Global Transcriptome and WGCNA Analysis Uncover Cultivar-Specific Molecular Signatures Associated with Low-Temperature Germination in Brassica napus L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. LTG Treatments and Sample Collection
2.3. RNA Sequencing and Identification of DEGs
2.4. qRT-PCR Analysis
2.5. GO and KEGG Pathway Enrichment Analysis
2.6. Coexpression Network Analysis for Construction of Modules
2.7. PPI-Network of BnaRGL2
2.8. Multiple Sequence Alignment and 3D Protein Structure
3. Results
3.1. Morphological Analysis of Two Rapeseed Genotypes with Contrasting LTG
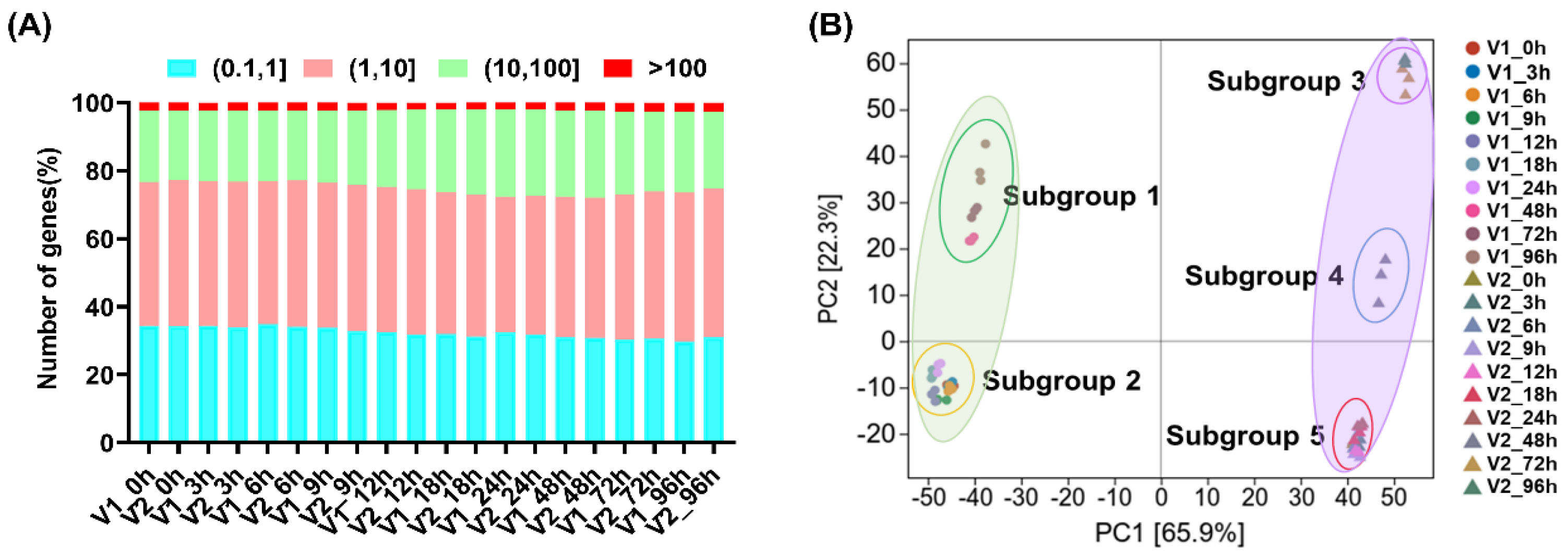
3.2. Global Transcriptome Analysis on LTG in Two Rapeseed Varieties with Contrasting Seed Germination Characteristics
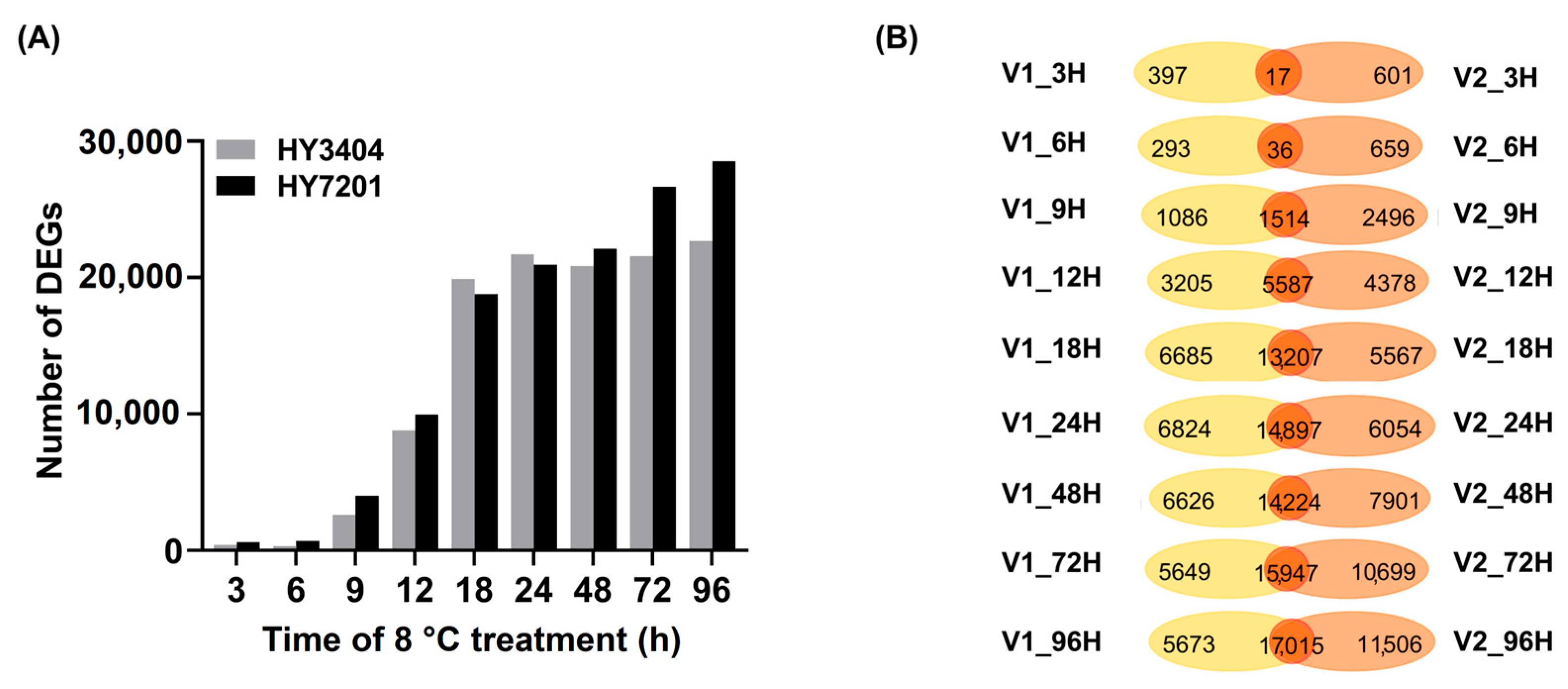
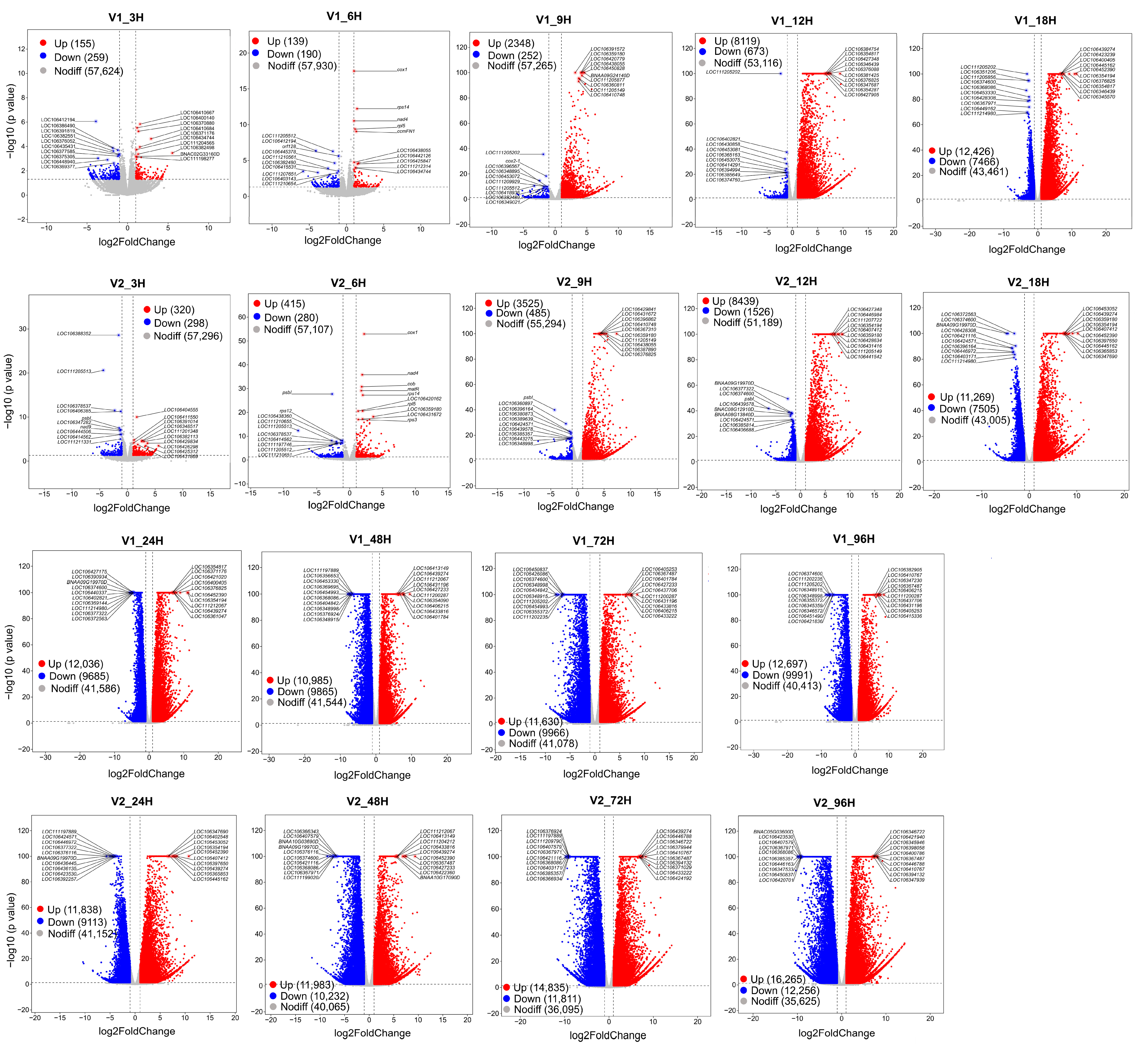
3.3. Identification of Genes with Differential Expression in LTG
3.4. GO and KEGG Analysis of Common and Time-Specific DEGs in LTG
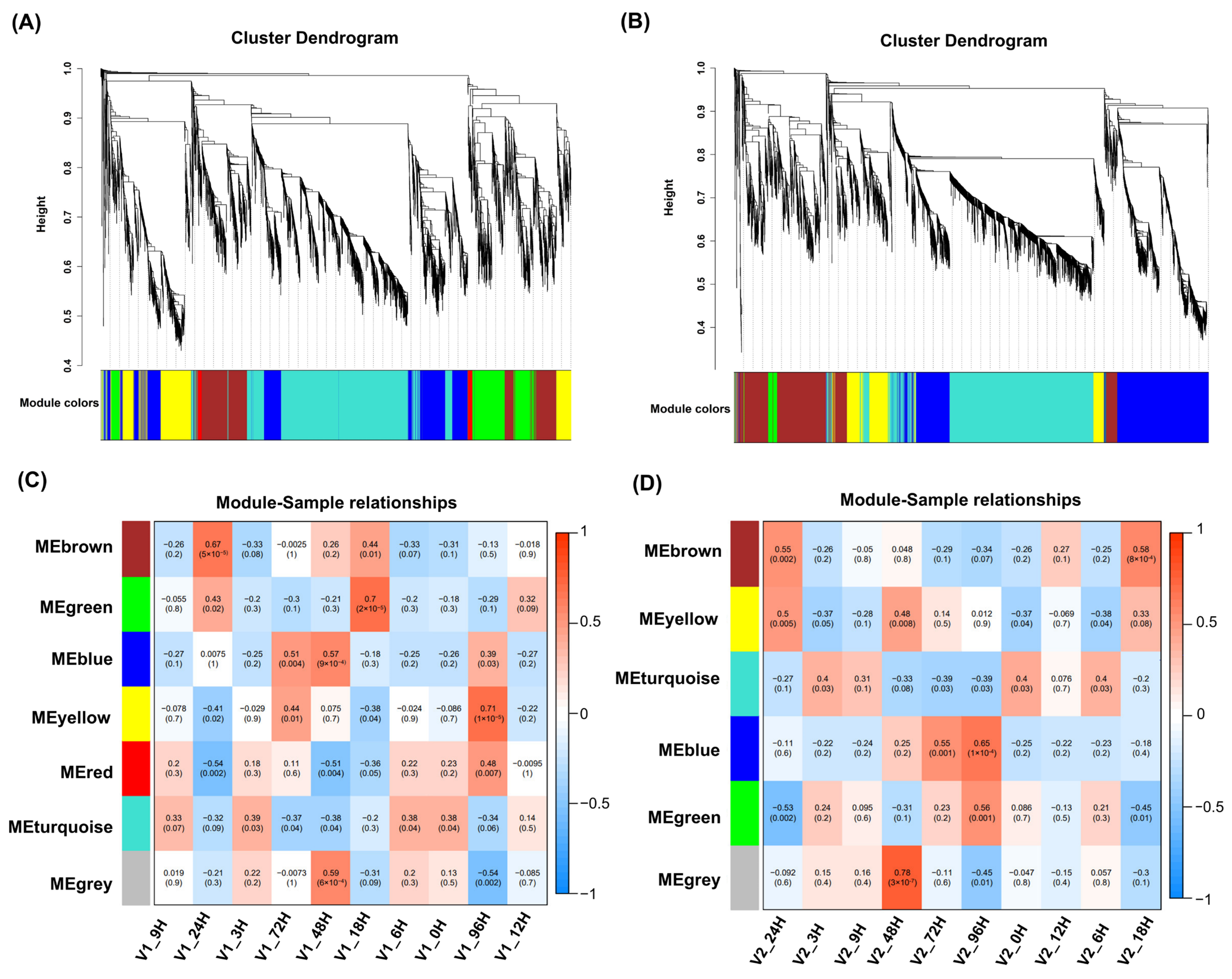
3.5. WGCNA Analysis and Module-Specific Hub Gene Association with LTG
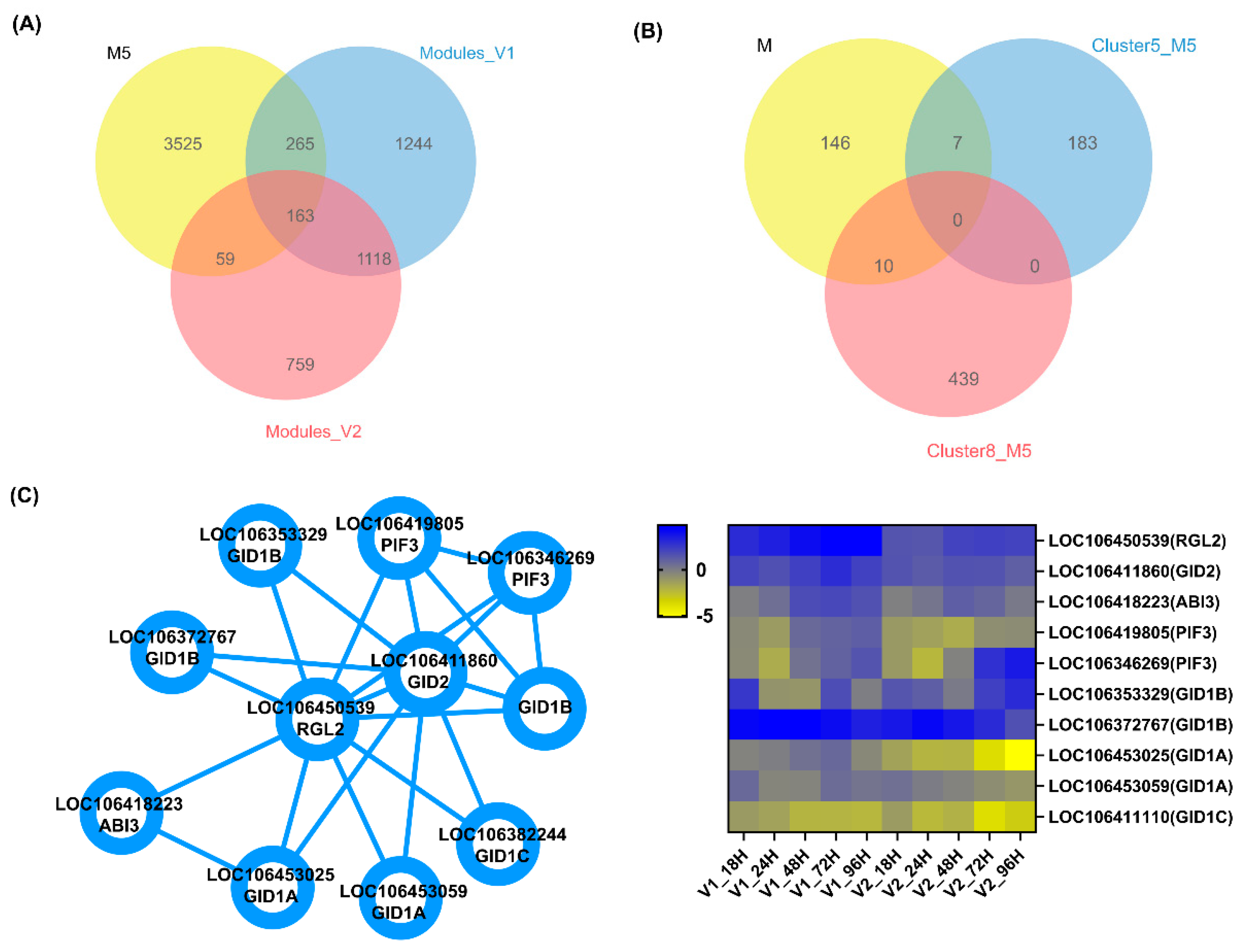
3.6. Genotype-Specific Candidate Genes Determining LTG
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LTG | Low-temperature germination |
| DEGs | Differentially Expressed Genes |
| WGCNA | Weighted gene co-expression network analysis |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GWAS | Genome-wide association studies |
| GA | Gibberellin |
| ABA | Abscisic Acid |
References
- FAO. Available online: https://www.fao.org/faostat/zh/#data/QCL/visualize (accessed on 3 September 2025).
- Tan, Z.; Han, X.; Dai, C.; Lu, S.; He, H.; Yao, X.; Che, P.; Yang, C.; Zhao, L.; Yang, Q.Y.; et al. Functional genomics of Brassica napus: Progresses, challenges, and perspectives. J. Integr. Plant Biol. 2024, 66, 484–509. [Google Scholar] [CrossRef]
- Hu, Q.; Hua, W.; Yin, Y.; Zhang, X.K.; Liu, L.L.; Shi, J.Q.; Zhao, Y.G.; Qin, L.; Chen, C.; Wang, H.Z. Rapeseed research and production in China. Crop J. 2017, 5, 127–135. [Google Scholar] [CrossRef]
- Wang, R.; Cheng, T.; Hu, L. Effect of wide-narrow row arrangement and plant density on yield and radiation use efficiency of mechanized direct-seeded canola in central China. Field Crops Res. 2015, 172, 42–52. [Google Scholar] [CrossRef]
- Wang, Z.; Han, Y.; Luo, S.; Rong, X.; Song, H.; Jiang, N.; Li, C.; Yang, L. Calcium peroxide alleviates the waterlogging stress of rapeseed by improving root growth status in a rice-rape rotation field. Front. Plant Sci. 2022, 13, 1048227. [Google Scholar] [CrossRef]
- Tian, Z.; Ji, Y.; Sun, L.; Xu, X.; Fan, D.; Zhong, H.; Liang, Z.; Gunther, F. Changes in production potentials of rapeseed in the Yangtze River Basin of China under climate change: A multi-model ensemble approach. J. Geogr. Sci. 2018, 28, 1700–1714. [Google Scholar] [CrossRef]
- Haj Sghaier, A.; Tarnawa, A.; Khaeim, H.; Kovacs, G.P.; Gyuricza, C.; Kende, Z. The effects of temperature and water on the seed germination and seedling development of rapeseed (Brassica napus L.). Plants 2022, 11, 2819. [Google Scholar] [CrossRef] [PubMed]
- Luo, T.; Zhang, Y.; Zhang, C.; Nelson, M.N.; Yuan, J.; Guo, L.; Xu, Z. Genome-wide association mapping unravels the genetic control of seed vigor under low-temperature conditions in rapeseed (Brassica napus L.). Plants 2021, 10, 426. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, W.; Jiang, M.; Yang, L.; Zhou, X. QTL mapping for low temperature germination in rapeseed. Sci. Rep. 2021, 11, 23382. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A. Physiological Processes in Plants Under Low Temperature Stress; Springer: Singapore, 2022; pp. 1–106. [Google Scholar]
- Shi, Y.; Ding, Y.; Yang, S. Molecular Regulation of CBF Signaling in Cold Acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shi, Y.; Yang, S. Regulatory networks underlying plant responses and adaptation to cold stress. Annu. Rev. Genet. 2024, 58, 43–65. [Google Scholar] [CrossRef]
- Li, D.; Zhang, C.; Dong, Z.; Yang, L.; Wang, H.; Wang, X.; Dirk, L.M.A.; Downie, A.B.; Zhao, T. ZmDREB1A Regulates myo-inositol-1-phosphate synthase 2 controlling maize germination at low temperatures. J. Agric. Food Chem. 2025, 73, 7562–7573. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, S.; Yuan, X.; Chen, J.; Tian, M.; Zhao, Z.; Guo, T.; Xiao, W. OsNAL11 and OsBURP12 affect rice seed germination at low temperature. Plant Cell Environ. 2025, 48, 6118–6139. [Google Scholar] [CrossRef]
- Wang, X.; Zou, B.; Shao, Q.; Cui, Y.; Lu, S.; Zhang, Y.; Huang, Q.; Huang, J.; Hua, J. Natural variation reveals that OsSAP16 controls low-temperature germination in rice. J. Exp. Bot. 2018, 69, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.A.; Zeeshan Ul Haq, M.; Zhang, L.; Wu, S.; Mushtaq, N.; Tahir, H.; Wang, Z. Transcriptomic insights into salt stress response in two pepper species: The role of MAPK and plant hormone signaling pathways. Int. J. Mol. Sci. 2024, 25, 9355. [Google Scholar] [CrossRef]
- Liu, E.; Xu, L.; Luo, Z.; Li, Z.; Zhou, G.; Gao, H.; Fang, F.; Tang, J.; Zhao, Y.; Zhou, Z.; et al. Transcriptomic analysis reveals mechanisms for the different drought tolerance of sweet potatoes. Front Plant Sci. 2023, 14, 1136709. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Fan, G.; Hu, Q.; Zhou, Y.; Guan, M.; Tong, C.; Li, J.; Du, D.; Qi, C.; Jiang, L.; et al. The high-quality genome of Brassica napus cultivar ‘ZS11’ reveals the introgression history in semi-winter morphotype. Plant J. 2017, 92, 452–468. [Google Scholar] [CrossRef]
- Wang, P.; Liu, F.; Wang, Y.; Chen, H.; Liu, T.; Li, M.; Chen, S.; Wang, D. Deciphering crucial salt-responsive genes in Brassica napus via statistical modeling and network analysis on dynamic transcriptomic data. Plant Physiol. Biochem. 2025, 220, 109568. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Z.; Ren, R.; Liu, Y.; Qian, L.; Guan, M.; Guan, C.; He, X. Transcriptome uncovers BnaFBH3-mediated regulatory networks associated with tolerant to abiotic stress in Brassica napus. Environ. Exp. Bot. 2023, 216, 105541. [Google Scholar] [CrossRef]
- Luo, T.; Xian, M.; Zhang, C.; Zhang, C.; Hu, L.; Xu, Z. Associating transcriptional regulation for rapid germination of rapeseed (Brassica napus L.) under low temperature stress through weighted gene co-expression network analysis. Sci. Rep. 2019, 9, 55. [Google Scholar] [CrossRef]
- Privitera, G.F.; Treccarichi, S.; Nicotra, R.; Branca, F.; Pulvirenti, A.; Piero, A.R.L.; Sicilia, A. Comparative transcriptome analysis of B. oleracea L. var. italica and B. macrocarpa Guss. genotypes under drought stress: De novo vs reference genome assembly. Plant Stress 2024, 14, 100657. [Google Scholar] [CrossRef]
- Singh, K.P.; Kumari, P.; Yadava, D.K. Development of de-novo transcriptome assembly and SSRs in allohexaploid Brassica with functional annotations and identification of heat-shock proteins for thermotolerance. Front Genet. 2022, 13, 958217. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Chen, Y.; Jiang, G.; Zhao, J.; Wang, W.; Hong, X. Integrated analysis of transcriptome and metabolome reveals insights for low-temperature germination in hybrid rapeseeds (Brassica napus L.). J. Plant Physiol. 2023, 291, 154120. [Google Scholar] [CrossRef]
- Wang, R.; Wu, G.; Zhang, J.; Hu, W.; Yao, X.; Jiang, L.; Zhu, Y. Integration of GWAS and transcriptome analysis to identify temperature-dependent genes involved in germination of rapeseed (Brassica napus L.). Front. Plant Sci. 2025, 16, 1551317. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, Y.; Zhang, J.; Qiu, X. Super-delta2: An enhanced differential expression analysis procedure for multi-group comparisons of RNA-seq data. Bioinformatics 2021, 37, 2627–2636. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, Y.; Zhang, J.; Qiu, X. A survey of quantitative real-time polymerase chain reaction internal reference genes for expression studies in Brassica napus. Anal. Biochem. 2010, 405, 138–140. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. cluster Profiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. TSWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Q.; He, Y.Q.; Liu, Y.X.; Wang, Z.F.; Zhao, J. JAZ proteins: Key regulators of plant growth and stress response. Crop J. 2024, 12, 1505–1516. [Google Scholar] [CrossRef]
- Ju, L.; Jing, Y.; Shi, P.; Liu, J.; Chen, J.; Yan, J.; Chu, J.; Chen, K.M.; Sun, J. JAZ proteins modulate seed germination through interaction with ABI5 in bread wheat and Arabidopsis. New Phytol. 2019, 223, 246–260. [Google Scholar] [CrossRef]
- Gomi, K.; Sasaki, A.; Itoh, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Kitano, H.; Matsuoka, M. GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J. 2004, 37, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Song, Y.; Bao, D.; Meng, L.Z.; Di, H.; Zhang, L.; Dong, L.; Zeng, X.; Zhang, J.Y.; Li, C.X.; et al. ZmBARK1 as a low-temperature tolerance gene in maize germination. Crop J. 2025, 73, 7562–7573. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Ogawa, M.; Kuwahara, A.; Hanada, A.; Kamiya, Y.; Yamaguchi, S. Activation of Gibberellin biosynthesis and response pathways by low temperature during imbibition of seeds. Plant Cell. 2004, 16, 367–378. [Google Scholar] [CrossRef]
- Li, Q.; Yang, A. Comparative studies on seed germination of two rice genotypes with different tolerances to low temperature. Environ. Exp Bot. 2020, 179, 104216. [Google Scholar] [CrossRef]
- Li, C.; Dong, S.; Beckles, D.M.; Miao, H.; Sun, J.; Liu, X.; Wang, W.; Zhang, S.; Gu, X. The qLTG1.1 candidate gene CsGAI regulates low temperature seed germination in cucumber. Theor. Appl. Genet. 2022, 135, 2593–2607. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, P.; Zou, C.; Chen, Z.; Yuan, G.; Gao, S.; Pan, G.; Shen, Y.; Ma, L. Comprehensive analysis of transcriptional data on seed germination of two maize inbred lines under low-temperature conditions. Plant Physiol. Biochem. 2023, 201, 107874. [Google Scholar] [CrossRef]
- Berglund, T.; Wallström, A.; Nguyen, T.V.; Laurell, C.; Ohlsson, A.B. Nicotinamide; antioxidative and DNA hypomethylation effects in plant cells. Plant Physiol. Biochem. 2017, 118, 551–560. [Google Scholar] [CrossRef]
- Meng, A.; Wen, D.; Zhang, C. Dynamic Changes in Seed Germination under Low-Temperature Stress in Maize. Int. J. Mol. Sci. 2022, 23, 5495. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, D.; Grin, I.R.; Zharkov, D.O.; Yu, B. Developing plant-derived DNA repair enzyme resources through studying the involvement of base excision repair DNA glycosylases in stress responses of plants. Physiol. Plant. 2025, 177, e70162. [Google Scholar] [CrossRef]
- Eastmond, P.J.; Graham, I.A. Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci. 2001, 6, 72–78. [Google Scholar] [CrossRef]
- Liu, J.; Yuan, X.; Tian, M.; Chen, J.; Chen, C.; Luo, Z.; Guo, T.; Huo, X.; Xiao, W. OsNAL11 and OsGASR9 Regulate the low-temperature germination of rice seeds by affecting GA content. Int. J. Mol. Sci. 2024, 25, 11291. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Jiang, Z.; Liu, S.; Lin, R. Interplay between REVEILLE1 and RGA-LIKE2 regulates seed dormancy and germination in Arabidopsis. New Phytol. 2020, 225, 1593–1605. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Molina, L.; Mongrand, S.; McLachlin, D.T.; Chait, B.T.; Chua, N.H. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002, 32, 317–328. [Google Scholar] [CrossRef]


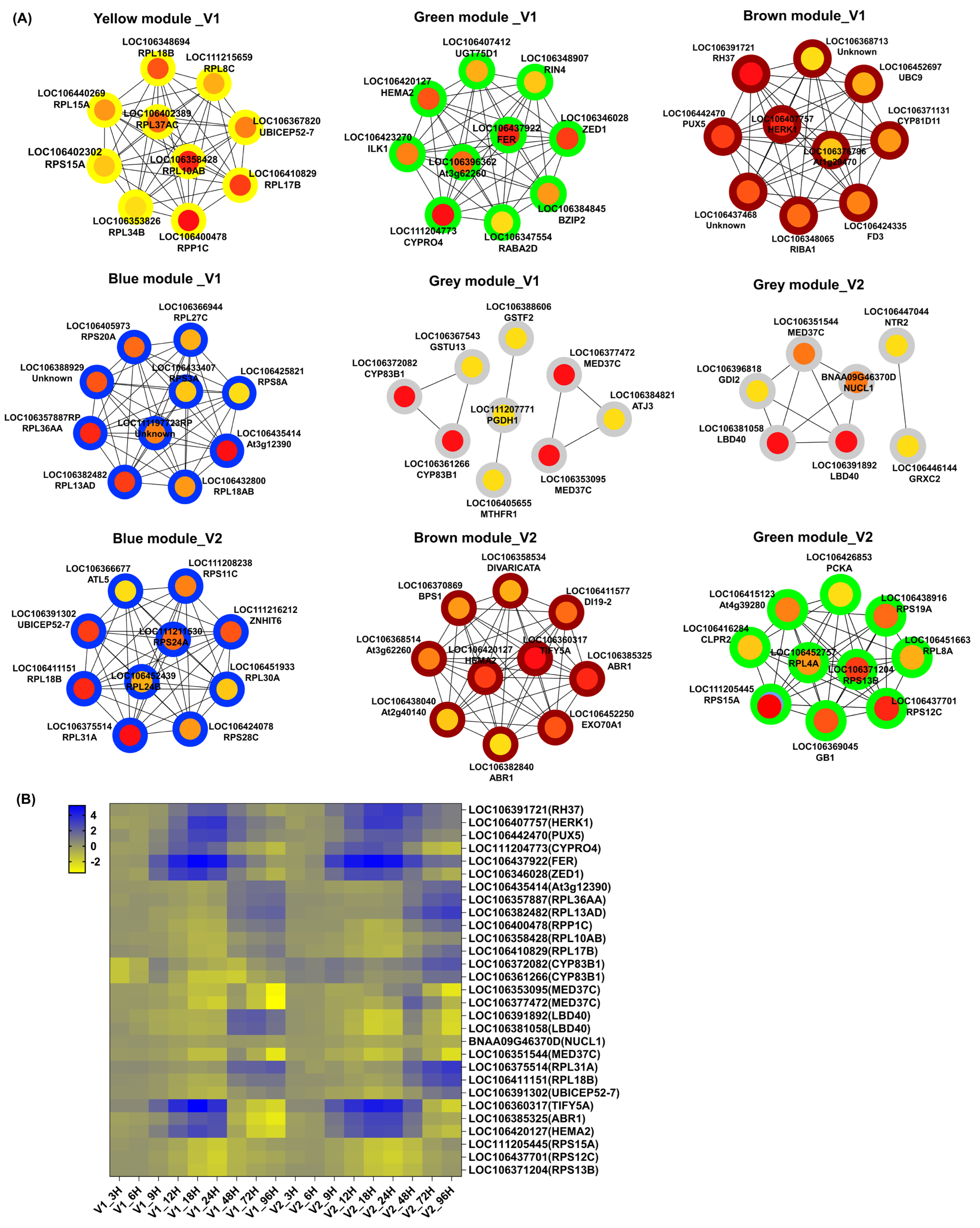

| Genotype | Modules | Time Specific | Gene ID | Function Prediction | Gene Name |
|---|---|---|---|---|---|
| HY3404 | Brown | 24 h | LOC106391721 | DEAD-box ATP-dependent RNA helicase 37, BnaC04g48400D | RH37 |
| LOC106407757 | receptor-like protein kinase HERK 1 | HERK1 | |||
| LOC106442470 | serine/threonine protein phosphatase 2A, BnaA03g41870D | PUX5 | |||
| Green | 18 h | LOC111204773 | Vacuolar import/degradation, Vid27 related protein, BnaC05g33690D | CYPRO4 | |
| LOC106437922 | receptor-like protein kinase FERONIA isoform X2 | FER | |||
| LOC106346028 | non-functional pseudokinase ZED1-like | ZED1 | |||
| Blue | 48 h & 72 h | LOC106435414 | nascent polypeptide-associated complex subunit alpha-like protein 1 | At3g12390 | |
| LOC106357887 | 60S ribosomal protein L144 | RPL36AA | |||
| LOC106382482 | 60S ribosomal protein L13a-4-like | RPL13AD | |||
| Yellow | 96 h | LOC106400478 | 60S acidic ribosomal protein family, BnaC05g00800D | RPP1C | |
| LOC106358428 | 60S ribosomal protein L10a-2-like | RPL10AB | |||
| LOC106410829 | 60S ribosomal protein L17-2-like, BnaA07g25010D | RPL17B | |||
| Grey | 48 h | LOC106372082 | cytochrome P450 83B1, BnaC08g05690D | CYP83B1 | |
| LOC106361266 | cytochrome P450 83B1 | CYP83B1 | |||
| LOC106353095 | probable mediator of RNA polymerase II transcription subunit 37c | MED37C | |||
| LOC106377472 | probable mediator of RNA polymerase II transcription subunit 37c | MED37C | |||
| HY7201 | Grey | 48 h | LOC106391892 | LOB domain-containing protein 40 | LBD40 |
| LOC106381058 | LOB domain-containing protein 40, BnaC02g17190D | LBD40 | |||
| BNAA09G46370D | RNA binding (RRM/RBD/RNP motifs) family protein, BnaA09g46370D | NUCL1 | |||
| LOC106351544 | probable mediator of RNA polymerase II transcription subunit 37c | MED37C | |||
| Blue | 72 h & 96 h | LOC106375514 | 60S ribosomal protein L31-1-like | RPL31A | |
| LOC106411151 | 60S ribosomal protein L18-2-like | RPL18B | |||
| LOC106391302 | ubiquitin-60S ribosomal protein L40-like | UBICEP52-7 | |||
| Brown | 18 h & 24 h | LOC106360317 | protein TIFY 5A-like, jasmonate zim domain protein 8 (JAZ8) | TIFY5A | |
| LOC106385325 | ethylene-responsive transcription factor ABR1-like, BnaC03g49530D | ABR1 | |||
| LOC106420127 | glutamyl-tRNA reductase 2, chloroplastic-like | HEMA2 | |||
| Green | 96 h | LOC111205445 | cytosolic ribosomal protein S15, BnaC05g02380D | RPS15A | |
| LOC106437701 | 40S ribosomal protein S12-2, BnaC03g17910D | RPS12C | |||
| LOC106371204 | 40S ribosomal protein S13-2-like | RPS13B |
| Clusters | Gene_ID | Chromosome | Function Prediction | Gene Name |
|---|---|---|---|---|
| 5 | LOC106450539 | NW_019168954.1 | DELLA protein RGL2 isoform X2 [Brassica napus] | RGL2 |
| 5 | LOC106391695 | NC_027770.2 | multifunctional methyltransferase subunit TRM112-like protein At1g22270 isoform X4 [Brassica napus], BnaC07g13840D | At1g22270 |
| 5 | LOC106361648 | NC_027764.2 | auxin-responsive protein SAUR64-like [Brassica napus] | SAUR64 |
| 5 | LOC106401784 | NC_027771.2 | protein LHY-like [Brassica napus] | LHY |
| 5 | LOC106363703 | NC_027765.2 | homeodomain-leucine zipper protein [Brassica napus] | ATHB-6 |
| 5 | LOC106417715 | NW_019169007.1 | F-box protein DOR-like [Brassica napus], BnaC04g13150D | DOR |
| 5 | BNAC08G30070D | NC_027774.2 | uncharacterized protein LOC106413893 [Brassica napus], BnaC08g30070D | - |
| 8 | LOC106392538 | NC_027770.2 | uncharacterized protein, BnaC04g03310D [Brassica napus] | ARL |
| 8 | LOC106367598 | NC_027765.2 | probable galacturonosyltransferase-like 8 [Brassica napus], BnaA09g29710D | GATL8 |
| 8 | LOC106443577 | NC_027759.2 | ethylene-responsive transcription factor ABI4-like [Brassica napus] | ABI4 |
| 8 | LOC106375427 | NC_027768.2 | selenoprotein H-like [Brassica napus], BnaC01g07140D | SELENOH |
| 8 | LOC106389643 | NW_019168773.1 | nuclear transport factor 2 [Brassica napus] | NTF2B |
| 8 | LOC111197801 | NC_027762.2 | probable trehalose-phosphate phosphatase J [Brassica napus] | TPPJ |
| 8 | LOC106411651 | NC_027773.2 | ribosomal L1 domain-containing protein 1-like [Brassica napus], BnaC04g23460D | RSL1D1 |
| 8 | LOC106426182 | NC_027775.2 | xylulose 5-phosphate/phosphate translocator, chloroplastic-like [Brassica napus], BnaC09g40320D | XPT |
| 8 | LOC106448658 | NC_027769.2 | 60S ribosomal protein L4-1 [Brassica rapa] | RPL4A |
| 8 | LOC106411239 | NC_027761.2 | uncharacterized protein LOC106411239 [Brassica napus] | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, L.; Meng, X.; Wang, W.; Li, H.; Zhou, X.; Zhu, J. Global Transcriptome and WGCNA Analysis Uncover Cultivar-Specific Molecular Signatures Associated with Low-Temperature Germination in Brassica napus L. Agronomy 2025, 15, 2529. https://doi.org/10.3390/agronomy15112529
Lei L, Meng X, Wang W, Li H, Zhou X, Zhu J. Global Transcriptome and WGCNA Analysis Uncover Cultivar-Specific Molecular Signatures Associated with Low-Temperature Germination in Brassica napus L. Agronomy. 2025; 15(11):2529. https://doi.org/10.3390/agronomy15112529
Chicago/Turabian StyleLei, Lei, Xianmin Meng, Weirong Wang, Hongwei Li, Xirong Zhou, and Jifeng Zhu. 2025. "Global Transcriptome and WGCNA Analysis Uncover Cultivar-Specific Molecular Signatures Associated with Low-Temperature Germination in Brassica napus L." Agronomy 15, no. 11: 2529. https://doi.org/10.3390/agronomy15112529
APA StyleLei, L., Meng, X., Wang, W., Li, H., Zhou, X., & Zhu, J. (2025). Global Transcriptome and WGCNA Analysis Uncover Cultivar-Specific Molecular Signatures Associated with Low-Temperature Germination in Brassica napus L. Agronomy, 15(11), 2529. https://doi.org/10.3390/agronomy15112529





