Sugarcane–Peanut Intercropping Enhances Farmland Productivity: A Multi-Omics Investigation into the Coordination of Zinc Homeostasis and Hormonal Signaling
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Determination of Heavy Metal Content in the Roots, Stems, and Leaves of Sugarcane
2.3. Determination of Metabolites and Endogenous Hormones Concentrations in the Roots, Stems, and Leaves of Sugarcane
2.4. Total RNA Extraction, Transcriptome Library Construction, Sequencing and Functional Annotation
2.5. Gene Expression Quantification and Differential Expression Analysis
2.6. GO Functional and KEGG Pathway Enrichment Analysis
2.7. Weighted Gene Co-Expression Network Analysis (WGCNA)
2.8. Verification of Zinc Pathways
2.9. Measurement of Sugarcane Agronomic Traits in Roots, Stems, and Leaves
3. Results
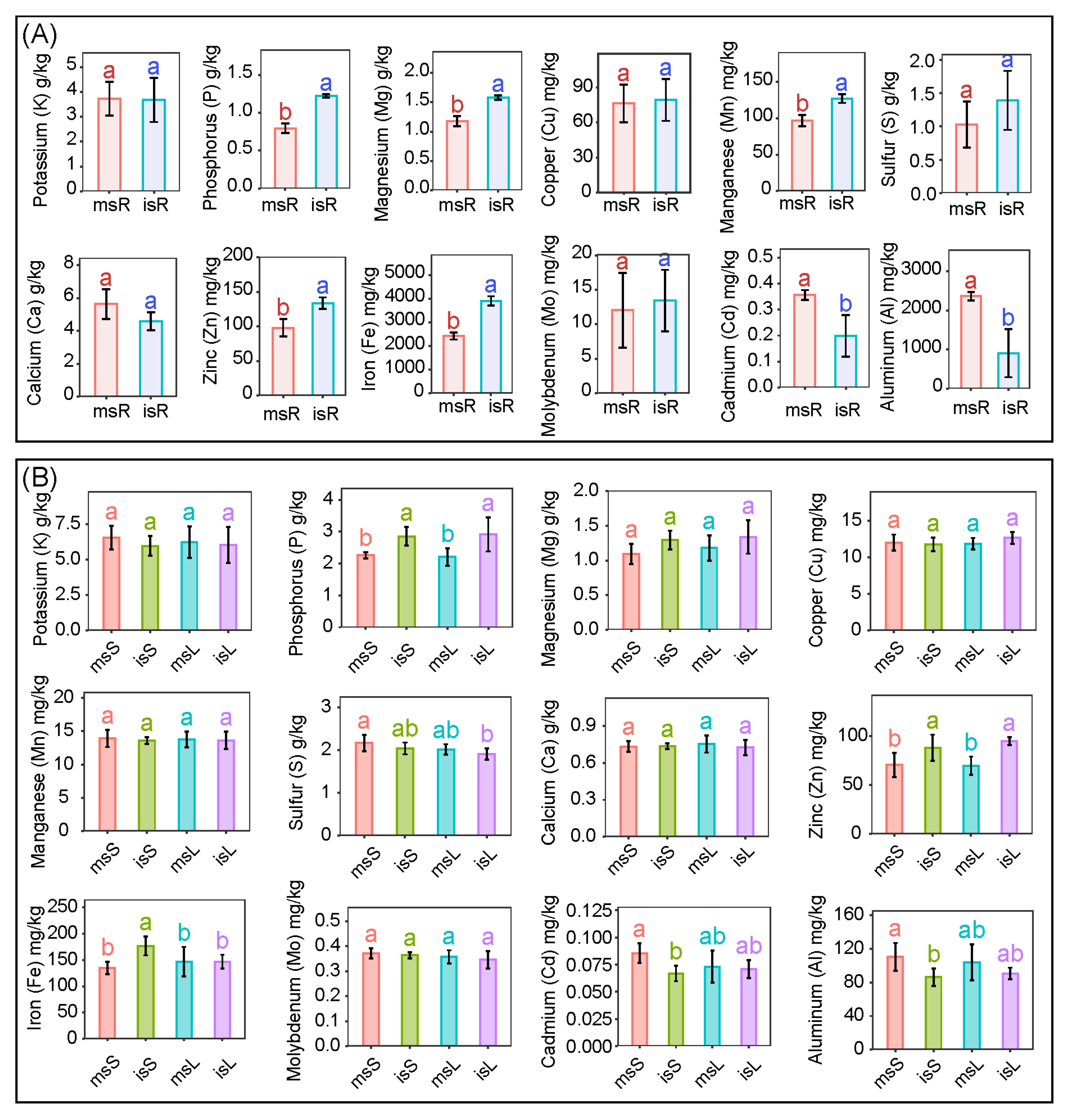
3.1. Accumulation of Metals Across Sugarcane Tissues Under Varied Cultivation Conditions
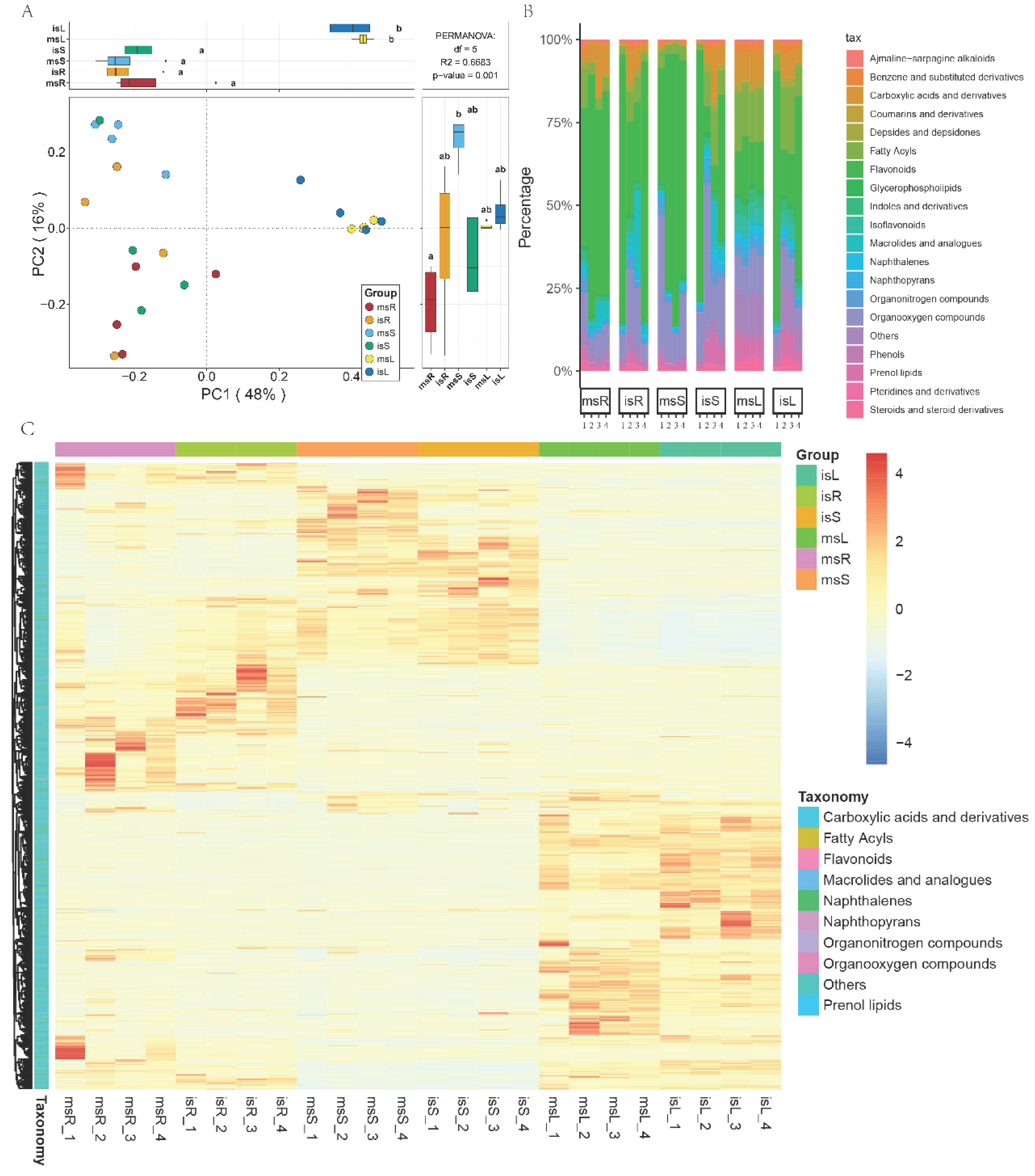
3.2. Analysis of Endogenous Metabolites in the Root, Stem, and Leaf of Sugarcane
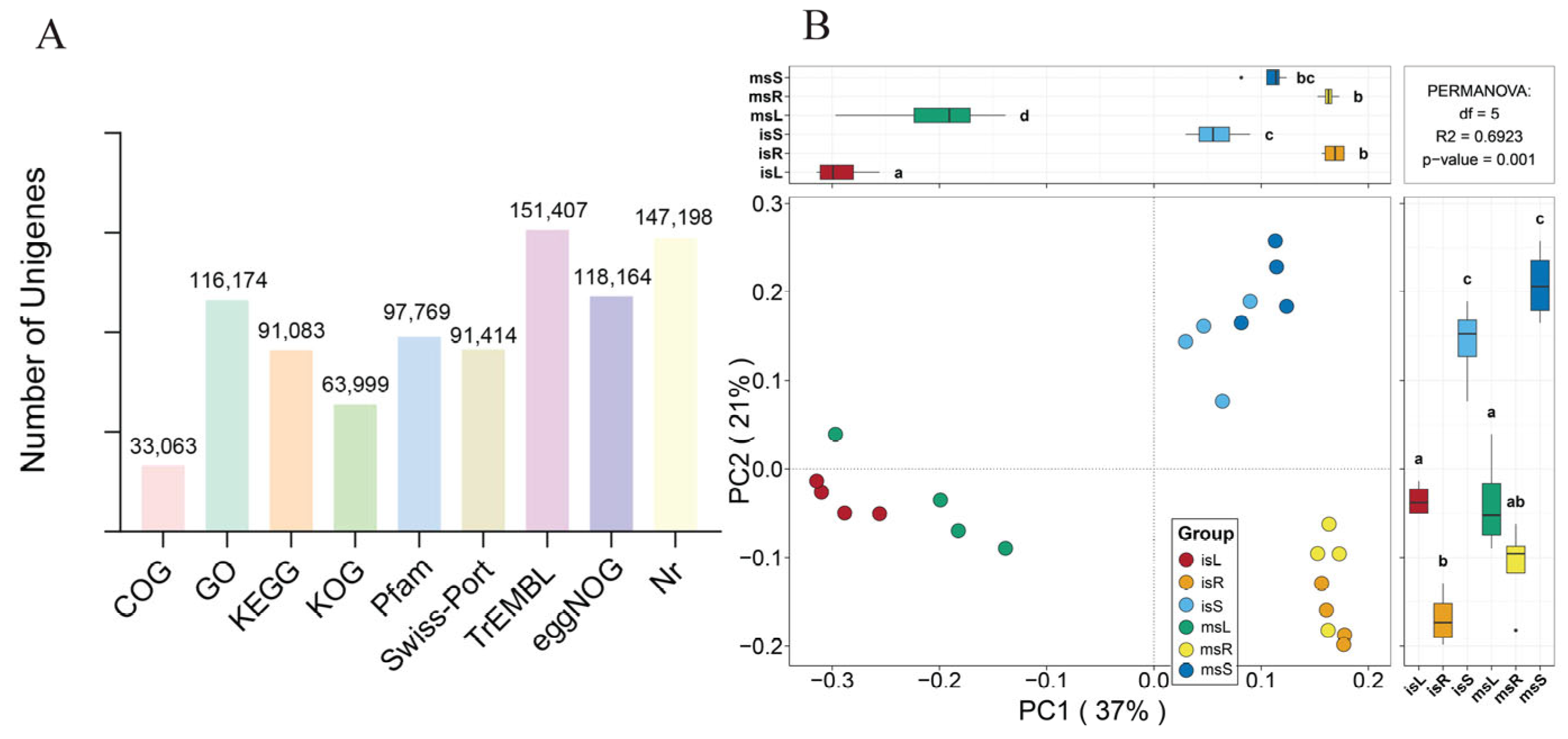
3.3. Transcriptome Sequencing and Assembly Analysis
3.4. Functional and Pathway Annotation of Genes
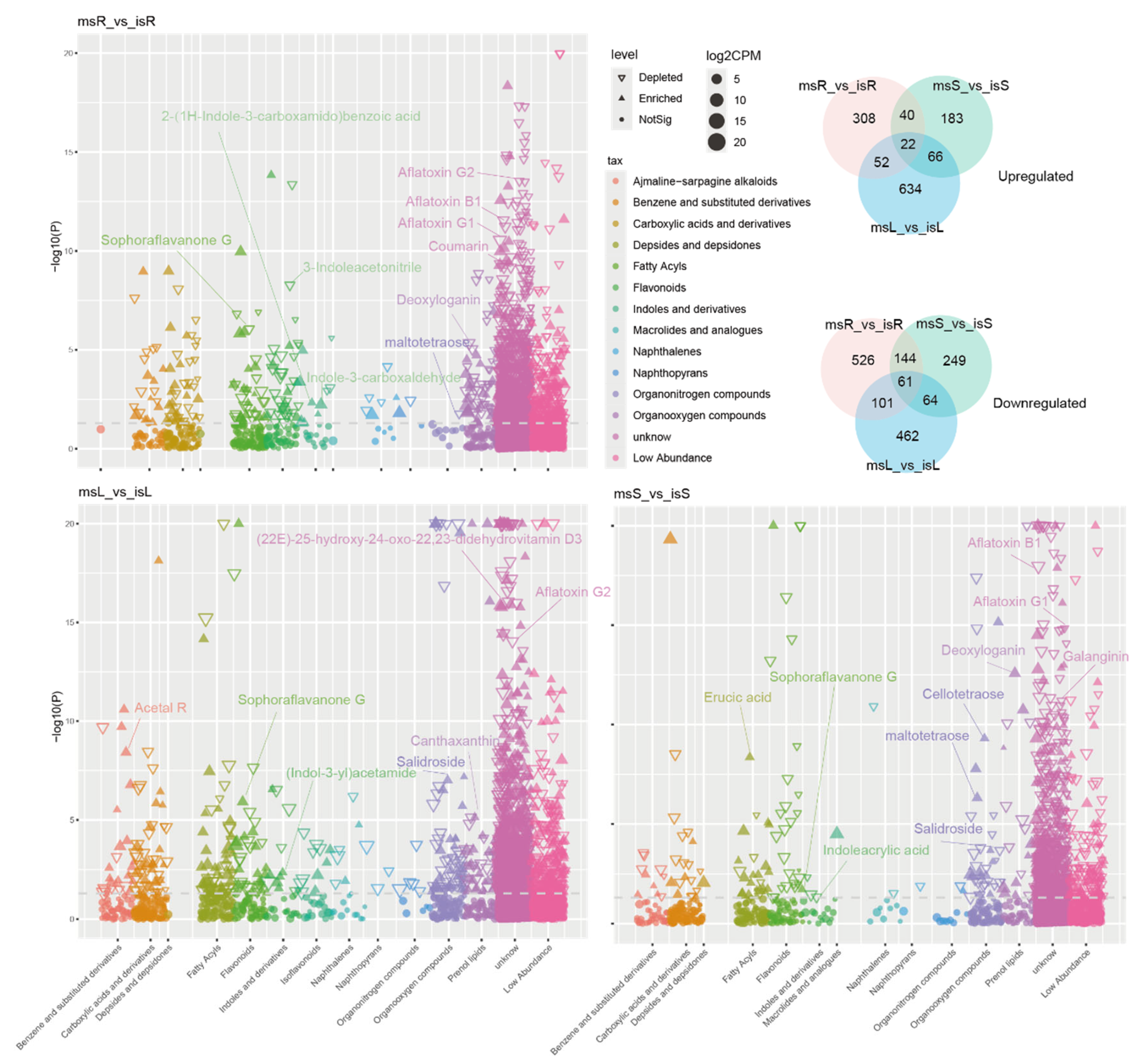
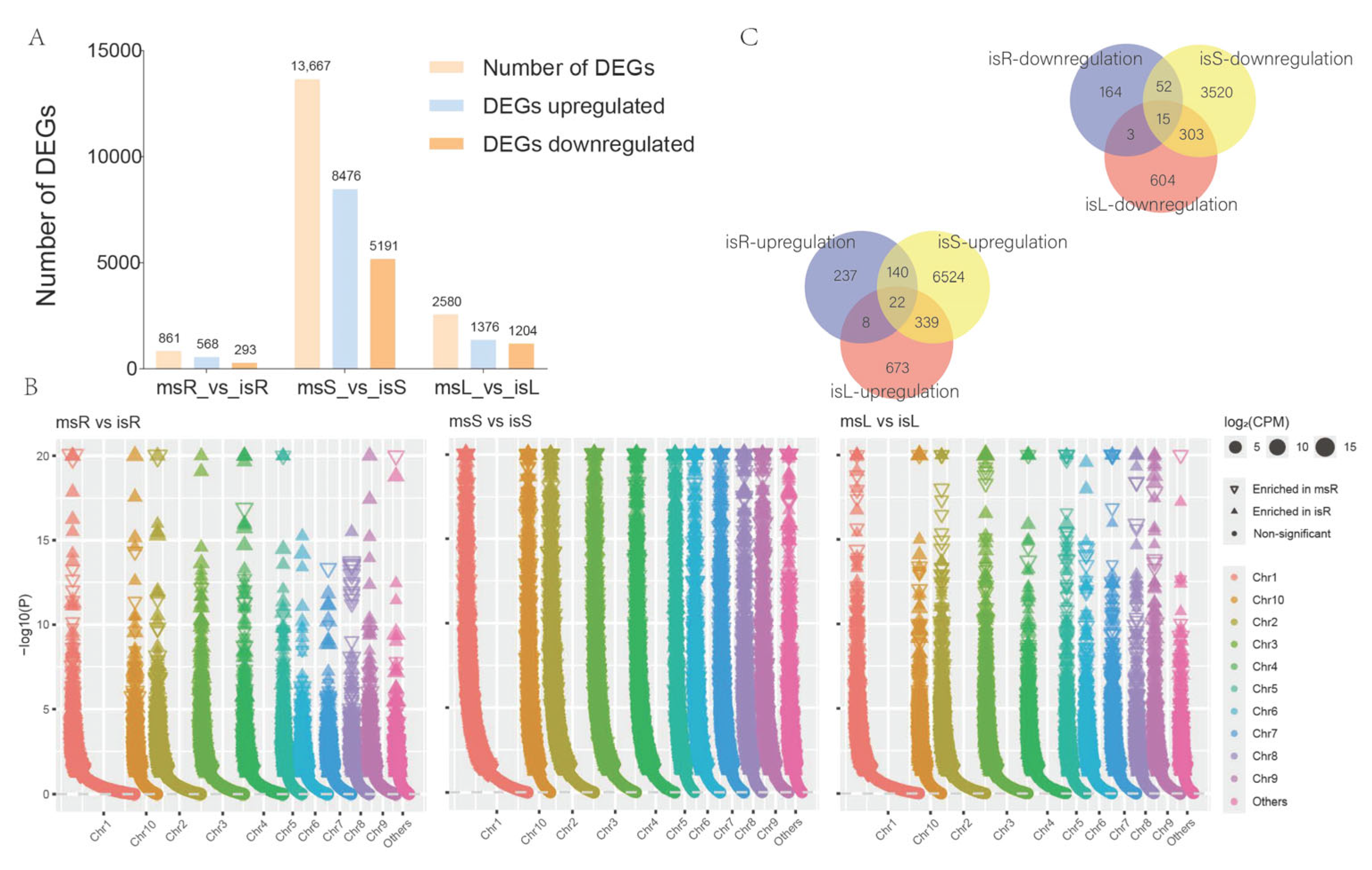
3.5. DEGs in Sugarcane Samples Under Different Cultivation Conditions
3.6. GO Functional Annotation and Enrichment Analysis of DEGs
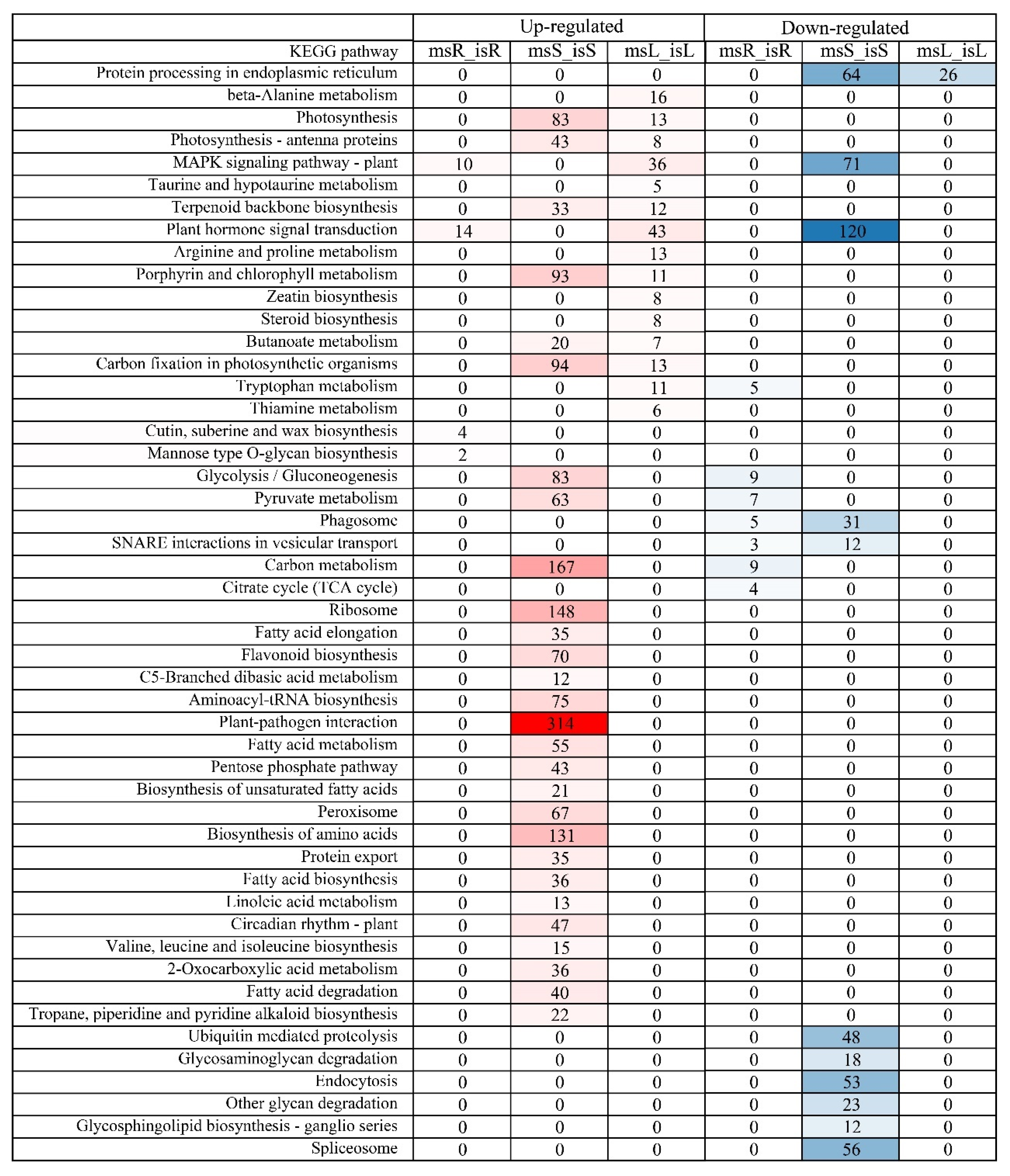
3.7. KEGG Pathway Enrichment Analysis
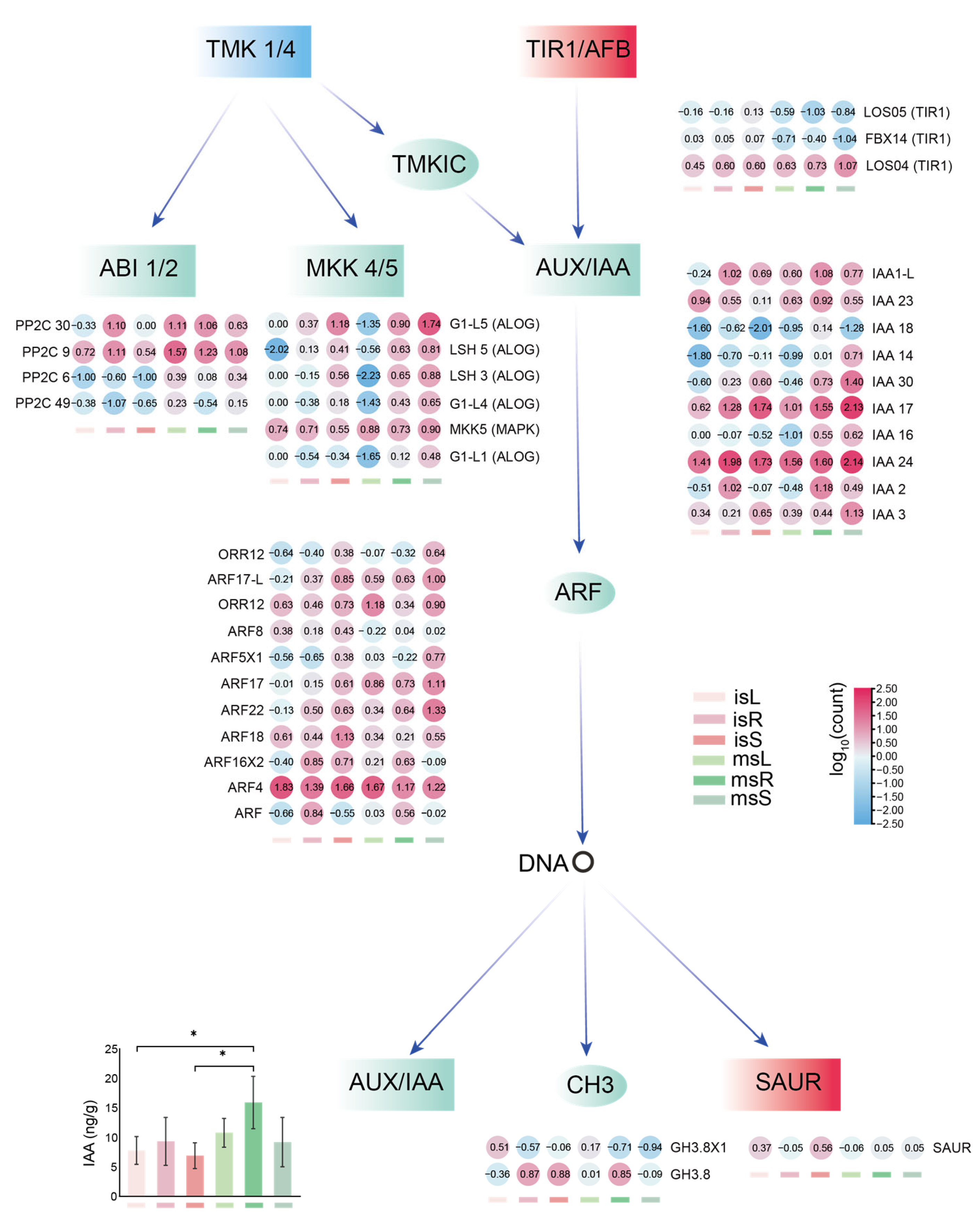
3.8. The Responses of Key Genes to Metal Transport and Hormone Signal Transduction Processes
3.9. Key Genes Involved in Plant Hormone Signal Transduction
3.10. Module Identification and Functional Enrichment Analysis in Weighted Gene Co-Expression Network Analysis
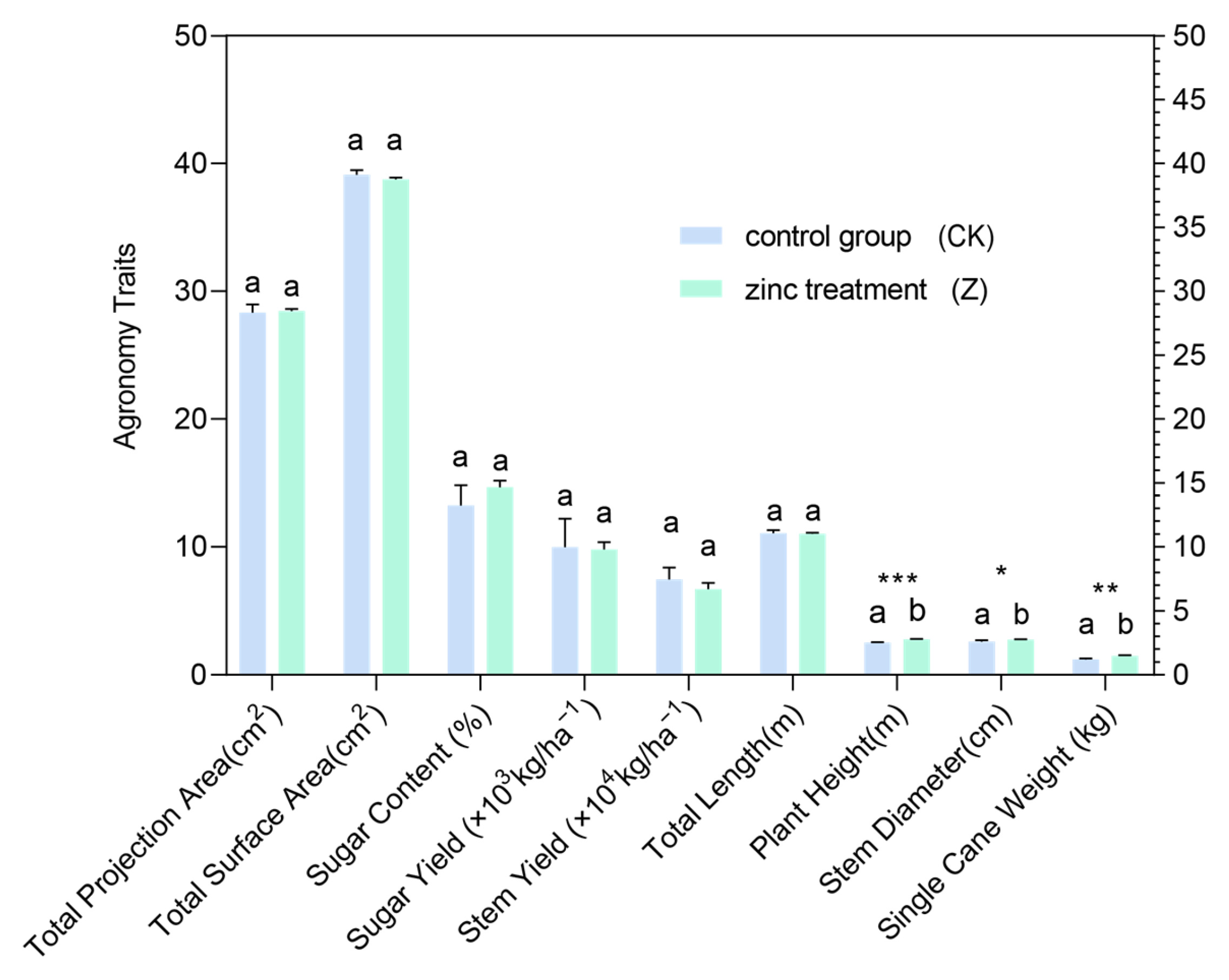
3.11. Zinc Regulates Agronomic Traits and Endogenous Hormone Levels in Sugarcane: An Integrative Assessment
4. Discussion
4.1. Intercropping Significantly Influences the Metal Profile, Metabolite Composition, and Gene Expression in Sugarcane
4.2. Strong Linkages Between Rhizosphere Metabolites and Microbiomes
4.3. Regulation of Metal Transport by Transcriptomic and Metabolomic Networks Promotes Plant Growth and Hormonal Homeostasis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, K.; Zhao, M.; Li, Y.; He, Y.; Han, X.; Ma, X.; Ma, F. Spatiotemporal Trends of the Carbon Footprint of Sugar Production in China. Sustain. Prod. Consum. 2024, 46, 502–511. [Google Scholar] [CrossRef]
- Brobbey, M.S.; Louw, J.; Görgens, J.F. Biobased propylene glycol production in a sugarcane biorefinery through lactic acid, glycerol, or sorbitol: A techno-economic and environmental evaluation of intermediates and downstream processing methods. Biochem. Eng. J. 2024, 205, 109292. [Google Scholar] [CrossRef]
- Islam, M.N.; Liza, A.A.; Dey, M.; Das, A.K.; Faruk, M.O.; Khatun, M.L.; Ashaduzzaman, M.; Xi, X. Bio-based composites from bagasse using carbohydrate enriched cross-bonding mechanism: A formaldehyde-free approach. Carbohydr. Polym. Technol. Appl. 2024, 7, 100467. [Google Scholar] [CrossRef]
- Ortiz, I.; Valdez-Vazquez, I.; Hernandez-Vazquez, A.; Olivares-Hernández, R.; Carillo- Reyes, J.; Alatriste-Mondragón, F.; Buitrón, G.; Razo-Flores, E. Process design and economic evaluation for methane and electricity production from Agave bagasse using different pretreatments. Energy Convers. Manag. 2024, 306, 118319. [Google Scholar] [CrossRef]
- Liu, J.; Yu, X.; Ma, J.; Deng, K.; Feng, X.; Zheng, J.; Fan, Y. Effects of replacing corn straw silage with sugarcane tip silage on growth performance, digestion and metabolism and meat quality of Hu sheep. J. Nanjing Agric. Univ. 2024, 47, 351–358. [Google Scholar]
- He, N.; Tian, Z.; Wang, W.; Huan, Q.; Jin, H.; Bai, S. Application of Sugarcane Biomass Carbon in Interface Solar Seawater Desalination. J. S. China Norm. Univ. Nat. Sci. Ed. 2024, 56, 44–52. [Google Scholar]
- Sherpa, S.W.; Ponnuchamy, M.; Kapoor, A.; Jacob, M.M.; Sivaraman, P. Facile removal of sulfamethoxazole antibiotic from contaminated water using bagasse-derived pyrolytic biocarbon: Parametric assessment, mechanistic insights and scale-up analysis. J. Water Process Eng. 2024, 60, 105110. [Google Scholar] [CrossRef]
- Liu, C.; Lu, K.; Wu, Z.; Liu, X.; Grag, A.; Qin, Y.; Mei, G.; Lv, C. Expansive soil improvement using industrial bagasse and low-alkali ecological cement. Constr. Build. Mater. 2024, 423, 135806. [Google Scholar] [CrossRef]
- Arafat, Y.; Shah, A.; Din, I.U.; Jamal, M.; Shah, M.; Kuchkarova, N.; Lin, W.; Lin, S.; Shao, H. Root interactions in wheat/fababean intercropping system enhanced growth performance and yield. Rhizosphere 2025, 33, 101007. [Google Scholar] [CrossRef]
- Sun, Y.; Wu, Z.; Hu, F.; Fan, H.; He, W.; Zhao, L.; Guo, C.; Bao, X.; Chai, Q.; Zhao, C. Maize and Pea Root Interactions Promote Symbiotic Nitrogen Fixation, Thereby Accelerating Nitrogen Assimilation and Partitioning in Intercropped Pea. Agronomy 2025, 15, 1615. [Google Scholar] [CrossRef]
- Xue, Y.; Xia, H.; Christie, P.; Zhang, Z.; Li, L.; Tang, C. Crop acquisition of phosphorus, iron and zinc from soil in cereal/legume intercropping systems: A critical review. Ann. Bot. 2016, 117, 363–377. [Google Scholar] [CrossRef]
- Zhang, L.; Tang, L.; Dong, Y.; Zheng, Y. Nitrogen Absorption of Crops Affected by Root Interaction in Maize and Soybean Intercropping. J. Yunnan Agric. Univ. Nat. Sci. 2016, 31, 1111–1119. [Google Scholar] [CrossRef]
- Lv, T.; Zhan, C.; Pan, Q.; Xu, H.; Fang, H.; Wang, M.; Matsumoto, H. Plant pathogenesis: Toward multidimensional understanding of the microbiome. iMeta 2023, 2, e129. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Song, Y. Toward understanding the genetic bases underlying plant-mediated “cry for help” to the microbiota. iMeta 2022, 1, e8. [Google Scholar] [CrossRef]
- Gao, Y.; Peng, K.; Bai, D.; Bai, X.; Bi, Y.; Chen, A.; Chen, B.; Chen, F.; Chen, J.; Chen, L.; et al. The Microbiome Protocols eBook initiative: Building a bridge to microbiome research. iMeta 2024, 3, e182. [Google Scholar] [CrossRef]
- Fallaj, N.; Pang, Z.; Zhou, Y.; Yuan, Z.; Lin, W. Sugarcane-peanut intercropping stimulatory effects on arbuscular mycorrhizal and entomopathogenic fungi promote soil health and crop productivity. Field Crops Res. 2023, 303, 109110. [Google Scholar] [CrossRef]
- Yuan, Z.; Liu, Q.; Pang, Z.; Fallah, N.; Liu, Y.; Hu, C.; Lin, W. Sugarcane Rhizosphere Bacteria Community Migration Correlates with Growth Stages and Soil Nutrient. Int. J. Mol. Sci. 2022, 23, 10303. [Google Scholar] [CrossRef]
- Singh, S.R.; Yadav, P.; Singh, D.; Shukla, S.K.; Tripathi, M.K.; Bahadur, L.; Mishra, A.; Kumar, S. Intercropping in Sugarcane Improves Functional Diversity, Soil Quality and Crop Productivity. Sugar Tech. 2021, 23, 794–810. [Google Scholar] [CrossRef]
- Cai, D.; Zhao, L.; Ren, Y. Effect of root separation on yield and interspecific competition in maize/soybean intercrops. J. Baoji Univ. Arts Sci. Nat. Sci. 2023, 43, 53–59. [Google Scholar] [CrossRef]
- Martin, L.B.B.; Fei, Z.; Giovannoni, J.J.; Rose, J.K.C. Catalyzing plant science research with RNA-seq. Front. Plant Sci. 2013, 4, 66. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Jiang, T. Transcriptome assembly and isoform expression level estimation from biased RNA-Seq reads. Bioinformatics 2012, 28, 2914–2921. [Google Scholar] [CrossRef]
- Aguirre-Noyola, J.L.; Rosenblueth, M.; Santiago-Martínez, M.G.; Martínez, R.E. Transcriptomic Responses of Rhizobium phaseoli to Root Exudates Reflect Its Capacity to Colonize Maize and Common Bean in an Intercropping System. Front. Microbiol. 2021, 12, 740818. [Google Scholar] [CrossRef]
- Dai, J.; Qiu, W.; Wang, N.; Nakanishi, H.; Zuo, Y. Comparative transcriptomic analysis of the roots of intercropped peanut and maize reveals novel insights into peanut iron nutrition. Plant Physiol. Biochem. 2018, 127, 516–524. [Google Scholar] [CrossRef]
- Ma, C.; Feng, Y.; Wang, J.; Zheng, B.; Wang, X.; Jiao, N. Integrative Physiological, Transcriptome, and Proteome Analyses Provide Insights into the Photosynthetic Changes in Maize in a Maize—Peanut Intercropping System. Plants 2024, 13, 65. [Google Scholar] [CrossRef]
- He, F.; Li, C.; Faisal, S.; Liu, F.; Duan, Y.; Wang, M.; Ruan, J.; Wei, M.; Jiang, H.; Ma, X. Transcriptome Analysis of Amorphophallus konjac with Different Root Barriers. Acta Agric. Boreali-Sin. 2023, 38, 65–74. [Google Scholar]
- Pu, L.; Mei, X.; Zhan, F.; Zu, Y.; Li, Y. Transcriptome Analysis of the Root of Sonchus asper under Cadmium and Lead Stress in the Intercropping System with RNA-Seq. J. Yunnan Agric. Univ. Nat. Sci. 2018, 33, 324–333. [Google Scholar]
- Liu, B.; Li, R.; Xu, W.; Zhang, Y.; Wng, G.; Tao, L.; Zhan, X.; Li, Z. Determination of five endogenous hormones in Zizania caduciflora by ultra-high performance liquid chromatography-tandem mass spectrometry. Agrochemicals 2022, 61, 280–284. [Google Scholar] [CrossRef]
- Xu, Z.; Jin, G.; Wu, T.; Wang, Y.; Chen, Y.; Kan, S. Determination of plant endogenous hormones in Scutellaria by high-performance liquid chromatography-mass spectrometry. Text. Aux. 2023, 40, 51–56. [Google Scholar]
- Yi, Y.; Tang, X.; Lin, X.; Liu, Y.; Lin, L.; Xu, C. Determination of 4 Kind of Plant Hormones in Sargassum by Ultra Performance Liquid Chromatography Tandem Mass Spectrometry. Chin. J. Anal. Chem. 2016, 44, 124–130. [Google Scholar]
- Zhao, Y.; Wang, Y.; Liu, H.; Wu, H.; Zou, D. Simultaneous Determination of Five Endogenous Plant Hormones in Lettuce Leaves by Ultra-Performance Liquid Chromatography Tandem Mass Spectrometry (UPLC-MSMS). North. Hortic. 2021, 16, 8–15. [Google Scholar]
- Zhong, D.; Ding, M.; Tang, F.; Mo, R.; Teng, Y. Simultaneous Determination of Four Endogenous Plant Hormones in Phyllostachys heterocycla cv pubescens Shoots by High-Performance Liquid Chromatography-Tandem Mass Spectrometry (HPLC-MSMS). Chin. J. Anal. Chem. 2013, 41, 1739–1743. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Deng, Y.; Li, J.; Wu, S.; Zhu, Y.; Chen, Y.; He, F. Integrated nr Database in Protein Annotation System and Its Localization. Comput. Eng. 2006, 32, 71–73. [Google Scholar]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S.; et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, R7. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Bulter, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Sherlock, G. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Eddy, S.R. Profile hidden Markov models. Bioinform. Rev. 1998, 14, 755–763. [Google Scholar] [CrossRef]
- Bateman, A.; Birney, E.; Cerruti, L.; Durbin, R.; Etwiller, L.; Eddy, S.R.; Griffiths-Jones, S.; Howe, K.L.; Marshall, M.; Sonnhammer, E.L.L. The Pfam Protein Families Database. Nucleic Acids Res. 2002, 30, 276–280. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van-Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Chitkala Devi, T.; Bharathalakshmi, M.; Kumari, M.B.G.S.; Naidu, N.V. Effect of Sources and Levels of Phosphorus with Zinc on Yield and Quality of Sugarcane. Sugar Tech. 2012, 14, 195–198. [Google Scholar] [CrossRef]
- Li, Q.; Liu, P.; Zhao, H.; Song, X.; Lin, H.; Shen, Y.; Li, L.; Wan, S. Effects of Maize Root Exudates on Allelopathy of Phenolic Acids in Soil of Continuous Cropping Peanut. J. Agric. Sci. Technol. 2020, 22, 119–130. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, J.; Rahman, M.K.U.; Gao, D.; Wei, Z.; Wu, F.; DiniAndreote, F. Interspecific plant interaction via root exudates structures the disease suppressiveness of rhizosphere microbiomes. Mol. Plant 2023, 16, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; He, X.; Baer, M.; Beirinckx, S.; Tian, T.; Moya, Y.A.T.; Zhang, X.; Deichmann, M.; Frey, F.P.; Bresgen, V.; et al. Plant flavones enrich rhizosphere Oxalobacteraceae to improve maize performance under nitrogen deprivation. Nat. Plants 2021, 7, 481–499. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Wang, Y.; Zhang, Y.; Fei, J.; Rong, X.; Peng, J.; Yin, L.; Luo, G. Intercropping enhances maize growth and nutrient uptake by driving the link between rhizosphere metabolites and microbiomes. New Phytol. 2024, 243, 1506–1521. [Google Scholar] [CrossRef]
- Yan, Z.; Wang, D.; Ding, L.; Cui, H.-Y.; Jin, H.; Yang, X.; Yang, J.; Qin, B. Mechanism of artemisinin phytotoxicity acton: Induction of reactive oxygen species and cell death in lettuce seedlings. Plant Physiol. Biochem. 2015, 88, 53–59. [Google Scholar] [CrossRef]
- Deng, J.; Guo, J.; Qin, W.; Chen, J.; He, Y.; Zhang, Q.; Vanholme, B.; Yang, W.; Liu, J. Shading stress promotes lignin biosynthesis in soybean seed coat and consequently extends seed longevity. Int. J. Biol. Macromol. 2025, 298, 139913. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Zhang, C.; van der Werf, W.; Ning, P.; Zhang, Z.; Wan, S.; Zhang, F. Intercropping modulates the accumulation and translocation of dry matter and nitrogen in maize and peanut. Field Crops Res. 2022, 284, 108561. [Google Scholar] [CrossRef]
- Leister, D.; Wang, L.; Kleine, T. Organellar Gene Expression and Acclimation of Plants to Environmental Stress. Front. Plant Sci. 2017, 8, 387. [Google Scholar] [CrossRef]
- Pang, Z.; Fallah, N.; Weng, P.; Zhou, Y.; Tang, X.; Tayyab, M.; Liu, Y.; Liu, Q.; Xiao, Y.; Hu, C.; et al. Sugarcane-Peanut Intercropping System Enhances Bacteria Abundance, Diversity, and Sugarcane Parameters in Rhizospheric and Bulk Soils. Front. Microbiol. 2022, 12, 815129. [Google Scholar] [CrossRef]
- Shukla, S.K.; Sharma, L.; Jaiswal, V.P.; Dwivedi, A.P.; Yadav, S.K.; Pathak, A.D. Diversification Options in Sugarcane-Based Cropping Systems for Doubling Farmers’ Income in Subtropical India. Sugar Tech. 2022, 24, 1212–1229. [Google Scholar] [CrossRef] [PubMed]
- Viaud, P.; Heuclin, B.; Letourmy, P.; Christina, M.; Versini, A.; Mansuy, A.; Chetty, J.; Naudin, K. Sugarcane yield response to legume intercropped: A meta-analysis. Field Crops Res. 2023, 295, 108882. [Google Scholar] [CrossRef]
- Mathieu, L.; Ballini, E.; Morel, J.; Méteignier, L. The root of plant-plant interactions Belowground special cocktails. Curr. Opin. Plant Biol. 2024, 80, 102547. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chen, S.; Shi, W.; Davd, S.R.; Li, S.; Yang, F.; Lin, Z. Transcriptome profiling reveals the effects of drought tolerance in Giant Juncao. BMC Plant Biol. 2021, 21, 2. [Google Scholar] [CrossRef]
- Wen, B.; Peng, X.; Tian, J.; Wu, Q.; Wang, H.; Tang, X. Cloning and expression of the FrWRKY28 gene from Fagopyrum tataricum under low phosphorus and hormone treatment. Acta Bot. Boreali-Occident. Sin. 2024, 44, 930–937. [Google Scholar]
- Lei, L.; Zhu, Q.; Xu, P.; Jing, Y. The intercropping and arbuscular mycorrhizal fungus decrease Cd accumulation in upland rice and improve phytoremediation of Cd-contaminated soil by Sphagneticola calendulacea (L.) Pruski. J. Environ. Manag. 2021, 298, 113516. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Giehl, R.F.H.; von Wire N, N. Nutrient-hormone relations Driving root plasticity in plants. Mol. Plant 2021, 15, 86–103. [Google Scholar] [CrossRef]
- Pan, Y.; Cheng, S. Research Progress on Oat Genomics Study. J. Plant Genet. Resour. 2021, 22, 304–308. [Google Scholar] [CrossRef]
- Cai, J.; Wang, Q.; Wu, Y.; Liu, H.; Zhu, C. Effects of phytohormone on tomato root growth under aluminum stress. J. Anhui Sci. Technol. Univ. 2024, 38, 39–46. [Google Scholar] [CrossRef]
- Ding, Z.; Fu, L.; Chen, G.; Yan, Y.; Tie, W.; Meng, X.; Yang, J.; Qiu, X.; Zhang, J.; Hu, W. Coordinated transcriptional regulation of carbohydrate-related pathways contributes to the difference of starch accumulation between starchy cassava and sugary cassava. Carbohydr. Polym. 2025, 354, 123314. [Google Scholar] [CrossRef]
- Mendes, J.O.T.; Ribeiro, P.R.; Sfor ç a, M.L.; Rocco, S.A.; Ferraz, C.G. Foliar application of gallic acid improves tolerance to cadmium stress in wheat: An NMR-based metabolomics approach. Plant Physiol. Biochem. 2025, 229, 110405. [Google Scholar] [CrossRef]
- Tsavkelova, E.; Oeser, B.; Oren-young, L.; Israeli, M.; Sasson, Y.; Tudzynski, B.; Sharon, A. Identification and functional characterization of indole-3-acetamide-mediated IAA biosynthesis in plant-associated Fusarium species. Fungal Genet. Biol. 2012, 49, 48–57. [Google Scholar] [CrossRef]
- Fang, Z.; Yang, Q.; Xie, J.; Du, S. The role and mechanism of cytokinin in phytoremediation of heavy metal contaminated soil. Acta Ecol. Sin. 2022, 42, 3056–3065. [Google Scholar] [CrossRef]
- Cui, S.; Zhang, Z.; Fu, X.; Liu, J.; Yang, H. Key Genes in Response to Drought Stress in Plant Hormone Signal Transduction Pathway of Oat. J. Triticeae Crops 2023, 43, 1384–1393. [Google Scholar]
- Feng, X.; Xu, Y.; Peng, L.; Yu, X.; Zhao, Q.; Feng, S.; Zhao, Z.; Li, F.; Hu, B. TaEXPB7-B, a beta-expansin gene involved in low-temperature stress and abscisic acid responses, promotes growth and cold resistance in Arabidopsis thaliana. J. Plant Physiol. 2019, 240, 153004. [Google Scholar] [CrossRef]
- Pirona, R.; Frugis, G.; Locatelli, F.; Mattana, M.; Genga, A.; Baldoni, E. Transcriptomic analysis reveals the gene regulatory networks involved in leaf and root response to osmotic stress in tomato. Front. Plant Sci. 2023, 14, 1155797. [Google Scholar] [CrossRef]
- Ueno, D.; Yamaji, N.; Kono, I.; Huang, C.; Ando, T.; Yano, M.; Ma, J. Gene limiting cadmium accumulation in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 16500–16505. [Google Scholar] [CrossRef]
- Hänsch, R.; Mendel, R.R. Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr. Opin. Plant Biol. 2009, 12, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhu, Q.; Zhang, Y.; Jiang, P.; Wang, Y.; Fei, J.; Rong, X.; Peng, J.; Wei, X.; Luo, G. Soil carbon storage and accessibility drive microbial carbon use efficiency by regulating microbial diversity and key taxa in intercropping ecosystems. Biol. Fert. Soils 2024, 60, 437–453. [Google Scholar] [CrossRef]
- Li, Y.; Yu, C.; Cheng, X.; Li, C.; Sun, J.; Zhang, F.; Lambers, H.; Li, L. Intercropping alleviates the inhibitory effect of N fertilization on nodulation and symbiotic N2 fixation of faba bean. Plant Soil 2009, 323, 295–308. [Google Scholar] [CrossRef]
- Bian, F.; Zhang, X.; Li, Q.; Huang, Z.; Zhong, Z. Enhancement of Phytoremediation of Heavy Metal Pollution Using an Intercropping System in Moso Bamboo Forests: Characteristics of Soil Organic Matter and Bacterial Communities. Forests 2023, 14, 1895. [Google Scholar] [CrossRef]
- Kama, R.; Liu, Y.; Zhao, S.; Hamani, A.K.M.; Song, J.; Cui, B.; Aidara, M.; Liu, C.; Li, Z. Combination of intercropping maize and soybean with root exudate additions reduces metal mobility in soil-plant system under wastewater irrigation. Ecotoxicol. Environ. Saf. 2023, 266, 115549. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhou, T.; Cheng, B.; Du, Y.; Qin, S.; Gao, Y.; Xu, M.; Lu, J.; Liu, T.; Li, S.; et al. Variable Light Condition Improves Root Distribution Shallowness and P Uptake of Soybean in Maize/Soybean Relay Strip Intercropping System. Plants 2020, 9, 1204. [Google Scholar] [CrossRef]
- Ulloa-Inostroza, E.M.; Córdova, C.; Campos, M.; Reyes Díaz, M. Methyl Jasmonate Improves Antioxidants, Protecting Photosynthetic Apparatus in Blueberry Plants under Water Deficit. Horticulturae 2024, 10, 259. [Google Scholar] [CrossRef]
- Andersen, I.K.L.; Dragsted, L.O.; Rasmussen, J.; Fomsgaard, I.S. Intercropping of Hordeum vulgare L. and Lupinus angustifolius L. causes the generation of prenylated flavonoids in Lupinus angustifolius L. J. Plant Interact. 2023, 18, 2255039. [Google Scholar] [CrossRef]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.; Touraine, B.; Moënne-Loccoz, Y.; Muller, D.; Legrndre, L.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013, 4, 356. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Wu, Q.; Yang, S.; Liu, J.; Zhao, Y.; Zhao, P.; Wang, L.; Lu, W.; Huang, D.; Zhang, Y.; et al. Dissection of genetic architecture for desirable traits in sugarcane by integrated transcriptomics and metabolomics. Int. J. Biol. Macromol. 2024, 280, 136009. [Google Scholar] [CrossRef]
- Clemens, S.; Aarts, M.G.M.; Thomine, S.B.; Verbruggen, N. Plant science: The key to preventing slow cadmium poisoning. Trends Plant Sci. 2013, 18, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.A.; Mishra, A.K.; Rehaman, A.; Gandhi, S.G.; Jan, A.T. Nitric oxide in plant stress: Rewilding and restoring signaling for enhancing plant growth and development. Biochim. Biophys. Acta 2025, 1869, 130837. [Google Scholar] [CrossRef]
- Maathuis, F.J. Physiological functions of mineral macronutrients Frans JM Maathuis. Curr. Opin. Plant Biol. 2009, 12, 250–258. [Google Scholar] [CrossRef]
- Sinclair, S.A.; Krämer, U. The zinc homeostasis network of land plants. Biochim. Biophys. Acta 2012, 18223, 1553–1567. [Google Scholar] [CrossRef]
- Somanath, N.; Singh, S.Y.; Radha, P.; Sunil, M.; Shamima, P.; Kirttiranjan, B.; Sreenivasa, R.K. Soil and foliar application of Zn enhances its biofortification, bioavailability and productivity in both biofortified and non-biofortified wheat varieties. J. Food Compos. Anal. 2023, 124, 105691. [Google Scholar] [CrossRef]
- Singh, T.; Saffeullah, P.; Umar, S. Foliar application of zinc oxide (ZnO) nanoparticles ameliorates growth, yield traits, osmolytes, cell viability, and antioxidant system of Brassica juncea (L.) Czern, grown in lead (Pb) stress. Chemosphere 2025, 370, 143950. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Guo, X.; Zhou, Y.; Wang, X.; Wang, T.; Li, T.; Li, P.; Yuan, Z.; Pang, Z. Sugarcane–Peanut Intercropping Enhances Farmland Productivity: A Multi-Omics Investigation into the Coordination of Zinc Homeostasis and Hormonal Signaling. Agronomy 2025, 15, 2510. https://doi.org/10.3390/agronomy15112510
Chen S, Guo X, Zhou Y, Wang X, Wang T, Li T, Li P, Yuan Z, Pang Z. Sugarcane–Peanut Intercropping Enhances Farmland Productivity: A Multi-Omics Investigation into the Coordination of Zinc Homeostasis and Hormonal Signaling. Agronomy. 2025; 15(11):2510. https://doi.org/10.3390/agronomy15112510
Chicago/Turabian StyleChen, Siqi, Xiang Guo, Yongmei Zhou, Xiao Wang, Tao Wang, Tengfei Li, Peiwu Li, Zhaonian Yuan, and Ziqin Pang. 2025. "Sugarcane–Peanut Intercropping Enhances Farmland Productivity: A Multi-Omics Investigation into the Coordination of Zinc Homeostasis and Hormonal Signaling" Agronomy 15, no. 11: 2510. https://doi.org/10.3390/agronomy15112510
APA StyleChen, S., Guo, X., Zhou, Y., Wang, X., Wang, T., Li, T., Li, P., Yuan, Z., & Pang, Z. (2025). Sugarcane–Peanut Intercropping Enhances Farmland Productivity: A Multi-Omics Investigation into the Coordination of Zinc Homeostasis and Hormonal Signaling. Agronomy, 15(11), 2510. https://doi.org/10.3390/agronomy15112510





