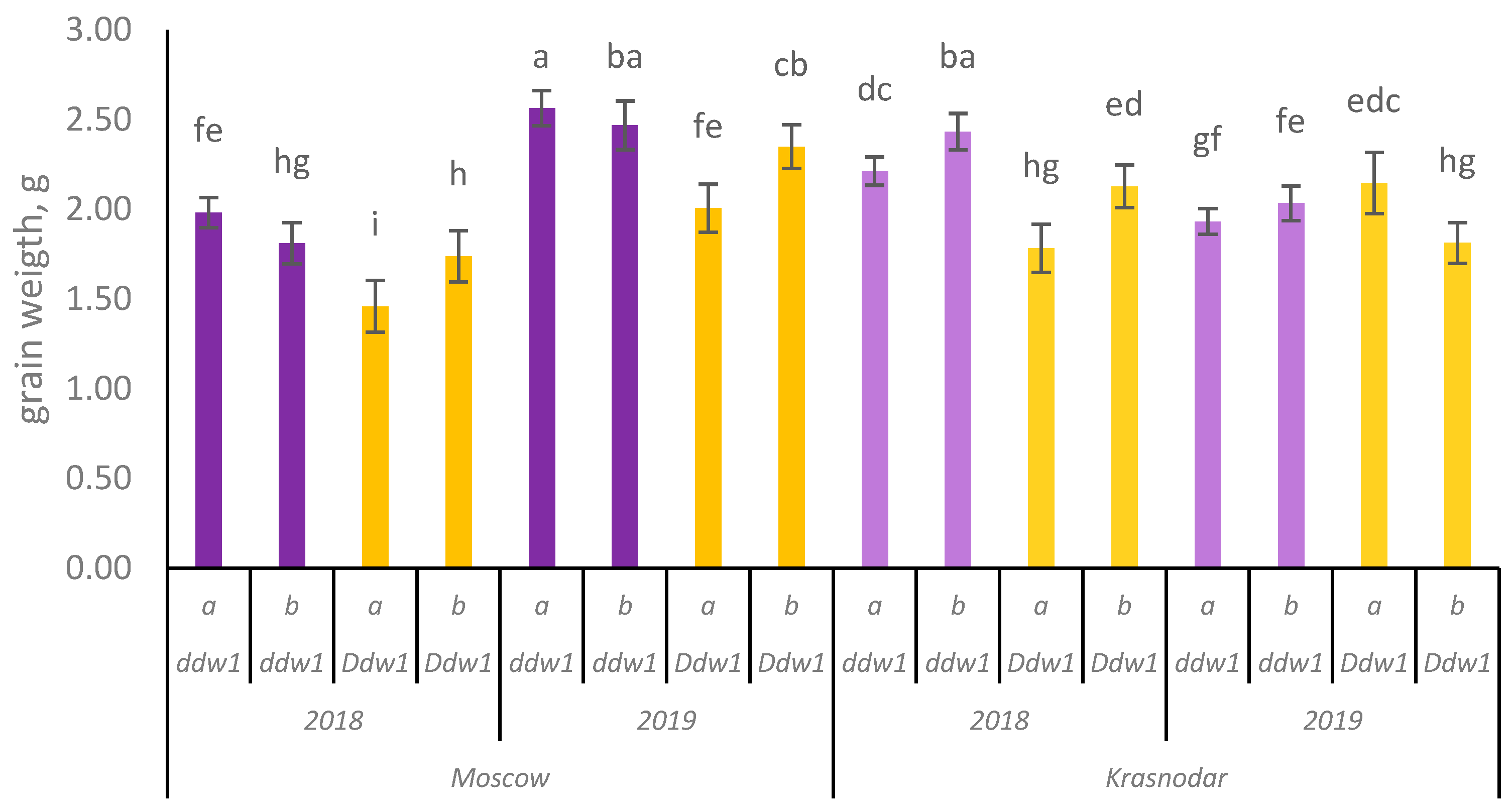
3.3.2. Effects of the GRF9-6A Gene
3.3.3. Interaction Between Ddw1 and GRF9-6A Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cui, L.; Xu, L.; Wang, H.; Fan, X.; Yan, C.; Zhang, Y.; Jiang, C.; Zhou, T.; Guo, Q.; Sun, Y.; et al. Evaluation of Dual-Purpose Triticale: Grain and Forage Productivity and Quality Under Semi-Arid Conditions. Agronomy 2025, 15, 881. [Google Scholar] [CrossRef]
- Derejko, A.; Studnicki, M.; Wójcik--Gront, E. Grain Yield Performance and Stability of Winter Wheat and Triticale Cultivars in a Temperate Climate. Crop Sci. 2021, 61, 3962–3971. [Google Scholar] [CrossRef]
- Baron, V.S.; Juskiw, P.E.; Aljarrah, M. Triticale as a Forage. In Triticale; Eudes, F., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 189–212. ISBN 978-3-319-22550-0. [Google Scholar]
- Ayalew, H.; Kumssa, T.T.; Butler, T.J.; Ma, X.-F. Triticale Improvement for Forage and Cover Crop Uses in the Southern Great Plains of the United States. Front. Plant Sci. 2018, 9, 1130. [Google Scholar] [CrossRef]
- Kobylyanskii, V. The Genetics of the Dominant Factor for Short Straw in Rye. Sov. Genet. 1972, 8, 12–17. [Google Scholar]
- Kroupin, P.; Chernook, A.; Karlov, G.; Soloviev, A.; Divashuk, M. Effect of Dwarfing Gene Ddw1 on Height and Agronomic Traits in Spring Triticale in Greenhouse and Field Experiments in a Non-Black Earth Region of Russia. Plants 2019, 8, 131. [Google Scholar] [CrossRef] [PubMed]
- Chernook, A.; Kroupin, P.; Karlov, G.; Soloviev, A.; Korshunova, A.; Rubets, V.; Igonin, V.; Divashuk, M. Effects of Rht-B1b and Ddw1 Dwarfing Genes in Two Connecting Populations of Spring Triticale under Greenhouse Experiment Conditions. Agriculture 2019, 9, 119. [Google Scholar] [CrossRef]
- Kalih, R.; Maurer, H.P.; Hackauf, B.; Miedaner, T. Effect of a Rye Dwarfing Gene on Plant Height, Heading Stage, and Fusarium Head Blight in Triticale (×Triticosecale Wittmack). Theor. Appl. Genet. 2014, 127, 1527–1536. [Google Scholar] [CrossRef]
- Divashuk, M.; Chernook, A.; Kroupina, A.; Vukovic, M.; Karlov, G.; Ermolaev, A.; Shirnin, S.; Avdeev, S.; Igonin, V.; Pylnev, V.; et al. TaGRF3-2A Improves Some Agronomically Valuable Traits in Semi-Dwarf Spring Triticale. Plants 2021, 10, 2012. [Google Scholar] [CrossRef] [PubMed]
- Miedaner, T.; Gruner, P.; Maurer, H.P. Verification of the Fusarium-Increasing Properties of the Dominant Dwarfing Gene Ddw1 in Triticale (×Triticosecale). Plant Breed. 2025, 144, 151–158. [Google Scholar] [CrossRef]
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Mueller-Roeber, B. Growth-Regulating Factors (GRFs): A Small Transcription Factor Family with Important Functions in Plant Biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef]
- Li, S.; Tian, Y.; Wu, K.; Ye, Y.; Yu, J.; Zhang, J.; Liu, Q.; Hu, M.; Li, H.; Tong, Y.; et al. Modulating Plant Growth–Metabolism Coordination for Sustainable Agriculture. Nature 2018, 560, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; He, Y.; Yang, L.; Lu, C.; Zhu, Y.; Sun, C.; Ma, D.; Yin, J. Genome-Wide Analysis of Growth-Regulating Factors (GRFs) in Triticum Aestivum. PeerJ 2021, 9, e10701. [Google Scholar] [CrossRef]
- Kroupin, P.Y.u.; Chernook, A.G.; Bazhenov, M.S.; Karlov, G.I.; Goncharov, N.P.; Chikida, N.N.; Divashuk, M.G. Allele Mining of TaGRF-2D Gene 5’-UTR in Triticum aestivum and Aegilops tauschii Genotypes. PLoS ONE 2020, 15, e0231704. [Google Scholar] [CrossRef]
- Bazhenov, M.; Chernook, A.; Bespalova, L.; Gritsay, T.; Polevikova, N.; Karlov, G.; Nazarova, L.; Divashuk, M. Alleles of the GRF3-2A Gene in Wheat and Their Agronomic Value. Int. J. Mol. Sci. 2021, 22, 12376. [Google Scholar] [CrossRef]
- Chernook, A.G.; Bazhenov, M.S.; Kroupin, P.Y.u.; Ermolaev, A.S.; Kroupina, A.Y.u.; Vukovic, M.; Avdeev, S.M.; Karlov, G.I.; Divashuk, M.G. Compensatory Effect of the ScGrf3-2R Gene in Semi-Dwarf Spring Triticale (x Triticosecale Wittmack). Plants 2022, 11, 3032. [Google Scholar] [CrossRef]
- Kroupin, P.Y.; Karlov, G.I.; Bespalova, L.A.; Salina, E.A.; Chernook, A.G.; Watanabe, N.; Bazhenov, M.S.; Panchenko, V.V.; Nazarova, L.A.; Kovtunenko, V.Y.; et al. Effects of Rht17 in Combination with Vrn-B1 and Ppd-D1 Alleles on Agronomic Traits in Wheat in Black Earth and Non-Black Earth Regions. BMC Plant Biol. 2020, 20, 304. [Google Scholar] [CrossRef]
- Komyshev, E.; Genaev, M.; Afonnikov, D. Evaluation of the SeedCounter, A Mobile Application for Grain Phenotyping. Front. Plant Sci. 2017, 7, 1990. [Google Scholar] [CrossRef] [PubMed]
- Doyle, P.J. DNA Protocols for Plants. In Molecular Techniques in Taxonomy; Hewitt, G.M., Johnston, A.W.B., Young, J.P.W., Eds.; NATO ASI Series; Springer: Berlin/Heidelberg, Germany, 1991; pp. 283–293. ISBN 978-3-642-83964-1. [Google Scholar]
- Alaux, M.; Rogers, J.; Letellier, T.; Flores, R.; Alfama, F.; Pommier, C.; Mohellibi, N.; Durand, S.; Kimmel, E.; Michotey, C.; et al. Linking the International Wheat Genome Sequencing Consortium Bread Wheat Reference Genome Sequence to Wheat Genetic and Phenomic Data. Genome Biol. 2018, 19, 1–10. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Nicholas, K.B. GeneDoc: Analysis and Visualization of Genetic Variation. EMBnet News 1997, 4, 14. [Google Scholar]
- SantaLucia, J. A Unified View of Polymer, Dumbbell, and Oligonucleotide DNA Nearest-Neighbor Thermodynamics. Proc. Natl. Acad. Sci. USA 1998, 95, 1460–1465. [Google Scholar] [CrossRef]
- Keller, O.; Kollmar, M.; Stanke, M.; Waack, S. A Novel Hybrid Gene Prediction Method Employing Protein Multiple Sequence Alignments. Bioinformatics 2011, 27, 757–763. [Google Scholar] [CrossRef]
- Andrews, S. FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 19 December 2019).
- Zaharia, M.; Bolosky, W.J.; Curtis, K.; Fox, A.; Patterson, D.; Shenker, S.; Stoica, I.; Karp, R.M.; Sittler, T. Faster and More Accurate Sequence Alignment with SNAP. arXiv 2011, arXiv:1111.5572. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-Based Variant Detection from Short-Read Sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Stanke, M.; Morgenstern, B. AUGUSTUS: A Web Server for Gene Prediction in Eukaryotes That Allows User-Defined Constraints. Nucleic Acids Res. 2005, 33, W465–W467. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Walkowiak, S.; Gao, L.; Monat, C.; Haberer, G.; Kassa, M.T.; Brinton, J.; Ramirez-Gonzalez, R.H.; Kolodziej, M.C.; Delorean, E.; Thambugala, D.; et al. Multiple Wheat Genomes Reveal Global Variation in Modern Breeding. Nature 2020, 588, 277–283. [Google Scholar] [CrossRef]
- Tateo, F.; Bononi, M.; Castorina, G.; Colecchia, S.A.; Benedetti, S.D.; Consonni, G.; Geuna, F. Whole-Genome Resequencing-Based Characterization of a Durum Wheat Landrace Showing Similarity to ‘Senatore Cappelli’. PLoS ONE 2023, 18, e0291430. [Google Scholar] [CrossRef] [PubMed]
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K.; et al. Wild Emmer Genome Architecture and Diversity Elucidate Wheat Evolution and Domestication. Science 2017, 357, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.-Q.; Ma, B.; Shi, X.; Liu, H.; Dong, L.; Sun, H.; Cao, Y.; Gao, Q.; Zheng, S.; Li, Y.; et al. Genome Sequence of the Progenitor of Wheat A Subgenome Triticum urartu. Nature 2018, 557, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.I.; Heuberger, M.; Schoen, A.; Koo, D.-H.; Quiroz-Chavez, J.; Adhikari, L.; Raupp, J.; Cauet, S.; Rodde, N.; Cravero, C.; et al. Einkorn Genomics Sheds Light on History of the Oldest Domesticated Wheat. Nature 2023, 620, 830–838. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating of the Human-Ape Splitting by a Molecular Clock of Mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s Conserved Domain Database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting Amino Acid Changes That Affect Protein Function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Yang, A.; Li, A.; Yang, B.; Zhang, B.; Hui, B.; Zheng, B.; Yu, B.; Gao, C.; Huang, C.; Lv, C.; et al. Qwen3 Technical Report. arXiv 2025, arXiv:2505.09388. [Google Scholar] [CrossRef]
- Tenhola-Roininen, T.; Tanhuanpää, P. Tagging the Dwarfing Gene Ddw1 in a Rye Population Derived from Doubled Haploid Parents. Euphytica 2010, 172, 303–312. [Google Scholar] [CrossRef]
- Litvinov, D.Y.; Chernook, A.G.; Kroupin, P.Y.; Bazhenov, M.S.; Karlov, G.I.; Avdeev, S.M.; Divashuk, M.G. A Convenient Co-Dominant Marker for Height-Reducing Ddw1 Allele Useful for Marker-Assisted Selection. Agriculture 2020, 10, 110. [Google Scholar] [CrossRef]
- Yu, Y.; Sun, F.; Chen, N.; Sun, G.; Wang, C.-Y.; Wu, D.-X. MiR396 Regulatory Network and Its Expression during Grain Development in Wheat. Protoplasma 2021, 258, 103–113. [Google Scholar] [CrossRef]
- Liebsch, D.; Palatnik, J.F. MicroRNA miR396, GRF Transcription Factors and GIF Co-Regulators: A Conserved Plant Growth Regulatory Module with Potential for Breeding and Biotechnology. Curr. Opin. Plant Biol. 2020, 53, 31–42. [Google Scholar] [CrossRef]
- Jatayev, S.; Sukhikh, I.; Vavilova, V.; Smolenskaya, S.E.; Goncharov, N.P.; Kurishbayev, A.; Zotova, L.; Absattarova, A.; Serikbay, D.; Hu, Y.; et al. Green Revolution ‘Stumbles’ in a Dry Environment: Dwarf Wheat with Rht Genes Fails to Produce Higher Grain Yield than Taller Plants under Drought. Plant Cell Environ. 2020, 43, 2355–2364. [Google Scholar] [CrossRef]
- Kronenberg, L.; Yates, S.; Boer, M.P.; Kirchgessner, N.; Walter, A.; Hund, A. Temperature Response of Wheat Affects Final Height and the Timing of Stem Elongation under Field Conditions. J. Exp. Bot. 2021, 72, 700–717. [Google Scholar] [CrossRef]
- Braun, E.-M.; Tsvetkova, N.; Rotter, B.; Siekmann, D.; Schwefel, K.; Krezdorn, N.; Plieske, J.; Winter, P.; Melz, G.; Voylokov, A.V.; et al. Gene Expression Profiling and Fine Mapping Identifies a Gibberellin 2-Oxidase Gene Co-Segregating with the Dominant Dwarfing Gene Ddw1 in Rye (Secale cereale L.). Front. Plant Sci. 2019, 10, 857. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.G.; Phillips, A.L.; Hedden, P. Molecular Cloning and Functional Expression of Gibberellin 2- Oxidases, Multifunctional Enzymes Involved in Gibberellin Deactivation. Proc. Natl. Acad. Sci. USA 1999, 96, 4698–4703. [Google Scholar] [CrossRef]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The Role of Gibberellin Signalling in Plant Responses to Abiotic Stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef]
- Kottmann, L.; Burzik, W.; Feike, T.; Siekmann, D.; Fromme, F.J.; Kucherova, V.; Hackauf, B. Gibberellin-Sensitive Dwarfing Gene Ddw1 Has No Negative Effect on the Root System of Field-Grown Winter Rye. Field Crops Res. 2023, 303, 109151. [Google Scholar] [CrossRef]
- Schomburg, F.M.; Bizzell, C.M.; Lee, D.J.; Zeevaart, J.A.D.; Amasino, R.M. Overexpression of a Novel Class of Gibberellin 2-Oxidases Decreases Gibberellin Levels and Creates Dwarf Plants. Plant Cell 2003, 15, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Ahmad, M.; Ahmed, M.; Iftikhar Hussain, M. Rising Atmospheric Temperature Impact on Wheat and Thermotolerance Strategies. Plants 2020, 10, 43. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Chu, Y.; Ma, G.; Zhang, Y.; Zhang, X.; Wang, M.; Lu, H.; Wang, L.; Kang, G.; Ma, D.; et al. Physiological Mechanisms Underlying Reduced Photosynthesis in Wheat Leaves Grown in the Field under Conditions of Nitrogen and Water Deficiency. Crop J. 2023, 11, 638–650. [Google Scholar] [CrossRef]
- Rosati, A.; Benincasa, P. Revisiting Source versus Sink Limitations of Wheat Yield during Grain Filling. Agron. J. 2023, 115, 3197–3205. [Google Scholar] [CrossRef]
- Kuijt, S.J.H.; Greco, R.; Agalou, A.; Shao, J.; ‘THoen, C.C.J.; Övernäs, E.; Osnato, M.; Curiale, S.; Meynard, D.; Van Gulik, R.; et al. Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX Families of Transcription Factors. Plant Physiol. 2014, 164, 1952–1966. [Google Scholar] [CrossRef]
- Lazzara, F.E.; Rodriguez, R.E.; Palatnik, J.F. Molecular Mechanisms Regulating GROWTH-REGULATING FACTORS Activity in Plant Growth, Development, and Environmental Responses. J. Exp. Bot. 2024, 75, 4360–4372. [Google Scholar] [CrossRef]
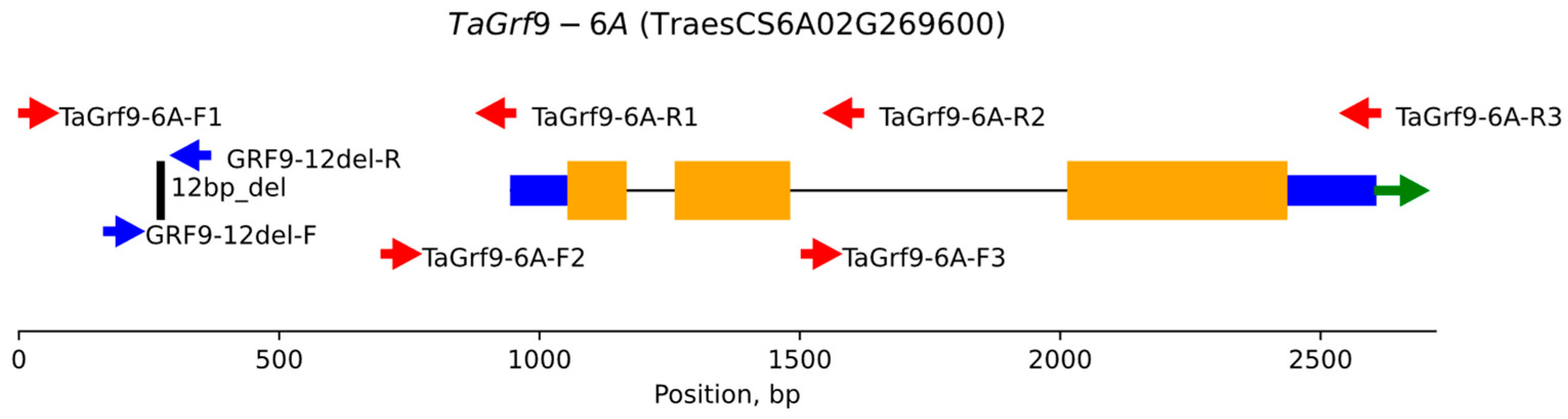

| Pair | Name | Sequence, 5′-3′ | Tm 1 | Amplicon Size, bp |
|---|---|---|---|---|
| 1 | GRF9-6A-F1 | CCTAAGGATGCCAATCCTACGAA | 60 | 953 |
| GRF9-6A-R1 | GAAGCGTCTTTAATGGGGTTCC | 60 | ||
| 2 | GRF9-6A-F2 | ATCTACTCCTCCTCGCTACTACC | 60 | 924 |
| GRF9-6A-R2 | ATCCCGGATTAACAAAAGACCGA | 60 | ||
| 3 | GRF9-6A-F3 | TCCTTCATCATCACCGCAAATCT | 60 | 1110 |
| GRF9-6A-R3 | AGCACTGTGCATAGAGGAACAAAT | 61 | ||
| 4 | GRF9-12del-F | CCGAGACATCGCTTTCATATTTGG | 60 | 203/191 |
| GRF9-12del-R | ACTATGTGCTTGCTTCTTGTGTC | 60 |
| Mutation | Amino-Acid Substitution | SIFT Prediction | ||
|---|---|---|---|---|
| Action | Score | Confidence | ||
| c.143C>G | p.(Pro48Arg) | Tolerated | 0.73 | Normal |
| c.562G>A | p.(Gly188Ser) | Tolerated | 0.20 | Normal |
| c.583G>A | p.(Val195Ile) | Tolerated | 1.00 | Normal |
| c.716G>A | p.(Gly239Asp) | Affect function | 0.01 | Low |
| Mutations in Non-Coding Regions | Region |
|---|---|
| chr6A:g.496018063_496018064delinsTT | promoter |
| chr6A:g.496018059C>T | promoter |
| chr6A:g.496018053A>G | promoter |
| chr6A:g.496018042C>T | promoter |
| chr6A:g.496018023C>T | promoter |
| chr6A:g.496017978A>C | promoter |
| chr6A:g.496017943C>T | promoter |
| chr6A:g.496017831_496017842del | promoter |
| chr6A:g.496017813T>G | promoter |
| chr6A:g.496017811G>A | promoter |
| chr6A:g.496017788G>A | promoter |
| chr6A:g.496017595T>C | promoter |
| chr6A:g.496017420G>A | promoter |
| chr6A:g.496017265G>C | promoter |
| chr6A:g.496015567A>G | 5′-UTR |
| Ddw1 Genotype | GRF9-6A Genotype | Valid N | |||
|---|---|---|---|---|---|
| Moscow | Krasnodar | ||||
| 2018 | 2019 | 2018 | 2019 | ||
| ddw1 | a | 306 | 308 | 255 | 302 |
| ddw1 | b | 183 | 203 | 152 | 153 |
| Ddw1 | a | 67 | 114 | 76 | 57 |
| Ddw1 | b | 80 | 160 | 74 | 138 |
| Trait | Moscow 2018 | Moscow 2019 | Krasnodar 2018 | Krasnodar 2019 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ddw1 | ddw1 | Ddw1 vs. ddw1 (%) | Ddw1 | ddw1 | Ddw1 vs. ddw1 (%) | Ddw1 | ddw1 | Ddw1 vs. ddw1 (%) | Ddw1 | ddw1 | Ddw1 vs. ddw1 (%) | |
| Plant height, cm | 54.7 e ± 7.8 | 80.8 c ± 12.3 | −26.1 (−32.3) | 57.1 e ± 8.4 | 84.6 b ± 13.3 | −27.5 (−32.5) | 64.7 d ± 8.5 | 94.8 a ± 13.9 | −30.1 (−31.8) | 95.9 a ± 19.9 | 94.2 a ± 20.3 | +1.7 (+1.8) |
| Grain weight per main spike, g | 1.61 d ± 0.64 | 1.92 c ± 0.76 | −0.31 (−16.1) | 2.20 b ± 0.77 | 2.53 a ± 0.92 | −0.33 (−13.0) | 1.95 c ± 0.59 | 2.30 b ± 0.66 | −0.35 (−15.2) | 1.92 c ± 0.66 | 1.97 c ± 0.63 | −0.05 (−2.5) |
| Grain number per main spike | 45.6 d ± 12.5 | 46.6 d ± 14.1 | −1.0 (−2.1) | 48.0 cd ± 13.0 | 48.3 c ± 14.4 | −0.3 (−0.6) | 57.1 b ± 12.6 | 57.1 b ± 13.1 | 0.0 (0.0) | 60.4 a ± 13.9 | 61.4 a ± 12.4 | −1.0 (−1.6) |
| 1000-kernel weight, g | 34.3 d ± 7.1 | 40.3 c ± 8.3 | −6.0 (−14.9) | 45.7 b ± 7.5 | 51.8 a ± 8.7 | −6.1 (−11.8) | 33.8 d ± 5.5 | 40.1 c ± 6.3 | −6.3 (−15.7) | 31.4 e ± 6.4 | 31.6 e ± 6.0 | −0.2 (−0.6) |
| Trait | Moscow 2018 | Moscow 2019 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ddw1 | ddw1 | Ddw1 | ddw1 | |||||||||
| GRF9-6Aa | GRF9-6Ab | b vs. a (%) | GRF9-6Aa | GRF9-6Ab | b vs. a (%) | GRF9-6Aa | GRF9-6Ab | b vs. a (%) | GRF9-6Aa | GRF9-6Ab | b vs. a (%) | |
| Grain weight per main spike, g | 1.46 i ± 0.60 | 1.74 h ± 0.64 | +0.28 (+19.2) | 1.98 fe ± 0.74 | 1.81 hg ± 0.79 | −0.17 (−8.6) | 2.01 fe ± 0.72 | 2.35 cb ± 0.78 | +0.34 (+16.9) | 2.56 a ± 0.88 | 2.47 ba ± 0.97 | −0.09 (−3.5) |
| 1000-kernel weight, g | 32.3 jih ± 6.6 | 36.0 g ± 7.1 | +3.7 (+11.5) | 40.5 fe ± 7.7 | 39.9 fe ± 9.2 | −0.6 (−1.5) | 44.0 d ± 7.2 | 46.9 c ± 7.5 | +2.9 (+6.6) | 51.2 b ± 7.6 | 52.7 a ± 10.1 | +1.5 (+2.9) |
| Heading time (days after sowing) | 62.9 a ± 7.3 | 56.6 cb ± 2.4 | −6.3 (−10.0) | 54.7 d ± 4.2 | 54.8 d ± 1.6 | +0.1 (+0.2) | 63.8 a ± 6.3 | 57.3 b ± 3.5 | −6.5 (−10.2) | 56.3 c ± 4.0 | 56.2 c ± 3.1 | −0.1 (−0.2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kroupin, P.Y.; Mokhov, T.D.; Panchenko, V.V.; Meglitskaya, Y.S.; Bespalova, L.A.; Chernook, A.G.; Bazhenov, M.S.; Kovtunenko, V.Y.; Kroupina, A.Y.; Karlov, G.I.; et al. The GRF9-6Ab Allele Compensates for the Pleiotropic Deficits of the Ddw1 Dwarfing Gene in Triticale. Agronomy 2025, 15, 2701. https://doi.org/10.3390/agronomy15122701
Kroupin PY, Mokhov TD, Panchenko VV, Meglitskaya YS, Bespalova LA, Chernook AG, Bazhenov MS, Kovtunenko VY, Kroupina AY, Karlov GI, et al. The GRF9-6Ab Allele Compensates for the Pleiotropic Deficits of the Ddw1 Dwarfing Gene in Triticale. Agronomy. 2025; 15(12):2701. https://doi.org/10.3390/agronomy15122701
Chicago/Turabian StyleKroupin, Pavel Yu., Timofey D. Mokhov, Vladimir V. Panchenko, Yana S. Meglitskaya, Ludmila A. Bespalova, Anastasiya G. Chernook, Mikhail S. Bazhenov, Victor Ya. Kovtunenko, Aleksandra Yu. Kroupina, Gennady I. Karlov, and et al. 2025. "The GRF9-6Ab Allele Compensates for the Pleiotropic Deficits of the Ddw1 Dwarfing Gene in Triticale" Agronomy 15, no. 12: 2701. https://doi.org/10.3390/agronomy15122701
APA StyleKroupin, P. Y., Mokhov, T. D., Panchenko, V. V., Meglitskaya, Y. S., Bespalova, L. A., Chernook, A. G., Bazhenov, M. S., Kovtunenko, V. Y., Kroupina, A. Y., Karlov, G. I., & Divashuk, M. G. (2025). The GRF9-6Ab Allele Compensates for the Pleiotropic Deficits of the Ddw1 Dwarfing Gene in Triticale. Agronomy, 15(12), 2701. https://doi.org/10.3390/agronomy15122701






