Abstract
Although the effects of soil type, plant genotype, and pathogen invasion on plant rhizosphere microbiomes have been preliminarily explored, their relative contributions and interactive influences on rhizobacterial community assembly remain unclear. In this study, we used tomato as a model to evaluate the individual and combined impacts of these three factors on rhizosphere bacterial community structure and function within a unified experimental framework. Microbiome-based analyses revealed that soil type was the predominant driver, explaining 53.1% of structural and 49.6% of functional variation, followed by tomato genotype (15.6% and 36.1%, respectively) and Fusarium oxysporum f. sp. lycopersici (Fol) inoculation (2.1% and 0.9%). Notably, the interaction between soil type and tomato genotype exerted a stronger influence than any other factor combination. Total nitrogen emerged as the key abiotic factor shaping the taxonomic composition of rhizobacterial communities, whereas soil pH played a dominant role in determining their functional profiles. Distinct tomato genotypes harbored rhizobacterial communities with divergent taxonomic and functional compositions. Although pathogen inoculation triggered the recruitment of beneficial microbes by the host plants, its impact on rhizobacterial community assembly was considerably weaker compared with the effects of soil type and tomato genotype. These findings provide a framework for understanding how soil, host, and pathogen collectively shape rhizobacterial communities and offer insights for optimizing microbiome management in crop production.
1. Introduction
Improving agricultural sustainability necessitates efficient resource utilization, with soil microbiomes offering substantial potential for sustainable intensive farming [1,2]. Soil microorganisms drive key biogeochemical cycles, effectively converting organic matter into plant-available nutrients, such as nitrogen fixation by rhizobia, phosphate solubilization by Bacillus, which promote nutrient cycling and enhance plant growth [3,4]. Comprehending the detailed connections between plants and their microbiomes is vital for reaching this goal. Plant roots exude diverse metabolites into the soil, enhancing microbial activity and attracting microorganisms to the rhizosphere [5,6]. The rhizosphere, often referred to as the plant’s ‘second genome,’ is vital for plant–microbe interactions because of its intricate microbial community [7]. These interactions can create positive feedback loops, where changes in the microbiome influence plant phenotype, and vice versa, with alterations in the plant’s phenotype reshaping the microbial community [8,9]. Rhizosphere bacteria are crucial for plant health, influencing stress resilience, nutrient uptake, and microbiome composition [10,11]. Understanding the factors shaping rhizosphere microbiome structure and function is key to strategically enhancing plant health and performance.
Soil, a primary source of plant-associated microbiomes, contains Earth’s most diverse and complex microbial communities, with microbial biomass carbon frequently surpassing 0.5 mg/g and hosting over 50,000 species/g [12]. The diversity and productivity of plants in land ecosystems are influenced by soil microbiomes [13], acting as reservoirs for root-associated microbes [14,15]. For example, the addition of biochar to soil can suppress the occurrence of bacterial wilt by improving soil health and enhancing the functional diversity of the rhizosphere microbiota [16]. Similarly, implementing practices like crop rotation supports a more diverse and stable soil microbiome, enhancing disease resistance and promoting sustainable agricultural production [17]. The chemical and physical characteristics of soil, including pH, nutrient levels, and organic material, are key factors shaping microbial community structure and, by regulating microbial competition and symbiotic dynamics, ultimately influence the plant microbiome [18,19]. For instance, the soil pH affects bacterial communities by directly imposing physiological constraints on soil bacteria and indirectly altering soil characteristics, such as nutrient availability [20]. In addition, the availability of nutrients such as nitrogen, phosphorus, and carbon directly influences microbial diversity, as microbes that can efficiently utilize specific nutrients dominate under those conditions [21].
Recent studies underscore the unique characteristics of root and rhizosphere microbiomes among various plant species and genotypes, emphasizing the intricate relationships between plants and the microorganisms in their soil [22,23,24,25]. Plant genotype is essential in directing the formation of microbial communities through its impact on the chemical composition and levels of root exudates [26,27,28]. For example, Wu et al. reported that flavonoid compounds secreted by roots can promote cabbage resistance to clubroot disease by recruiting beneficial microorganisms [29]. Additionally, plant immune responses [30] and other ecologically relevant phenotypes exhibit significant genetic variation, making changes in microbiome structure due to plant genotype a common occurrence [31,32]. For example, plants can selectively recruit beneficial microorganisms by altering their immune responses [33]. Additionally, the root architecture of different genotypes can influence the spatial distribution of rhizosphere microorganisms. Plants with deeper or more fibrous roots can alter the availability of carbon and nutrients in different soil layers, thereby affecting microbial diversity and the presence of specific functional groups [34].
In addition to soil type and genotype, pathogen invasion is a major biotic factor that significantly affects microbiome assembly [35,36]. Studies in crops such as wheat [37,38], sugar beet [39], Arabidopsis [40], and chili pepper [41] have shown that pathogen-infected plants can selectively recruit specific beneficial microbes through a process known as the “cry-for-help” strategy, where plants modulate their microbiomes to enhance defense or protect future generations. For example, plants recruit beneficial microorganisms by activating the salicylic acid signaling pathway after pathogen infection [40].
Although soil type, plant genotype, and pathogen stress are recognized as key determinants of rhizosphere microbiome assembly, their relative contributions and interactive effects remain poorly understood. This knowledge gap hinders our ability to optimize agricultural practices for improved crop health and disease resistance. In this study, we used tomato as a model system owing to its global agronomic relevance and high susceptibility to Fusarium oxysporum f. sp. lycopersici (Fol). In a common garden experiment, we cultivated two tomato genotypes across three native soils with and without Fol inoculation, with the objectives of (1) assessing the relative contributions and combined effects of soil type, tomato genotype, and Fol invasion in shaping the tomato rhizobacterial community, and (2) investigating how these factors influence the assembly of rhizobacterial communities. By addressing these interactions, our study aims to provide crucial insights into the mechanisms driving rhizosphere microbiome composition, with significant implications for improving disease management and crop resilience in agriculture.
2. Materials and Methods
2.1. Experiment Design and Collection of Samples
To investigate the effects of soil type, tomato genotype, and pathogen inoculation on the assembly of tomato rhizobacterial communities, we grew two tomato genotypes in three different soils, with and without inoculation of the pathogen, to evaluate the relative and combined contributions of these three factors to rhizobacterial community assembly. Soil samples were collected from three open-field locations in China: Guangzhou (GZ), Guangdong (113°42′3″ E, 23°16′42″ N); Harbin (HEB), Heilongjiang (126°43′1″ E, 45°43′1″ N); and Hangzhou (HZ), Zhejiang (120°11′34.8″ E, 30°13′45.75″ N). Two tomato genotypes, Moneymaker (MM) and LA2157, were used in this study. In addition, the tomato Fusarium wilt pathogen Fusarium oxysporum f. sp. lycopersici (Fol) was used for root inoculation. The Fol strain was obtained from the Institute of Vegetables and Flowers at the Chinese Academy of Agricultural Sciences. The experiment was conducted under greenhouse conditions. Tomato seeds were sterilized by being dipped in 70% ethanol for 60 s, followed by 2.5% sodium hypochlorite for 3 min, and thoroughly washed with sterile deionized water. After sterilization, the tomato seeds were placed at 25 °C to germinate. Upon germination, the seedlings were sown in a sterilized commercial substrate. Once the seedlings developed two cotyledons, they were transplanted into the collected natural soil. To improve soil water retention and aeration, a mixed substrate consisting of commercial growing medium, vermiculite, and natural soil in a 4:3:2 ratio was used for transplanting, ensuring optimal growth conditions for the plants [24]. Seedlings with two true leaves were root-inoculated with Fol using a root-drench method. Spores were harvested from a 3-day-old Potato Lactose (PL) medium, counted using a hemocytometer, and diluted to a final concentration of 106 spores/mL. Every pot received 5 mL of the Fol suspension, with non-inoculated plants acting as controls. The plants were maintained in a growth chamber at 28 °C under a 12 h light/12 h dark photoperiod, with daily watering to maintain soil moisture. For each treatment, 36 pots (10 × 10 × 12 cm) were used, with one plant per pot. Fifteen days post-inoculation, rhizosphere soil was gathered [42]. In each treatment, tomato plants exhibiting wilt symptoms were considered successfully infected by Fol. Each treatment included nine biological replicates, with each replicate comprising a pooled sample from three randomly chosen individual samples. Rhizosphere soil was harvested by carefully uprooting the plants and gently tapping the roots to shake off the soil, as described by Wei et al. [43]. The rhizosphere soil sample was stored at −80 °C. Soil physicochemical properties were determined using the loose soil adjacent to the rhizosphere. Soil pH and electrical conductivity (EC) were determined in a 1:2.5 (w/v) soil–water suspension, whereas soil organic matter (SOM) and total nitrogen (TN) were analyzed following the K2Cr2O7-H2SO4 oxidation method and the semi-micro Kjeldahl method, respectively [44].
2.2. Total DNA Extraction and Amplicon Sequencing
Genomic DNA from bacteria was extracted using the FastDNA Spin Kit for Soil (MP Biomedicals, Santa Ana, CA, USA) following the manufacturer’s guidelines. The V3–V4 region of the bacterial 16S rRNA gene was PCR-amplified using primers 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′). The thermal cycling protocol started with an initial denaturation at 98 °C for 2 min, followed by 25 cycles comprising 15 s of denaturation at 98 °C, 30 s of annealing at 55 °C, and 30 s of extension at 72 °C, and concluded with a final extension at 72 °C for 5 min. PCR products were purified using Agencourt AMPure Beads and quantified with the Invitrogen PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific Inc., Waltham, MA USA). After quantification, the amplicons were pooled in equal volumes and sequenced using paired-end (2 × 300) on the Illumina MiSeq PE300 platform (Illumina Inc., San Diego, CA, USA).
2.3. Amplicon Sequencing Data Analysis
Amplicon sequencing data were processed following the instructions and pipeline structure of EasyAmplicon [45,46]. Specifically, raw paired-end reads of bacterial 16S rRNA genes were first quality-controlled using fastp [47,48] to remove reads with average quality scores below Q20. Paired-end reads were merged using vsearch [49], and sequences with mismatches or low overlap were discarded. Primer sequences were trimmed from both ends. Using unoise3 [50] with its default settings, biological reads were organized into zero-radius operational taxonomic units (zOTUs) at 100% sequence similarity. The taxonomic classification was conducted using USEARCH against the SILVA reference database (version 13_8) [51]. To avoid contamination from non-bacterial sequences, zero-radius operational taxonomic units (zOTUs) classified as Mitochondria, Cyanobacteria, or Chloroplast were excluded. To prevent bias, zOTUs with fewer than two sequences were also excluded. A zOTU table was constructed from the remaining sequences (-zotus) and rarefied to 20,000 reads for further analysis (single_rarefaction.py).
2.4. Diversity Analysis
Alpha diversity at the zOTU level was evaluated using the Richness and Shannon diversity indices. These indices were calculated with QIIME v1.9.1 (alpha_diversity.py) [52], and differences among treatments were analyzed using t-tests or one-way ANOVA followed by Tukey’s post hoc test. Moreover, Shannon diversity values were treated as the response variable in a random forest (RF) regression model to quantify the relative contributions of the soil type, plant genotype, and Fol inoculation. Bray–Curtis dissimilarity was used to assess beta diversity at the zOTU level. Principal Coordinate Analysis (PCoA) was performed based on the Bray–Curtis dissimilarity matrices using the vegan [53] package in R, and the results were visualized with ImageGP 2 (https://www.bic.ac.cn/BIC, accessed on 26 August 2025) [54]. The ‘vegan’ package was used to perform PERMANOVA with 999 permutations to assess overall treatment differences.
2.5. Taxonomic Composition
The taxonomic profiles were analyzed at the phylum, class, genus, and zOTU levels across three experimental factors: plant genotype (MM and LA2157), soil type (GZ, HEB, and HZ), and pathogen treatment (Fol and CK). Based on relative abundances, the ten most dominant phyla and classes and the twenty most dominant genera were identified, and their taxonomic distributions were evaluated accordingly. To enhance clarity in the figure, only the top ten genera were labeled. Taxonomic composition was visualized using the R package ggplot2 [55], and differences among treatments were assessed by two-way ANOVA.
2.6. Functional Prediction and Quantitative Validation of Rhizobacterial Communities
To estimate the functional potential of the tomato rhizosphere, we utilized the FAPROTAX platform v.1.2.3 [56], a tool that links prokaryotic taxa to specific metabolic and ecological functions, based on available data from cultured strains. To compare the overall functional profiles across different groups, unconstrained PCoA combined with PERMANOVA was applied. We specifically examined functions related to nitrogen and organic compound metabolism [57], comparing their potentials among the groups utilizing two-way ANOVA and Tukey post hoc tests or Sidak’s test. To further evaluate the robustness of the functional predictions, real-time quantitative PCR (qPCR) was conducted on uninoculated soil samples from each soil type to quantify the abundance of the target functional genes. All reactions were performed on an MA-6000 Real-Time PCR System (Molarray, Suzhou, China). Primer sequences, adopted from previously published studies [58,59,60,61], are provided in Table S1. DNA from the collected soil samples was extracted with the FastDNA Spin Kit for Soil (MP Biomedicals, Santa Ana, CA, USA) according to the manufacturer’s instructions. For qPCR assays, each 20 μL reaction mixture contained 10 μL of 2× SYBR qPCR Master Mix (Vazyme, Nanjing, China), 1 μL each of forward and reverse gene-specific primers, 1 μL of DNA template, and 1 μL of ddH2O. Plasmids harboring the target amplicon were constructed from the extracted genomic DNA and used as quantitative standards; copy numbers were calculated from plasmid concentration and insert size. Ten-fold serial dilutions of the plasmid standards were amplified in triplicate to generate standard curves (Table S2). Statistical analysis of the qPCR-derived functional gene copy numbers was performed using one-way ANOVA followed by Tukey’s post hoc test via the Wekemo Bioincloud platform [62].
2.7. Network Construction
To investigate the effects of soil type, tomato genotype, and Fol inoculation on rhizosphere bacterial interaction networks, co-occurrence networks were constructed using the integrated Network Analysis Pipeline 2.0 (iNAP2.0) platform (https://inap.denglab.org.cn/, accessed on 21 July 2025) [63]. For each treatment group, only zOTUs present in all samples and with a relative abundance greater than 0.01% were retained for network construction. Missing values were replaced with a small constant (0.01) in blanks with paired valid values. Prior to correlation analysis, abundance data were subjected to a logarithmic transformation to reduce the influence of extreme values. A similarity matrix was then generated using the Pearson correlation coefficient, which served as the input for Random Matrix Theory (RMT)-based cutoff scanning [64]. The RMT algorithm scans decreasing similarity thresholds and applies Poisson regression to determine the transition point. Although RMT can automatically determine the cutoff, a uniform threshold of 0.74 was applied across all networks to ensure comparability among treatments. The global topological properties of the networks, as well as the within-module connectivity (Zi) and among-module connectivity (Pi) of each node, were calculated in iNAP2.0. Based on Zi–Pi scores, putative keystone species were further identified. Networks were visualized using Gephi 0.92 [65]. To quantify the effect sizes of soil type, tomato genotype, and pathogen inoculation, we calculated Euclidean distances based on network topological properties. Five key metrics—number of nodes, number of edges, average degree, average clustering coefficient, and average path distance—were selected to construct the Euclidean distance matrix. The average pairwise distance among the three soil-derived networks was defined as the effect of soil type, the distance between the two genotype networks represented the effect of genotype, and the distance between pathogen-inoculated and control networks was used to indicate the effect of pathogen inoculation.
2.8. Redundancy Analysis
The relationships between rhizobacterial taxonomic and functional compositions and soil physicochemical properties were assessed using redundancy analysis (RDA) implemented in the ‘vegan’ R package (v4.3.3). The overall and individual contributions of soil properties to rhizobacterial communities were tested using PERMANOVA with 999 permutations.
3. Results
3.1. Quality Metrics of Sequencing Analysis
Tomato genotypes LA2157 and Moneymaker (MM) were planted in soils collected from Guangzhou (GZ), Harbin (HEB), and Hangzhou (HZ), with Fusarium oxysporum f. sp. lycopersici (Fol) inoculation. DNA was extracted from 108 samples and amplified with primers for the V3–V4 regions of the bacterial 16S rRNA gene. High-throughput sequencing generated 17,786,266 bacterial raw reads. Following quality filtering and the exclusion of chimeric and plant-derived sequences, 9,216,675 high-quality bacterial reads were retained. As shown by the rarefaction curves, the sequencing depth was adequate to capture the microbial diversity within each sample (Figure S1). The resampled zOTU table facilitated further microbial community analysis (Table S3).
3.2. Diversity of Rhizobacterial Communities Is Driven by Soil Type, Tomato Genotype and Fol Inoculation
The bacterial community diversity in the tomato rhizosphere was assessed by measuring α-diversity using the Richness and Shannon indices at the zOTU level, with samples categorized by soil type, tomato genotype, and Fol inoculation treatments. Bacterial diversity exhibited distinct distribution patterns across various soils, with significant differences in both Richness and Shannon indices, ordered as GZ > HZ > HEB (Richness, p < 0.001; Shannon, p < 0.001; Figure 1A,D; Table S4). Tomato genotypes notably influenced α-diversity metrics, with MM showing greater Richness and Shannon diversity compared to LA2157 (Richness, p < 0.001; Shannon, p < 0.001; Figure 1B,E; Table S4). After Fol inoculation, no significant differences were detected in Richness and Shannon diversity (Figure 1C,F; Table S4). To further assess the relative contribution of soil type, genotype, and Fol inoculation to microbial diversity, we used the Shannon diversity index as the response variable and applied a random forest regression model. The results showed that soil type was the most influential factor affecting community diversity (%IncMSE = 30.85%), followed by plant genotype (%IncMSE = 21.48%), while Fol inoculation had minimal or even negative importance (%IncMSE = −4.10%, Figure S2).
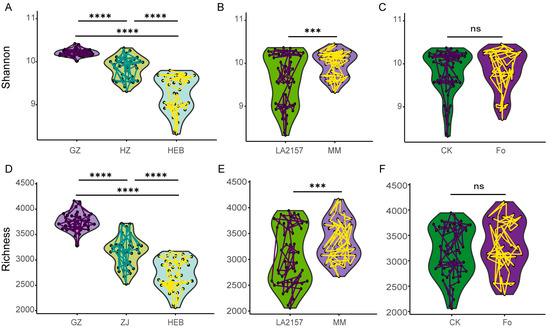
Figure 1.
Alpha diversity of tomato rhizosphere microbiomes. Shannon and Richness indices of microbiota across various soil types ((A,D); n = 36). Shannon and Richness indices of the microbiota in two tomato genotypes ((B,E); n = 54). Bar charts depicting the influence of pathogen Fol inoculation on the microbiota’s Shannon and Richness indices ((C,F); n = 54). LA2157 and MM represent the tomato genotypes LA2157 and Moneymaker, respectively. GZ, Guangzhou; HZ, Hangzhou; HEB, Harbin. CK refers to the control treatment without pathogen Fol inoculation, and Fo refers to the treatment with Fol inoculation. *** and **** indicate significant difference at p < 0.001 and p < 0.0001 (t test or Tukey’s HSD test), respectively.
Unconstrained principal coordinate analysis (PCoA) further revealed the integrated effects of tomato genotype, soil type, and Fol inoculation on the rhizobacterial community structure. PCoA showed significant separation along the axes of genotype, soil type, and Fol inoculation (Figure 2; Table S5). As hypothesized, soil type was identified as the main factor influencing the rhizobacterial community structure, accounting for the highest variance in bacterial community composition (R2 = 0.531, p < 0.05; Figure 2A). This result underscores the dominant role of edaphic factors in shaping microbial communities in the rhizosphere. Tomato genotype was a secondary factor (R2 = 0.156, p < 0.05; Figure 2B), indicating that genetic differences in the host plants also contribute to community composition, though to a lesser extent compared to soil type. Fol inoculation was the least influential factor (R2 = 0.021, p < 0.05; Figure 2C), suggesting that while inoculation with Fol may have some effect, it is not a major driver of rhizobacterial community composition in this study. A significant interaction effect between genotype and soil type was observed (R2 = 0.048, p < 0.05; Table S5), highlighting the combined influence of these factors in shaping microbial community structure. However, interactions between Fol and other factors, such as genotype and soil type, appeared to have relatively minor effects, with R2 values of 0.0156 and 0.0135, respectively. These findings suggest that soil type and tomato genotype play more prominent roles in determining rhizobacterial communities, with Fol inoculation exerting a comparatively smaller influence. Additionally, the three-way interaction between genotype, Fol inoculation, and soil type demonstrated only a minimal effect (R2 = 0.0117, p < 0.05), reinforcing the primary roles of soil and genotype in this context. Among the different soil types, HEB exhibited the highest variability, followed by HZ and GZ (p < 0.001; Figure 2D). The LA2157 genotype’s bacterial communities showed significantly more variability than those of the MM genotype (p < 0.001; Figure 2E), as assessed by β-dispersion using Bray–Curtis dissimilarity. Moreover, the variability of rhizobacterial communities in tomato plants inoculated with Fol was significantly greater than in non-inoculated plants (p < 0.001; Figure 2F). These results suggest that soil type, genotype, and Fol inoculation synergistically influence the diversity of the tomato rhizobacterial community, with soil type being the dominant factor shaping bacterial community structure.
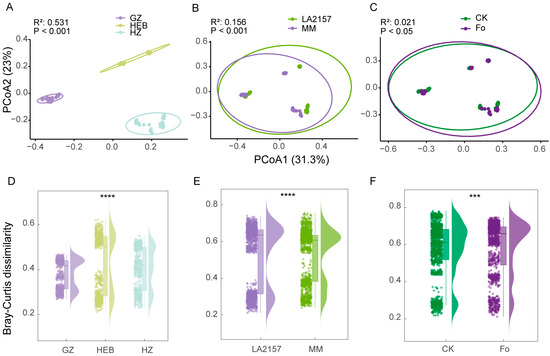
Figure 2.
Patterns of beta diversity in the microbiomes of tomato rhizospheres. PCoA analysis of microbial communities in each microhabitat for the three soil types ((A), n = 36), two tomato genotypes ((B), n = 54), and Fol pathogen inoculation versus control treatments ((C), n = 54). Different groups are highlighted by ellipses and point colors. PERMANOVA tests were employed to evaluate the significance of differences in microbial communities between groups. Beta-dispersion analysis of rhizobacterial communities for soil types (D), different genotypes (E), and pathogen inoculation treatments (F) based on Bray–Curtis dissimilarity (Wilcoxon rank-sum test). LA2157 and MM represent the tomato genotypes LA2157 and Moneymaker, respectively. GZ, Guangzhou; HZ, Hangzhou; HEB, Harbin. CK refers to the control treatment without pathogen Fol inoculation, and Fo refers to the treatment with Fol inoculation. *** and **** indicate significant difference at p < 0.001 and p < 0.0001 (t test or Tukey’s HSD test), respectively.
3.3. Core Microbiota Response to Soil Type, Tomato Genotype, and Fol Inoculation
The observed significant effects of soil type, tomato genotype, and Fol inoculation on both α- and β-diversity of the tomato rhizosphere microbiota prompted us to further investigate how bacterial taxa responded in terms of relative abundance. Soil type, tomato genotype, and Fol inoculation significantly influenced the composition of the rhizobacterial community (Figure 3, Figure 4 and Figure 5 and Table S6). Across the three soil types (GZ, HEB, and HZ), the dominant bacterial phyla were Proteobacteria, Actinobacteriota, Bacteroidota, and Acidobacteriota. Notably, Proteobacteria and Bacteroidota were more enriched in HEB (62.6% and 16.5%) and HZ (63.0% and 15.4%) compared with GZ (57.7% and 9.8%). Actinobacteriota and Acidobacteriota exhibited significantly higher relative abundances in GZ (10.9% and 5.2%) than in HEB (8.4% and 2.9%) and HZ (6.1% and 3.7%) (Tukey’s HSD test, p < 0.05; Figure 3A,B; Table S6). At the class level, Alphaproteobacteria, Gammaproteobacteria, Bacteroidia, and Actinobacteria were dominant across all three soil types, but their relative abundances varied. Alphaproteobacteria was significantly enriched in HEB (43.1%) compared to GZ (34.7%) and HZ (35.7%). Gammaproteobacteria exhibited the highest relative abundance in HZ (32.1%), which was significantly greater than in GZ (30.6%) and HEB (23.9%). In contrast, Bacteroidia was significantly enriched in HEB (17.3%) and HZ (16.2%) compared with GZ (10.8%), while no significant difference was observed between HEB and HZ. Actinobacteria was significantly enriched in GZ (7.2%) than in HEB (5.2%) and HZ (3.1%). Additionally, Gemmatimonadetes (4.5%) and Bacilli (3.5%), were significantly enriched in GZ, while Acidimicrobiia (2.2%), Bdellovibrionia (1.2%) and Vicinamibacteria (1.3%) were reduced in HEB (Tukey’s HSD test, p < 0.05; Figure 3C,D; Table S6). At the genus level, different soil types harbored distinct genera: Pseudomonas (20.2%), Flavobacterium (10.0%), and Devosia (8.1%) were significant enriched in HZ; Massilia (12.4%), Caulobacter (10.7%), Pedobacter (6.9%), Rhizobium (5.6%), Asticcacaulis (5.1%), Novosphingobium (4.4%), Dyadobacter (3.2%) and Noviherbaspirillum (2.7%) were significant enriched in HEB; and Cupriavidus (8.9%), Bacillus (6.5%), Comamonas (4.6%), Mucilaginibacter (4.3%) and Ramlibacter (3.2%) were significant enriched in GZ (Tukey’s HSD test, p < 0.05; Figure 3E,F; Table S6). At the zOTU level, 339 zOTUs were enriched in GZ, 216 zOTUs were enriched in HEB, and 254 zOTUs were enriched in HZ (Tukey’s HSD test, p < 0.05; Table S6).
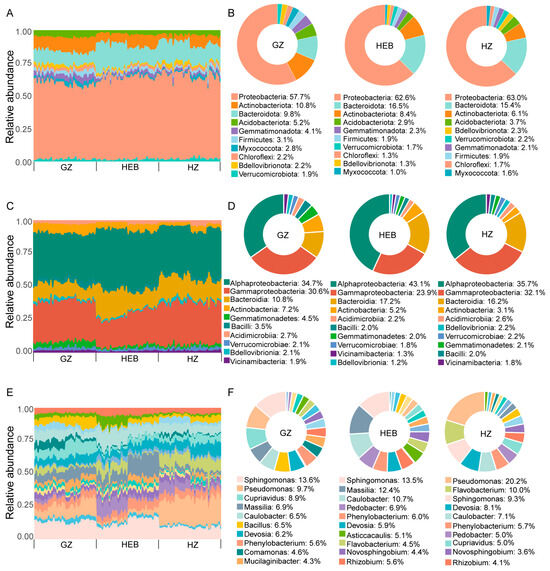
Figure 3.
Taxonomic composition of the tomato microbiome across different soil types. (A) A bar plot of phyla abundances in GZ, HEB, and HZ. (B) A pie plot of the top 10 phyla of microbiota in GZ, HEB, and HZ. (C) Relative proportions of the most frequent classes in GZ, HEB, and HZ. (D) A pie plot of the top 10 classes of microbiota in GZ, HEB, and HZ. (E) Relative proportions of the most frequent genera in GZ, HEB, and HZ. (F) A pie plot of the top 10 genera of microbiota in GZ, HEB, and HZ. GZ, Guangzhou; HZ, Hangzhou; HEB, Harbin. Tukey’s HSD test, n = 36.
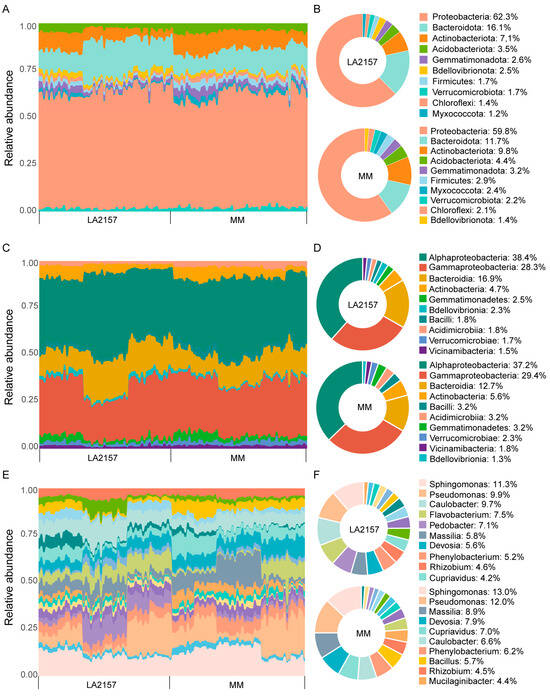
Figure 4.
The taxonomic profile of the tomato microbiome in the two genotypes. (A) A bar chart showing the abundance of phyla in the LA2157 and MM genotypes. (B) A pie plot of the top 10 phyla of microbiota in the LA2157 and MM genotypes. (C) A bar chart showing the abundance of classes in the LA2157 and MM genotypes. (D) A pie plot of the top 10 classes of microbiota in the LA2157 and MM genotypes. (E) Relative proportions of the most frequent genera in the LA2157 and MM genotypes. (F) A pie plot of the top 10 genera of microbiota in the LA2157 and MM genotypes. LA2157 and MM represent the tomato genotypes LA2157 and Moneymaker, respectively. Sidak’s test, n = 54.
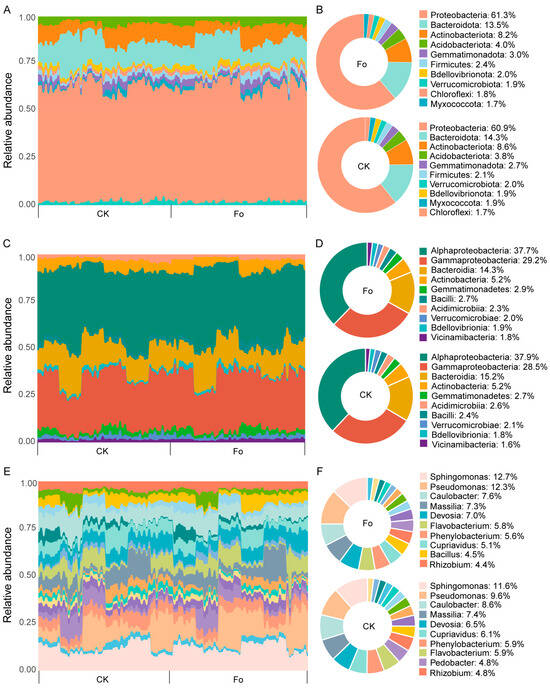
Figure 5.
Changes in the taxonomic composition of the tomato microbiome induced by Fol inoculation. (A) A bar plot of phyla abundances present in Fol inoculated and CK groups. (B) A pie plot of the top 10 phyla of microbiota in Fol inoculated and CK groups. (C) A bar plot of class abundances present in Fol inoculated and CK groups. (D) A pie plot of the top 10 classes of microbiota in Fol inoculated and CK groups. (E) Relative proportions of the most frequent genera in Fol inoculated and CK groups. (F) A pie plot of the top 10 genera of microbiota in Fol inoculated and CK groups. CK refers to the control treatment without pathogen Fol inoculation, and Fo refers to the treatment with Fol inoculation. Sidak’s test, n = 54.
Tomato genotypes LA2157 and MM displayed unique patterns of microbial composition. Proteobacteria, Bacteroidota, Actinobacteriota, and Acidobacteriota were the dominant phyla in both LA2157 and MM. Notably, Proteobacteria (62.4%) and Bacteroidota (16.1%) were enriched in LA2157, whereas Actinobacteriota (7.1%) and Acidobacteriota (3.5%) were more abundant in MM (Sidak’s test, p < 0.05; Figure 4A,B; Table S6). The dominant classes in both genotypes were Alphaproteobacteria, Gammaproteobacteria, Bacteroidia, and Actinobacteria. In LA2157, Alphaproteobacteria (38.4%), Bacteroidia (16.9%), and Bdellovibrionia (2.3%) were significantly more abundant, whereas Gammaproteobacteria (28.3%), Actinobacteria (4.7%), Bacilli (1.8%), and Acidimicrobiia (1.8%) were significantly less abundant compared to MM (Sidak’s test, p < 0.05; Figure 4C,D; Table S6). At the genus level, Caulobacter (9.7%), Flavobacterium (7.5%), Pedobacter (7.1%), Asticcacaulis (4.1%), Bdellovibrio (3.7%), Comamonas (3.4%), and Noviherbaspirillum (2.6%) were significantly enriched in LA2157. In contrast, Sphingomonas (13.0%), Pseudomonas (12.0%), Massilia (8.9%), Devosia (7.9%), Cupriavidus (7.0%), Phenylobacterium (6.2%), Bacillus (5.7%), and Mucilaginibacter (4.4%) were significantly more abundant in MM (Sidak’s test, p < 0.05; Figure 4E,F; Table S6). At the zOTU level, 51 zOTUs were significantly enriched in LA2157, whereas 45 zOTUs were significantly enriched in MM (Sidak’s test, p < 0.05; Table S6).
Fol inoculation also significantly influenced the composition of the tomato rhizobacterial community; however, its effect was considerably smaller than that of soil type and tomato genotype. At the phylum level, Fol inoculation led to a significant decrease in the relative abundance of Bacteroidota (Sidak’s test, p < 0.05; Figure 5A,B; Table S6). At the class level, the abundance of Bacteroidia was significantly reduced following Fol inoculation (Sidak’s test, p < 0.05; Figure 5C,D; Table S6). At the genus level, Fol inoculation significantly enriched Sphingomonas (12.7%), Pseudomonas (12.3%), and Bacillus (4.5%) in the tomato rhizosphere (Sidak’s test, p < 0.05; Figure 5E,F; Table S6). At the zOTU level, 21 zOTUs were enriched in the CK treatment, whereas 24 zOTUs were enriched under Fol inoculation (Sidak’s test, p < 0.05; Table S6). These findings demonstrate that soil type, tomato genotype, and Fol inoculation reshaped the composition of the tomato rhizosphere microbiota.
3.4. Drivers of Rhizobacterial Functional Profiles Under Soil Type, Tomato Genotype, and Fol Inoculation
The ecological functional profiles of tomato rhizospheres were annotated using the Functional Annotation of Prokaryotic Taxa (FAPROTAX) tool. PCoA coupled with PERMANOVA revealed that soil type, genotype, and Fol inoculation significantly shaped the functional features of the tomato rhizosphere (Figure S3). Consistent with previous results, soil type emerged as the primary factor, explaining the largest proportion of variance in the functional profile of the bacterial community (R2 = 0.496, p < 0.05; Figure S3; Table S5). Tomato genotype was identified as a secondary factor (R2 = 0.361, p < 0.05; Figure S3; Table S5), followed by Fol inoculation (R2 = 0.009, p < 0.05; Figure S3; Table S5). Notably, the rhizosphere in HZ exhibited significantly higher functional potentials associated with denitrification, nitrate, nitrite, and nitrogen respiration, ligninolysis, and fermentation. In contrast, the rhizosphere in GZ showed elevated functional potentials related to nitrate reduction, aromatic hydrocarbon, and hydrocarbon degradation. Moreover, GZ and HZ exhibited significantly greater functional potential for aromatic-compound degradation than HEB, whereas the difference between GZ and HZ was not significant (Figure 6A,B). Furthermore, the genotype LA2157 demonstrated higher functional potentials for denitrification, nitrate, nitrite, and nitrogen respiration, aromatic compound, aromatic hydrocarbon, and hydrocarbon degradation compared to MM. On the other hand, functions related to nitrate reduction and fermentation were more pronounced in the MM rhizosphere (Figure 6C,D). Ultimately, Fol inoculation reduced the functional potentials for nitrate reduction and aromatic compound degradation, while increasing the functional potential related to ligninolysis (Figure 6E,F).
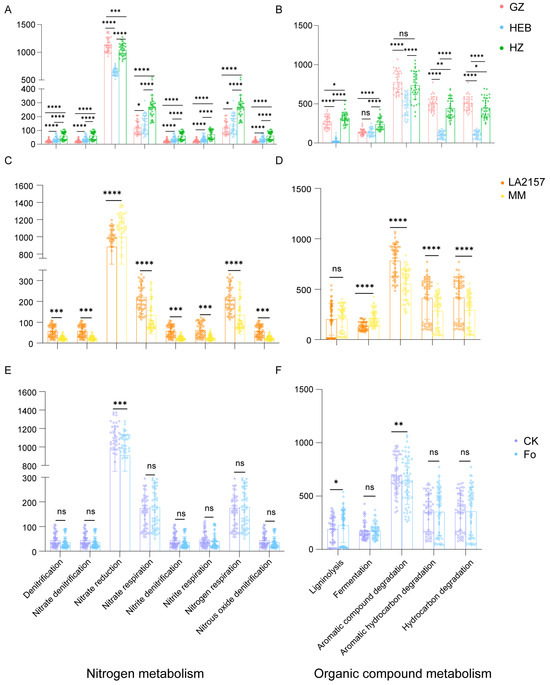
Figure 6.
Functional signatures in the tomato rhizosphere. The functional differences analysis of microbial communities in each microhabitat for the three soil types ((A,B); n = 36), two tomato genotypes ((C,D); n = 54), and Fol pathogen inoculation versus control treatments ((E,F); n = 54). LA2157 and MM represent the tomato genotypes LA2157 and Moneymaker, respectively. GZ, Guangzhou; HZ, Hangzhou; HEB, Harbin. CK refers to the control treatment without pathogen Fol inoculation, and Fo refers to the treatment with Fol inoculation. *, **, *** and **** indicate significant difference at p < 0.05, p < 0.01, p < 0.001 and p < 0.0001 (Sidak’s test or Tukey’s HSD test), respectively.
To verify the robustness of the functional predictions, we performed real-time quantitative PCR on DNA extracted from the three soil types, the factor exerting the greatest influence on the rhizobacterial community. Copy numbers of genes associated with nitrogen metabolism and organic compound metabolism were quantified for GZ, HEB, and HZ soils. The qPCR results confirmed the predictions: the HZ rhizosphere contained significantly higher abundances of denitrification genes (nirS, nirK, and nosZ) than either GZ or HEB, whereas the aromatic-compound degradation gene c230 was significantly enriched in both GZ and HZ relative to HEB, with no significant difference between GZ and HZ (Figure S4).
3.5. Co-Occurrence Networks of Rhizobacterial Are Driven by Soil Type, Tomato Genotype, and Fol Inoculation
To further evaluate the effects of soil type, tomato genotype, and Fol inoculation on the complex interactions among rhizobacterial taxa, we performed co-occurrence network analysis. The rhizobacterial networks of GZ, HEB, and HZ differed markedly in structure. The GZ, HEB, and HZ networks comprised 96 nodes and 496 edges, 152 nodes and 2798 edges, and 151 nodes and 1686 edges, respectively (Figure 7A–C; Table S7). Other topological properties assessing overall network structure, including average degree, average clustering coefficient, and average path distance, also showed substantial differences. The HEB network exhibited the highest average degree (36.82) and average clustering coefficient (0.69), as well as the lowest average path distance (1.96) compared with GZ and HZ (Table S7). Proteobacteria, Actinobacteriota, Bacteroidota, and Firmicutes dominated the nodes in all three soil networks, accounting for over 90% of total nodes. Notably, nodes classified as Chloroflexi were unique to GZ, while Acidobacteriota nodes were only present in HZ (Figure 7D). Tomato genotype also strongly influenced the rhizobacterial co-occurrence networks. The LA2157 network contained 129 nodes and 1360 edges, whereas MM had 106 nodes and 371 edges (Figure 7E,F; Table S7). Compared with MM, LA2157 displayed a higher average degree (21.09), a higher average clustering coefficient (0.67), and a lower average path distance (2.47) (Table S7). Most nodes in both genotypes were assigned to Proteobacteria, Bacteroidota, Actinobacteriota, Firmicutes, and Bdellovibrionota. However, LA2157 harbored unique nodes identified as Verrucomicrobiota, while MM possessed unique nodes assigned to Acidobacteriota and Gemmatimonadota (Figure 7G). Fol inoculation also altered the rhizobacterial co-occurrence network. Following pathogen inoculation, the number of nodes increased from 98 to 99, while the number of edges decreased from 405 to 392 (Figure 7H,I; Table S7). Other network properties also changed: average degree (CK: 8.27; Fol: 7.92), average clustering coefficient (CK: 0.57; Fol: 0.48), and average path distance (CK: 3.31; Fol: 3.04) (Table S7). Additionally, a new node identified as Bdellovibrionota appeared in the Fol-inoculated network, whereas nodes assigned to Acidobacteriota disappeared (Figure 7J). In microbial networks, keystone taxa can be identified as hub nodes based on within-module degree (Zi) and among-module connectivity (Pi). We observed 10 hub nodes in the GZ network, whereas HEB and HZ had none. Genotype also influenced hub node composition: MM contained one hub node, while LA2157 had none. Following Fol inoculation, the hub node shifted from zOTU_28 (Dyadobacter) to zOTU_107 (Sphingomonas) and zOTU_156 (Allorhizobium–Neorhizobium–Pararhizobium–Rhizobium) (Figure S5; Table S7).
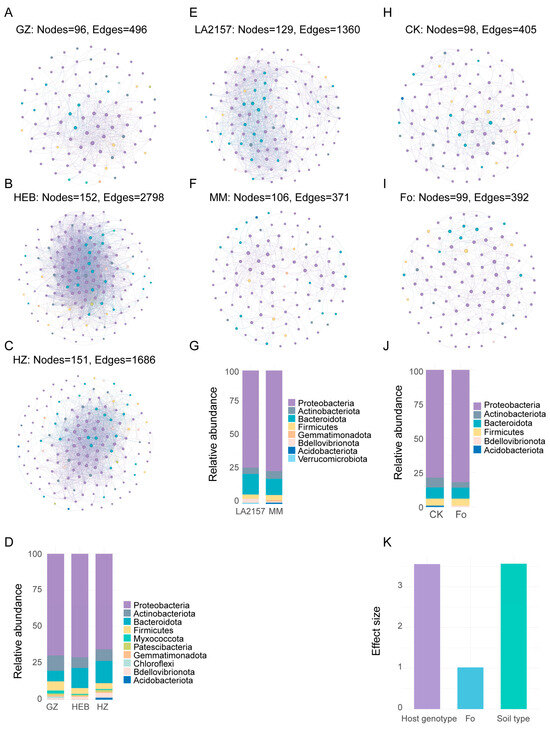
Figure 7.
Tomato rhizobacterial interaction networks. Overview of microbial co-occurrence networks for GZ (A), HEB (B) and HZ (C). (D) Taxonomic composition of network nodes in GZ, HEB, and HZ. Overview of microbial co-occurrence networks for LA2157 (E) and MM (F). (G) Taxonomic composition of network nodes in LA2157 and MM. Overview of microbial co-occurrence networks for Fol inoculated (H) and CK groups (I). (J) Taxonomic composition of network nodes in Fol-inoculated and CK groups. (K) Relative effects of soil type, plant genotype, and pathogen inoculation on network topology. CK refers to the control treatment without pathogen Fol inoculation, and Fo refers to the treatment with Fol inoculation.
Furthermore, we quantitatively assessed the effects of soil type, tomato genotype, and Fol inoculation on rhizobacterial co-occurrence networks by calculating Euclidean distances among networks within each factor. Among the tested factors, soil type exerted the strongest influence on network topology (effect size = 3.57), followed closely by tomato genotype (effect size = 3.56), whereas the effect of pathogen inoculation was comparatively weak (effect size = 1.02) (Figure 7K).
3.6. Relationships Between Rhizobacterial Taxonomic and Functional Composition and Soil Physicochemical Properties
Given that soil type was identified as the primary driver of rhizobacterial community structure and function, we further explored the underlying mechanisms. Significant differences were observed in soil pH, soil organic matter (SOM), and total nitrogen (TN) among the three soil types (GZ, HEB, and HZ) (p < 0.05), whereas electrical conductivity (EC) did not differ significantly (Table S8). Redundancy analysis (RDA) indicated that soil pH, SOM, and TN were the main factors driving variation in microbial community composition (p < 0.01), with the first two constrained axes explaining 23.91% and 17.3% of the variance, respectively. The effect sizes followed the order TN > pH > SOM, while EC had no significant effect on community composition (Figure 8A). Similarly, RDA of rhizobacterial functional composition revealed that soil pH, SOM, and TN were also the key drivers of functional variation (p < 0.01), with the first two axes explaining 47.33% and 10.67% of the variance, respectively. In this case, the effect sizes followed the order pH > SOM > TN, whereas EC had no significant influence on functional composition (Figure 8B).
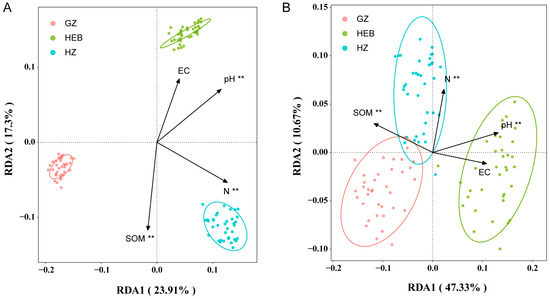
Figure 8.
Relationship between rhizobacterial taxonomic composition (A) and functional composition (B) and soil physicochemical properties indicated by redundancy analysis (RDA). PERMANOVA test, **, p < 0.01. n = 36.
4. Discussion
The rhizobacterial community is crucial for plant health, affecting growth, stress tolerance, and disease resistance. Geographical location, plant genetic variation, and pathogen infection significantly influence the structure and function of the rhizobacterial community. Nevertheless, systematic evaluations of these factors and their divergent roles in shaping microbial communities are limited. This study sought to address this gap by investigating the interactions between soil types, tomato genotypes, and Fusarium oxysporum f. sp. lycopersici (Fol) inoculation in the tomato rhizobacterial community. Our results demonstrate that soil type is the primary determinant of rhizobacterial community structure and function, whereas genotype and pathogen invasion exert secondary influences. Among soil physicochemical properties, total nitrogen (TN), pH, and soil organic matter (SOM) were identified as the key abiotic factors shaping both the taxonomic and functional composition of the rhizobacterial community. In different soil types, tomato genotypes, and with or without Fol inoculation, Proteobacteria, Bacteroidota, Actinobacteriota, and Acidobacteriota consistently dominate the tomato rhizosphere. These phyla may represent microbial groups highly adapted to their respective environments. These phyla play crucial roles in nutrient cycling, including the regulation of carbon, nitrogen, iron, and sulfur cycles [66,67,68,69]. These findings provide new insights into the roles of soil type, genotype, and pathogen infection in structuring tomato rhizosphere bacterial communities.
This study highlights the dominant role of soil type in shaping tomato rhizobacterial communities. Under controlled greenhouse conditions, where water availability and other environmental factors were kept consistent, the observed differences in plant growth and microbial communities could be largely attributed to soil-specific properties. We found that soil type accounted for the largest proportion of variation in α- and β-diversity, community functional composition, and co-occurrence network structure. These results are consistent with previous studies indicating that soil factors are key determinants of plant rhizosphere microbial communities [70,71]. For example, Edwards et al. demonstrated that soil type determines the microbial taxa that rice roots can recruit by providing a unique microbial reservoir, with its influence exceeding that of rice genotype [70]. Bonito et al. showed that soil origin directly governs the presence and abundance of microbial taxa and alters the associations among functional groups by reshaping bacterial–fungal co-occurrence patterns, with this effect being stronger than that of plant genotype [71]. Our study further revealed that soil pH, TN, and SOM significantly influenced both community structure and function, with TN exerting the strongest effect on community composition and pH having the greatest impact on functional composition. Zhang and Wang et al. demonstrated that TN and SOM contents were significantly correlated with the abundances of Proteobacteria and Bacteroidota [72,73], which in turn affected the abundances of rhizobacterial taxa related to nitrogen and organic matter metabolism, such as Pseudomonas, Caulobacter, Rhizobium, and Flavobacterium. Soil pH affects microbial metabolism and biogeochemical cycles, such as carbon, nitrogen, and sulfur processes. Soil pH can be altered by microbial activities like nitrification, leading to acidification or alkalinization, which in turn shapes microbial diversity and functionality [20]. Xu et al. also demonstrated that pH is a key factor influencing soil bacterial functional activity [74]. Overall, these findings indicate that the physicochemical environment and nutrient status provided by the soil are the primary drivers determining rhizosphere microbial colonization and interactions. Therefore, manipulating soil nutrient levels and pH may serve as an effective strategy to optimize rhizosphere microbial communities and enhance crop health. Appropriate pH levels can be maintained through methods such as applying lime in acidic soils or using organic amendments, thereby stabilizing microbial communities and promoting plant health [75]. Additionally, ensuring balanced nutrient availability, especially nitrogen and organic matter, supports beneficial microbes by providing both immediate nutrients and a stable environment for microbial activity, thereby enhancing plant resilience and productivity [42,76]. Yu et al. demonstrated that the addition of organic fertilizer can promote the diversity of the pakchoi rhizosphere community and increase the weight of both leaves and roots [77].
Although soil type is considered the primary driver of rhizobacterial community assembly in plants, tomato genotype, as a secondary factor, also plays a significant role in shaping the tomato rhizosphere microbiome. Our results indicate that different tomato genotypes exhibit distinct α- and β-diversity patterns and recruit different bacterial taxa. The tomato genotype accounted for 15.6% of the variation in rhizobacterial community structure. Plants selectively modulate the abundance and diversity of rhizobacteria through root exudates, promoting the proliferation of beneficial taxa while suppressing potentially harmful ones, thereby shaping a rhizosphere microbial community favorable for their own growth [28]. We observed that the dominant genera recruited differed between genotypes and possessed potential plant growth-promoting traits. For instance, LA2157 preferentially recruited Caulobacter and Flavobacterium, whereas MM enriched Sphingomonas and Pseudomonas, all of which are considered potentially beneficial [78,79,80]. Caulobacter has been shown to promote watermelon seedling growth and enhance tolerance to heavy metals [81]. Flavobacterium plays a key role in suppressing Ralstonia solanacearum-induced bacterial wilt in tomato [82]. Additionally, previous studies have demonstrated that Sphingomonas is significantly enriched in the rhizosphere of healthy wheat, and re-inoculation with Sphingomonas markedly promotes wheat growth and health [83]. Lv et al. reported that Pseudomonas can enhance banana resistance to Fusarium wilt by modulating jasmonic acid (JA) and salicylic acid (SA) levels [84]. Genotype-dependent effects have also been noted in other species of plants, such as wheat [85], beans [86], barley [27], and maize [87], where plant root exudates mediate microbiome assembly in a genotype-dependent manner. Moreover, plant genotype explained 36.1% of the variation in bacterial community function, particularly reflected in significant differences in nitrogen and organic matter metabolism. These functional disparities are likely driven by genotype-specific root exudates that recruit microbes with corresponding metabolic capabilities [88,89]. In addition, rhizobacteria communities of different tomato genotypes exhibited distinct interaction networks, likely resulting from genotype-specific root exudates that alter microbial resource competition [90].
Although Fol inoculation did not significantly affect α-diversity of the tomato rhizobacterial community, certain specific bacterial taxa showed increased abundance, such as Sphingomonas, Bacillus, and Pseudomonas. This observation aligns with previous studies showing that plants under stress employ a “cry-for-help” strategy to recruit beneficial microbes to cope with adverse conditions [91,92]. Wen et al. reported that after inoculation with Fusarium oxysporum f. sp. cucumerinum, cucumber recruits Sphingomonas and Bacillus to protect against pathogen infection [93]. Our previous study also demonstrated that disease-resistant cabbage varieties recruit Pseudomonas under Fusarium oxysporum f. sp. conglutinans infection to enhance pathogen resistance [24]. Moreover, Fol inoculation only caused minor changes in rhizobacterial functional composition and network structure, likely due to the selective recruitment of disease-suppressive microbes induced by Fol [94]. Notably, following Fol inoculation, the network hub zOTUs shifted from zOTU_28 (Dyadobacter) to zOTU_107 (Sphingomonas) and zOTU_156 (Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium), both of which are potential beneficial bacteria [95,96]. These results indicate that plants can maintain rhizosphere health under pathogen stress by selectively recruiting beneficial microbes.
5. Conclusions
This study systematically examined the effects of soil type, tomato genotype, and pathogen invasion on the assembly of tomato rhizobacterial communities. Our results indicate that soil type is the primary determinant shaping both the structure and function of rhizobacterial communities, exerting a stronger influence than genotype or pathogen invasion. Further analyses revealed that soil physicochemical properties, particularly TN, pH, and SOM, are key abiotic drivers underlying differences in microbial community composition and metabolic potential. Meanwhile, tomato genotype and pathogen invasion, as secondary factors, also significantly influenced community assembly and ecological function. These findings not only advance our understanding of plant–soil–microbe interactions but also provide a theoretical basis for optimizing rhizosphere microbiomes through soil management and genotype selection to enhance crop health and promote sustainable agriculture.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy15112517/s1, Figure S1: Alpha rarefaction curves for bacteria were plotted at the species level using the “observed zOTU richness”; Figure S2: Random forest evaluation of the relative importance of soil type, plant genotype, and Fusarium oxysporum f. sp. lycopersici inoculation in shaping the tomato rhizosphere bacterial community; Figure S3: Differences in functional composition among different microhabitats for the three soil types, two tomato genotypes, and Fol pathogen inoculation versus control treatments; Figure S4: Effect of soil type on copy numbers of denitrification genes (nirS, nirK, nosZ) and the aromatic-compound degradation gene c230; Figure S5: Node categorization for the identification of potential keystone taxa in rhizosphere networks; Table S1: Primer sequences utilized in this study; Table S2: Genes standard curve lines for qPCR; Table S3 zOTU representative sequences, taxonomy annotation and zOTU table; Table S4: Alpha diversity estimators of samples; Table S5: PERMANOVA analysis using the Adonis function was performed on all bacterial 16S samples; Table S6: Differential abundance analysis of bacterial taxa; Table S7: Topology properties of the co-occurrence networks; Table S8: The physicochemical properties of the GZ, HEB and HZ soils.
Author Contributions
Conceptualization, X.L. and B.X.; methodology, X.L., X.P. and J.Z.; investigation, X.P., C.P., Y.W., M.X., J.L., J.Z. and Y.L.; data curation, X.L., X.P. and C.P.; formal analysis, C.P. and J.L.; software, Y.W. and Y.L.; validation, Y.W., J.L., J.Z., Y.L. and Z.M.; resources, Z.M.; writing—original draft preparation, X.P.; writing—review and editing, X.P. and X.L.; supervision, X.L., B.X. and Z.M.; funding acquisition, X.L., B.X. and M.X. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by the Sanya Science and Technology Innovation Project (2022KJCX99), the Startup Fund from the Nankai University to XZL (030/C029215002), and the National Key R&D program of China (2022YFD1400700).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw sequencing data have been submitted to the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/, accessed on 10 July 2025) under the accession number PRJNA1196329.
Conflicts of Interest
The authors declare that they have no competing interest.
References
- Bano, S.; Wu, X.; Zhang, X. Towards sustainable agriculture: Rhizosphere microbiome engineering. Appl. Microbiol. Biotechnol. 2021, 105, 7141–7160. [Google Scholar] [CrossRef] [PubMed]
- Xun, W.; Liu, Y.; Ma, A.; Yan, H.; Miao, Y.; Shao, J.; Zhang, N.; Xu, Z.; Shen, Q.; Zhang, R. Dissection of rhizosphere microbiome and exploiting strategies for sustainable agriculture. New Phytol. 2024, 242, 2401–2410. [Google Scholar] [CrossRef]
- Shameem M, R.; Sonali J, M.I.; Kumar, P.S.; Rangasamy, G.; Gayathri, K.V.; Parthasarathy, V. Rhizobium mayense sp. Nov., an efficient plant growth-promoting nitrogen-fixing bacteria isolated from rhizosphere soil. Environ. Res. 2023, 220, 115200. [Google Scholar] [CrossRef]
- Valencia-Marin, M.F.; Chávez-Avila, S.; Guzmán-Guzmán, P.; Orozco-Mosqueda, M.d.C.; de los Santos-Villalobos, S.; Glick, B.R.; Santoyo, G. Survival strategies of Bacillus spp. in saline soils: Key factors to promote plant growth and health. Biotechnol. Adv. 2024, 70, 108303. [Google Scholar] [CrossRef]
- Hu, L.; Robert, C.A.M.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.; Manzo, D.; Chervet, N.; Steinger, T.; van der Heijden, M.G.A.; et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 2018, 9, 2738. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, J.; Khashi u Rahman, M.; Gao, D.; Wei, Z.; Wu, F.; Dini-Andreote, F. Interspecific plant interaction via root exudates structures the disease suppressiveness of rhizosphere microbiomes. Mol. Plant 2023, 16, 849–864. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- de la Fuente Cantó, C.; Simonin, M.; King, E.; Moulin, L.; Bennett, M.J.; Castrillo, G.; Laplaze, L. An extended root phenotype: The rhizosphere, its formation and impacts on plant fitness. Plant J. 2020, 103, 951–964. [Google Scholar] [CrossRef]
- Pérez-Jaramillo, J.E.; Carrión, V.J.; Bosse, M.; Ferrão, L.F.V.; de Hollander, M.; Garcia, A.A.F.; Ramírez, C.A.; Mendes, R.; Raaijmakers, J.M. Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J. 2017, 11, 2244–2257. [Google Scholar] [CrossRef] [PubMed]
- de Faria, M.R.; Costa, L.S.A.S.; Chiaramonte, J.B.; Bettiol, W.; Mendes, R. The rhizosphere microbiome: Functions, dynamics, and role in plant protection. Trop. Plant Pathol. 2021, 46, 13–25. [Google Scholar] [CrossRef]
- Pantigoso, H.A.; Newberger, D.; Vivanco, J.M. The rhizosphere microbiome: Plant–microbial interactions for resource acquisition. J. Appl. Microbiol. 2022, 133, 2864–2876. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Van Der Heijden, M.G.A.; Bardgett, R.D.; Van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Lorente, A.I.; Romero, D.; Molina-Santiago, C. Unravelling the impact of environmental factors in shaping plant microbiomes. Microb. Biotechnol. 2024, 17, e14504. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Li, C.; Ahmed, W.; Li, D.; Yu, L.; Xu, L.; Xu, T.; Zhao, Z. Biochar suppresses bacterial wilt disease of flue-cured tobacco by improving soil health and functional diversity of rhizosphere microorganisms. Appl. Soil Ecol. 2022, 171, 104314. [Google Scholar] [CrossRef]
- Feng, C.; Yi, Z.; Qian, W.; Liu, H.; Jiang, X. Rotations improve the diversity of rhizosphere soil bacterial communities, enzyme activities and tomato yield. PLoS ONE 2023, 18, e0270944. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Chenu, C.; Kappler, A.; Rillig, M.C.; Fierer, N. The interplay between microbial communities and soil properties. Nat. Rev. Microbiol. 2024, 22, 226–239. [Google Scholar] [CrossRef]
- Sokol, N.W.; Slessarev, E.; Marschmann, G.L.; Nicolas, A.; Blazewicz, S.J.; Brodie, E.L.; Firestone, M.K.; Foley, M.M.; Hestrin, R.; Hungate, B.A.; et al. Life and death in the soil microbiome: How ecological processes influence biogeochemistry. Nat. Rev. Microbiol. 2022, 20, 415–430. [Google Scholar] [CrossRef] [PubMed]
- Xiong, R.; He, X.; Gao, N.; Li, Q.; Qiu, Z.; Hou, Y.; Shen, W. Soil pH amendment alters the abundance, diversity, and composition of microbial communities in two contrasting agricultural soils. Microbiol. Spectr. 2024, 12, e04165-23. [Google Scholar] [CrossRef]
- Paudel, D.; Wang, L.; Poudel, R.; Acharya, J.P.; Victores, S.; de Souza, C.H.L.; Rios, E.; Wang, J. Elucidating the effects of organic vs. conventional cropping practice and rhizobia inoculation on rhizosphere microbial diversity and yield of peanut. Environ. Microbiome 2023, 18, 60. [Google Scholar] [CrossRef]
- Brown, S.P.; Grillo, M.A.; Podowski, J.C.; Heath, K.D. Soil origin and plant genotype structure distinct microbiome compartments in the model legume Medicago truncatula. Microbiome 2020, 8, 139. [Google Scholar] [CrossRef]
- Bziuk, N.; Maccario, L.; Straube, B.; Wehner, G.; Sørensen, S.J.; Schikora, A.; Smalla, K. The treasure inside barley seeds: Microbial diversity and plant beneficial bacteria. Environ. Microbiome 2021, 16, 20. [Google Scholar] [CrossRef]
- Ping, X.; Khan, R.A.A.; Chen, S.; Jiao, Y.; Zhuang, X.; Jiang, L.; Song, L.; Yang, Y.; Zhao, J.; Li, Y.; et al. Deciphering the role of rhizosphere microbiota in modulating disease resistance in cabbage varieties. Microbiome 2024, 12, 160. [Google Scholar] [CrossRef] [PubMed]
- Stopnisek, N.; Shade, A. Persistent microbiome members in the common bean rhizosphere: An integrated analysis of space, time, and plant genotype. ISME J. 2021, 15, 2708–2722. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Jia, H.; Ran, L.; Wu, F.; Liu, J.; Schlaeppi, K.; Dini-Andreote, F.; Wei, Z.; Zhou, X. Fusaric acid mediates the assembly of disease-suppressive rhizosphere microbiota via induced shifts in plant root exudates. Nat. Commun. 2024, 15, 5125. [Google Scholar] [CrossRef] [PubMed]
- Pacheco-Moreno, A.; Bollmann-Giolai, A.; Chandra, G.; Brett, P.; Davies, J.; Thornton, O.; Poole, P.; Ramachandran, V.; Brown, J.K.M.; Nicholson, P.; et al. The genotype of barley cultivars influences multiple aspects of their associated microbiota via differential root exudate secretion. PLoS Biol. 2024, 22, e3002232. [Google Scholar] [CrossRef]
- Yang, K.; Fu, R.; Feng, H.; Jiang, G.; Finkel, O.; Sun, T.; Liu, M.; Huang, B.; Li, S.; Wang, X.; et al. RIN enhances plant disease resistance via root exudate-mediated assembly of disease-suppressive rhizosphere microbiota. Mol. Plant 2023, 16, 1379–1395. [Google Scholar] [CrossRef]
- Wu, J.; Hu, S.; Chen, J.; Zhou, L.; Yang, S.; Zhou, N.; Wu, L.; Niu, G.; Zhang, Y.; Ren, X.; et al. Soil microbial legacy mediated by buckwheat flavonoids enhances cabbage resistance to clubroot disease. Microbiome 2025, 13, 176. [Google Scholar] [CrossRef]
- Tzipilevich, E.; Russ, D.; Dangl, J.L.; Benfey, P.N. Plant immune system activation is necessary for efficient root colonization by auxin-secreting beneficial bacteria. Cell Host Microbe 2021, 29, 1507–1520.e1504. [Google Scholar] [CrossRef]
- Liu, Y.; Wilson, A.J.; Han, J.; Hui, A.; O’Sullivan, L.; Huan, T.; Haney, C.H. Amino acid availability determines plant immune homeostasis in the rhizosphere microbiome. mBio 2023, 14. [Google Scholar] [CrossRef]
- Quiza, L.; Tremblay, J.; Pagé, A.P.; Greer, C.W.; Pozniak, C.J.; Li, R.; Haug, B.; Hemmingsen, S.M.; St-Arnaud, M.; Yergeau, E. The effect of wheat genotype on the microbiome is more evident in roots and varies through time. ISME Commun. 2023, 3, 32. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, J.; Carvalhais, L.C.; Percy, C.D.; Prakash Verma, J.; Schenk, P.M.; Singh, B.K. Evidence for the plant recruitment of beneficial microbes to suppress soil-borne pathogens. New Phytol. 2021, 229, 2873–2885. [Google Scholar] [CrossRef]
- Galindo-Castañeda, T.; Hartmann, M.; Lynch, J.P. Location: Root architecture structures rhizosphere microbial associations. J. Exp. Bot. 2024, 75, 594–604. [Google Scholar] [CrossRef]
- Chapelle, E.; Mendes, R.; Bakker, P.A.H.M.; Raaijmakers, J.M. Fungal invasion of the rhizosphere microbiome. ISME J. 2016, 10, 265–268. [Google Scholar] [CrossRef]
- Fernández-González, A.J.; Cardoni, M.; Gómez-Lama Cabanás, C.; Valverde-Corredor, A.; Villadas, P.J.; Fernández-López, M.; Mercado-Blanco, J. Linking belowground microbial network changes to different tolerance level towards Verticillium wilt of olive. Microbiome 2020, 8, 11. [Google Scholar] [CrossRef]
- Liu, X.; Ao, K.; Yao, J.; Zhang, Y.; Li, X. Engineering plant disease resistance against biotrophic pathogens. Curr. Opin. Plant Biol. 2021, 60, 101987. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Y.-F.; Cui, H.-L.; Zhou, R.; Li, L.; Duan, G.-L.; Zhu, Y.-G. Distinctive structure and assembly of phyllosphere microbial communities between wild and cultivated rice. Microbiol. Spectr. 2023, 11, e04371-22. [Google Scholar] [CrossRef]
- Carrión, V.J.; Perez-Jaramillo, J.; Cordovez, V.; Tracanna, V.; de Hollander, M.; Ruiz-Buck, D.; Mendes, L.W.; van Ijcken, W.F.J.; Gomez-Exposito, R.; Elsayed, S.S.; et al. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 2019, 366, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Vismans, G.; Yu, K.; Song, Y.; de Jonge, R.; Burgman, W.P.; Burmølle, M.; Herschend, J.; Bakker, P.A.H.M.; Pieterse, C.M.J. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018, 12, 1496–1507. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Xiong, C.; Gao, C.; Tsui, C.K.M.; Wang, M.-M.; Zhou, X.; Zhang, A.-M.; Cai, L. Disease-induced changes in plant microbiome assembly and functional adaptation. Microbiome 2021, 9, 187. [Google Scholar] [CrossRef]
- Jin, X.; Bai, Y.; Khashi u Rahman, M.; Kang, X.; Pan, K.; Wu, F.; Pommier, T.; Zhou, X.; Wei, Z. Biochar stimulates tomato roots to recruit a bacterial assemblage contributing to disease resistance against Fusarium wilt. iMeta 2022, 1, e37. [Google Scholar] [CrossRef]
- Wei, Z.; Yang, X.; Yin, S.; Shen, Q.; Ran, W.; Xu, Y. Efficacy of Bacillus-fortified organic fertiliser in controlling bacterial wilt of tomato in the field. Appl. Soil Ecol. 2011, 48, 152–159. [Google Scholar] [CrossRef]
- Ferreira, A.C.C.; Leite, L.F.C.; de Araújo, A.S.F.; Eisenhauer, N. Land-use type effects on soil organic carbon and microbial properties in a semi-arid region of northeast brazil. Land Degrad. Dev. 2016, 27, 171–178. [Google Scholar] [CrossRef]
- Liu, Y.-X.; Chen, L.; Ma, T.; Li, X.; Zheng, M.; Zhou, X.; Chen, L.; Qian, X.; Xi, J.; Lu, H.; et al. EasyAmplicon: An easy-to-use, open-source, reproducible, and community-based pipeline for amplicon data analysis in microbiome research. iMeta 2023, 2, e83. [Google Scholar] [CrossRef]
- Yousuf, S.; Luo, H.; Zeng, M.; Chen, L.; Ma, T.; Li, X.; Zheng, M.; Zhou, X.; Chen, L.; Xi, J.; et al. Unveiling microbial communities with EasyAmplicon: A user-centric guide to perform amplicon sequencing data analysis. iMetaOmics 2024, 1, e42. [Google Scholar] [CrossRef]
- Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta 2023, 2, e107. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, R.B.; Simpson, G.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community ecology package. R package version. 2.0-10. J. Stat. Softw. 2013, 48, 1–21. [Google Scholar]
- Chen, T.; Liu, Y.-X.; Chen, T.; Yang, M.; Fan, S.; Shi, M.; Wei, B.; Lv, H.; Cao, W.; Wang, C.; et al. ImageGP 2 for enhanced data visualization and reproducible analysis in biomedical research. iMeta 2024, 3, e239. [Google Scholar] [CrossRef] [PubMed]
- Ginestet, C. ggplot2: Elegant graphics for data analysis. J. R. Stat. Soc. Ser. A Stat. Soc. 2011, 174, 245–246. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Li, C.; Gillings, M.R.; Zhang, C.; Chen, Q.; Zhu, D.; Wang, J.; Zhao, K.; Xu, Q.; Leung, P.H.; Li, X.; et al. Ecology and risks of the global plastisphere as a newly expanding microbial habitat. Innov. 2024, 5, 100543. [Google Scholar] [CrossRef]
- Michotey, V.; Méjean, V.; Bonin, P. Comparison of methods for quantification of cytochrome cd1-denitrifying bacteria in environmental marine samples. Appl. Environ. Microbiol. 2000, 66, 1564–1571. [Google Scholar] [CrossRef]
- Hallin, S.; Lindgren, P.-E. PCR Detection of genes encoding nitrite reductase in denitrifying bacteria. Appl. Environ. Microbiol. 1999, 65, 1652–1657. [Google Scholar] [CrossRef]
- Henry, S.; Bru, D.; Stres, B.; Hallet, S.; Philippot, L. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 2006, 72, 5181–5189. [Google Scholar] [CrossRef] [PubMed]
- Sei, K.; Asano, K.-I.; Tateishi, N.; Mori, K.; Ike, M.; Fujita, M. Design of PCR primers and gene probes for the general detection of bacterial populations capable of degrading aromatic compounds via catechol cleavage pathways. J. Biosci. Bioeng. 1999, 88, 542–550. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, G.; Jiang, S.; Liu, Y.-X. Wekemo Bioincloud: A user-friendly platform for meta-omics data analyses. iMeta 2024, 3, e175. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Peng, X.; Zhang, Z.; Gu, S.; He, Q.; Shen, W.; Wang, Z.; Wang, D.; Hu, Q.; Li, Y.; et al. iNAP: An integrated network analysis pipeline for microbiome studies. iMeta 2022, 1, e13. [Google Scholar] [CrossRef]
- Deng, Y.; Jiang, Y.-H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef] [PubMed]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the International AAAI Conference on Web and Social Media, San Jose, CA, USA, 17–20 May 2009. [Google Scholar]
- Zhang, X.; Ma, Y.-N.; Wang, X.; Liao, K.; He, S.; Zhao, X.; Guo, H.; Zhao, D.; Wei, H.-L. Dynamics of rice microbiomes reveal core vertically transmitted seed endophytes. Microbiome 2022, 10, 216. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Liu, X.; Wang, L.; Khalid, M.; Wang, X.; Jiang, L.; Wang, F.; Pang, Z.; Peng, Y.; Zhao, X. Integrated metagenomic and soil chemical analyses revealed shifts of microbial nutrient cycling with poplar plantation age. Front. Plant Sci. 2025, 16, 1513281. [Google Scholar] [CrossRef]
- Yu, L.; Li, D.; Zhang, Y.; Wang, Y.; Yao, Q.; Yang, K. An optimal combined slow-release nitrogen fertilizer and urea can enhance the decomposition rate of straw and the yield of maize by improving soil bacterial community and structure under full straw returning system. Front. Microbiol. 2024, 15, 1358582. [Google Scholar] [CrossRef]
- Gonçalves, O.S.; Fernandes, A.S.; Tupy, S.M.; Ferreira, T.G.; Almeida, L.N.; Creevey, C.J.; Santana, M.F. Insights into plant interactions and the biogeochemical role of the globally widespread Acidobacteriota phylum. Soil Biol. Biochem. 2024, 192, 109369. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef]
- Bonito, G.; Benucci, G.M.N.; Hameed, K.; Weighill, D.; Jones, P.; Chen, K.-H.; Jacobson, D.; Schadt, C.; Vilgalys, R. Fungal-bacterial networks in the populus rhizobiome are impacted by soil properties and host genotype. Front. Microbiol. 2019, 10, 481. [Google Scholar] [CrossRef]
- Zhang, G.; Chu, X.; Zhu, H.; Zou, D.; Li, L.; Du, L. The response of soil nutrients and microbial community structures in long-term tea plantations and diverse agroforestry intercropping systems. Sustainability 2021, 13, 7799. [Google Scholar] [CrossRef]
- Wang, X.; Riaz, M.; Babar, S.; Eldesouki, Z.; Liu, B.; Xia, H.; Li, Y.; Wang, J.; Xia, X.; Jiang, C. Alterations in the composition and metabolite profiles of the saline-alkali soil microbial community through biochar application. J. Environ. Manag. 2024, 352, 120033. [Google Scholar] [CrossRef]
- Xu, P.; Gao, M.; Li, Y.; Ye, J.; Su, J.; Li, H. Combined effects of acidification and warming on soil denitrification and microbial community. Front. Microbiol. 2025, 16, 1572497. [Google Scholar] [CrossRef]
- Tang, Z.; Zhang, L.; He, N.; Liu, Z.; Ma, Z.; Fu, L.; Wang, H.; Wang, C.; Sui, G.; Zheng, W. Influence of planting methods and organic amendments on rice yield and bacterial communities in the rhizosphere soil. Front. Microbiol. 2022, 13, 918986. [Google Scholar] [CrossRef]
- Li, L.; Hu, Z.; Tan, G.; Fan, J.; Chen, Y.; Xiao, Y.; Wu, S.; Zhi, Q.; Liu, T.; Yin, H.; et al. Enhancing plant growth in biofertilizer-amended soil through nitrogen-transforming microbial communities. Front. Plant Sci. 2023, 14, 1259853. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, Q.; Kang, J.; Xu, N.; Zhang, Z.; Deng, Y.; Gillings, M.; Lu, T.; Qian, H. Effects of organic fertilizers on plant growth and the rhizosphere microbiome. Appl. Environ. Microbiol. 2024, 90, e01719–e01723. [Google Scholar] [CrossRef]
- Ikeda, S.; Okazaki, K.; Takahashi, H.; Tsurumaru, H.; Minamisawa, K. Seasonal shifts in bacterial community structures in the lateral root of sugar beet grown in an andosol field in japan. Microbes Environ. 2023, 38, ME22071. [Google Scholar] [CrossRef] [PubMed]
- Shukla, V.; Shaikh, N. Study of the effect of consortia of PGPR on the growth of Trichosanthes cucumerina in a hydroponic system. J. Appl. Hortic. 2023, 25, 112–116. [Google Scholar] [CrossRef]
- Guo, S.; Geisen, S.; Mo, Y.; Yan, X.; Huang, R.; Liu, H.; Gao, Z.; Tao, C.; Deng, X.; Xiong, W.; et al. Predatory protists impact plant performance by promoting plant growth-promoting rhizobacterial consortia. ISME J. 2024, 18, wrae180. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.; Sun, L.; Ding, X.; Sun, D.; Liu, J.; Wang, W. Complete genome sequence of Caulobacter flavus RHGG3T, a type species of the genus Caulobacter with plant growth-promoting traits and heavy metal resistance. 3 Biotech 2019, 9, 42. [Google Scholar] [CrossRef]
- Kwak, M.-J.; Kong, H.G.; Choi, K.; Kwon, S.-K.; Song, J.Y.; Lee, J.; Lee, P.A.; Choi, S.Y.; Seo, M.; Lee, H.J.; et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 2018, 36, 1100–1109. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, H.; Liu, H.; Wu, C.; Wan, Y.; Zhu, L.; Yang, J.; Cai, P.; Chen, J.; Ge, T. Combating wheat yellow mosaic virus through microbial interactions and hormone pathway modulations. Microbiome 2024, 12, 200. [Google Scholar] [CrossRef]
- Lv, N.; Tao, C.; Ou, Y.; Wang, J.; Deng, X.; Liu, H.; Shen, Z.; Li, R.; Shen, Q. Root-associated antagonistic Pseudomonas spp. contribute to soil suppressiveness against banana Fusarium wilt disease of banana. Microbiol. Spectr. 2023, 11, e03525-22. [Google Scholar] [CrossRef]
- Yue, H.; Sun, X.; Wang, T.; Zhang, A.; Han, D.; Wei, G.; Song, W.; Shu, D. Host genotype-specific rhizosphere fungus enhances drought resistance in wheat. Microbiome 2024, 12, 44. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Jaramillo, J.E.; de Hollander, M.; Ramírez, C.A.; Mendes, R.; Raaijmakers, J.M.; Carrión, V.J. Deciphering rhizosphere microbiome assembly of wild and modern common bean (Phaseolus vulgaris) in native and agricultural soils from Colombia. Microbiome 2019, 7, 114. [Google Scholar] [CrossRef]
- Lopes Lucas, D.; Wang, P.; Futrell Stephanie, L.; Schachtman Daniel, P. Sugars and jasmonic acid concentration in root exudates affect maize rhizosphere bacterial communities. Appl. Environ. Microbiol. 2022, 88, e00971-22. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Paulitz, T.C.; Steinberg, C.; Alabouvette, C.; Moënne-Loccoz, Y. The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 2009, 321, 341–361. [Google Scholar] [CrossRef]
- Kelly, C.; Haddix, M.; Byrne, P.; Cotrufo, M.F.; Schipanski, M.; Kallenbach, C.; Wallenstein, M.; Fonte, S. Long-term compost amendment modulates wheat genotype differences in belowground carbon allocation, microbial rhizosphere recruitment and nitrogen acquisition. Soil Biol. Biochem. 2022, 172, 108768. [Google Scholar] [CrossRef]
- Hu, S.; He, R.; Wang, W.; Zhao, D.; Zeng, J.; Huang, R.; Duan, M.; Yu, Z. Composition and co-occurrence patterns of Phragmites australis rhizosphere bacterial community. Aquat. Ecol. 2021, 55, 695–710. [Google Scholar] [CrossRef]
- Wang, Z.; Song, Y. Toward understanding the genetic bases underlying plant-mediated “cry for help” to the microbiota. iMeta 2022, 1, e8. [Google Scholar] [CrossRef]
- Xuan, P.; Ma, H.; Deng, X.; Li, Y.; Tian, J.; Li, J.; Ma, E.; Xu, Z.; Xiao, D.; Bezemer, T.M.; et al. Microbiome-mediated alleviation of tobacco replant problem via autotoxin degradation after long-term continuous cropping. iMeta 2024, 3, e189. [Google Scholar] [CrossRef]
- Wen, T.; Ding, Z.; Thomashow, L.S.; Hale, L.; Yang, S.; Xie, P.; Liu, X.; Wang, H.; Shen, Q.; Yuan, J. Deciphering the mechanism of fungal pathogen-induced disease-suppressive soil. New Phytol. 2023, 238, 2634–2650. [Google Scholar] [CrossRef]
- Liu, L.; Huang, X.; Zhang, J.; Cai, Z.; Jiang, K.; Chang, Y. Deciphering the relative importance of soil and plant traits on the development of rhizosphere microbial communities. Soil Biol. Biochem. 2020, 148, 107909. [Google Scholar] [CrossRef]
- Zhang, W.; Mao, G.; Zhuang, J.; Yang, H. The co-inoculation of Pseudomonas chlororaphis H1 and Bacillus altitudinis Y1 promoted soybean [Glycine max (L.) Merrill] growth and increased the relative abundance of beneficial microorganisms in rhizosphere and root. Front. Microbiol. 2023, 13, 1079348. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Zhang, D.; Huang, B.; Song, Z.; Ren, L.; Hao, B.; Liu, J.; Zhu, J.; Fang, W.; Yan, D.; et al. Organic fertilizer improves soil fertility and restores the bacterial community after 1,3-dichloropropene fumigation. Sci. Total Environ. 2020, 738, 140345. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).