Abstract
Modern crop breeding requires efficient tools for genetic identity and parentage verification to manage large-scale programs. To address this, we present GIPA (Genomic Identity and Parentage Analysis), a high-performance toolkit designed for these tasks. GIPA integrates key innovations: a sliding-window algorithm enhances accuracy by correcting genotyping errors, an intelligent system classifies samples by heterozygosity to streamline parentage analysis, and an integrated engine generates intuitive chromosome-level heatmaps. We demonstrate its utility in a soybean backcrossing scenario, where it identified a donor line with 98.02% genomic identity to the recipient, providing a strategy to significantly shorten the breeding program. In maize, its parentage module accurately identified the known parents of commercial hybrids with match scores exceeding 97%, validating its use for variety authentication and quality control. By transforming complex SNP data into clear, quantitative, and visual insights, GIPA provides a robust solution that accelerates data-driven decision-making in plant breeding.
1. Introduction
The advent of high-throughput sequencing technologies has revolutionized plant breeding, enabling the transition from phenotype-based selection to more precise genomics-assisted breeding (GAB) strategies [1,2,3]. Single Nucleotide Polymorphisms (SNPs) have emerged as the marker of choice due to their abundance, genome-wide distribution, and amenability to automated genotyping platforms like genotyping-by-sequencing (GBS) and SNP arrays [4,5,6]. These markers are foundational to advanced breeding methods, including marker-assisted selection (MAS) [7], genomic selection (GS) [8], and genome-wide association studies (GWAS) [9]. The application of these technologies is critical for ensuring global food security, particularly in staple crops like soybean and maize [10].
Soybean (Glycine max) and maize (Zea mays) were selected as case studies for this work due to their immense global importance as primary sources of food, feed, and industrial raw materials. They also present contrasting genomic landscapes, providing a robust framework for validating GIPA’s performance. Maize possesses a large, complex genome (~2.3 Gb) with high levels of genetic diversity and repetitive elements [11],, whereas soybean has a more compact, paleopolyploid genome (~1.1 Gb) with lower diversity stemming from a significant population bottleneck during domestication [12]. Despite these differences, both are functionally diploid, which simplifies the initial validation of Mendelian inheritance logic within the toolkit and makes them ideal models for demonstrating GIPA’s utility across varied genomic contexts.
Despite these advances, challenges persist in large-scale breeding programs, including the accurate verification of genetic identity and the unambiguous determination of parentage. Accidental mislabeling, cross-contamination, or seed mixing can occur at various stages of breeding and seed production. A rapid and accurate method to verify the genetic identity of a given sample against a reference database is essential for maintaining the integrity of breeding materials. Otherwise, misidentification of breeding materials can lead to costly errors, wasting years of effort and resources [13]. This is particularly critical in backcross breeding, a cornerstone technique for introgressing specific traits, such as disease resistance or transgenes, from a donor parent into an elite recipient (recurrent) parent [14]. The goal of backcrossing is to recover the recurrent parent’s genetic background as quickly as possible while retaining the target gene. This requires meticulous tracking and selection of progeny with the highest genomic similarity to the recurrent parent [15]. Furthermore, the global commercialization of genetically modified (GM) crops necessitates stringent stewardship and regulatory compliance, making the ability to rapidly confirm the genetic background of new varieties paramount [16]. Similarly, in hybrid seed production, verifying the parentage of F1 hybrids is essential for quality control and intellectual property (IP) protection [17]. Traditional methods are often laborious, whereas computational methods can accurately simulate Mendel’s law of segregation [18].
While various bioinformatics tools exist for population genetics and relationship inference, such as PLINK [19] or TASSEL [20], they often require significant computational expertise and may not offer an integrated solution tailored specifically to the routine workflows of plant breeders. Phylogenetic analysis, while powerful for exploring evolutionary relationships, can be cumbersome and visually complex for the simple task of identifying the single most identical individual from a large panel [21]. Furthermore, prominent parentage analysis software, such as COLONY (v2.0.7.2) [22], and SEQUOIA (v3.0.3) [23] have proven highly effective in population ecology and animal breeding, but their likelihood models rely on population allele frequencies and Hardy–Weinberg assumptions, which do not fit the deterministic F1 hybrid produced from two homozygous inbred lines. There is a pressing need for a user-friendly, efficient, and robust tool that combines identity analysis, parentage verification, and error correction within a single framework.
To bridge this gap, we developed GIPA (Genomic Identity and Parentage Analysis), a command-line toolkit specifically tailored to the needs of modern crop breeders. GIPA offers a unified platform for both identity and parentage analysis, incorporating several innovative features:
- Dual-Functionality: Seamlessly switch between identity verification and parentage discovery.
- Advanced Error Correction: A sliding-window algorithm minimizes the impact of sporadic genotyping errors on final calculations.
- Intelligent Sample Classification: Automatically distinguishes inbred from hybrid lines based on heterozygosity, refining the parentage search space.
- Integrated High-Quality Visualization: Generates intuitive, chromosome-level heatmaps that provide a clear visual representation of genomic similarity, surpassing the abstract nature of phylogenetic trees for this application.
We validate the performance and practical utility of GIPA using case studies in soybean (Glycine max) and maize (Zea mays), demonstrating its potential to accelerate breeding programs and improve quality control.
2. Materials and Methods
2.1. Software Architecture and Implementation
GIPA (v1.0.0) is implemented in Python 3 (v3.7) and leverages several core scientific libraries, including Pysam (v0.19.0) for efficient VCF file parsing [24], Pandas (v1.3.0) and NumPy (v1.21.0) for data manipulation [25,26], and Matplotlib (v3.5.0)/Seaborn (v0.11.0) for visualization [27]. This implementation ensures cross-platform compatibility, allowing GIPA to run natively on Linux, macOS, and Windows operating systems with minimal setup. The software is organized into a main executable (gipa.py) and modular helper scripts for data parsing and visualization, promoting code maintainability and extensibility.
2.2. Identity Analysis Module
The identity analysis module quantifies the genetic similarity between a query sample and one or more reference samples. Input data is a standard Variant Call Format (VCF) file. For each SNP locus, GIPA compares the genotype of the query sample with each reference sample. The comparison yields one of three outcomes: ‘1’ for a perfect match (e.g., 0/0 vs. 0/0 or 0/1 vs. 0/1), ‘0’ for a mismatch, and ‘/’ for loci where at least one sample has a missing genotype call (e.g., ‘./.’). The overall identity score is calculated as the ratio of matched SNPs to the total number of compared (non-missing) SNPs. This analysis is performed for the entire genome and for each chromosome individually.
To account for genotyping errors, GIPA employs a sliding window correction algorithm [28]. First, SNPs are sorted by chromosomal position to establish genomic context. For each sample, the algorithm then iterates through every SNP site in the initial comparison vector (composed of matches ‘1’ and mismatches ‘0’). At each site, it examines a local window of size W (default: 5 SNPs, set by the --filter-window parameter). Within this window, it calculates the frequency of matches and mismatches from non-missing data points. If the frequency of one state exceeds a dominant threshold (≥60%) and the window contains sufficient data, the central site’s value is overridden to match this local consensus. This process is repeated for a specified number of passes (default: 2, set by --filter-times) to progressively eliminate isolated, likely erroneous signals. The final identity score is then calculated from the corrected vector, providing a more robust measure of genetic identity.
2.3. Parentage Analysis Module
The parentage analysis module is designed to identify the most likely pair of parents for a query hybrid from a panel of candidates. 1. Automated Sample Classification. GIPA calculates the genome-wide heterozygosity rate for each candidate parent. Using the distribution of these rates, it intelligently identifies a threshold to classify samples as either ‘Inbred’ (low heterozygosity) or ‘Hybrid’ (high heterozygosity). This is achieved by finding the largest gap in the sorted heterozygosity rates, a method robust to variations between different species and datasets. The heuristic algorithm first sorts all heterozygosity values in ascending order. It then calculates the difference between each adjacent pair of values. The position of the largest difference is selected as the optimal threshold to separate the low-heterozygosity ‘Inbred’ group from the high-heterozygosity ‘Hybrid’ group. This heuristic approach is robust as it does not rely on predefined thresholds, which can vary significantly between different species, populations, and marker densities. This step allows GIPA to automatically exclude biologically unlikely parental combinations, such as two hybrid lines, thereby increasing accuracy and computational efficiency. 2. Mendelian Inheritance Validation. For each valid parental combination (Inbred × Inbred or Inbred × Hybrid), GIPA evaluates every SNP locus against the query hybrid’s genotype based on Mendelian inheritance rules (Table 1). The rules cover all possible diploid genotype combinations. 3. Parentage Match Score Calculation. The final match score for each parental combination is calculated similarly to the identity score, based on the ratio of matched SNPs to the total number of informative SNPs. The combinations are then ranked to reveal the most likely parents.

Table 1.
Mendelian inheritance rules for diploid genotypes used in GIPA’s parentage validation. ‘A’ and ‘B’ represent different alleles.
2.4. Visualization Module
GIPA’s visualization engine generates two types of high-resolution (300 DPI) heatmaps. 1. Single-Sample Chromosome-level Heatmap: For a given sample (or parental combination), this plot displays all chromosomes as horizontal bars, scaled by their relative lengths. The chromosome is segmented into windows of user-defined size (e.g., 50 kb), and each window is colored according to its SNP match rate, using a Red-Yellow-Blue color scale. This provides an ideogram-like overview of genomic similarity. 2. Multi-Sample Comparison Heatmap: This plot compares multiple samples (rows) across a single chromosome (x-axis, segmented into windows). It allows for direct visual comparison of the genomic similarity patterns of the top candidate samples, facilitating the identification of shared or distinct genomic regions.
2.5. Usage and Parameters
GIPA is operated via the command line. The main parameters are listed in Table 2.

Table 2.
Key command-line parameters for GIPA.
3. Results
To validate the performance and practical utility of GIPA, we conducted two case studies in soybean and maize, representing common and critical tasks in modern crop breeding programs.
3.1. Case Study 1: Identity Analysis for Soybean Backcross Breeding
To accelerate the introgression of a transgene located on Chr01 into a new elite soybean variety, we used GIPA to identify the most genetically similar donor from a panel of 20 existing transgenic lines. The objective was to minimize the number of subsequent backcross generations.
GIPA’s identity analysis identified TianLong1 as the top candidate, sharing a 98.02% whole-genome identity with the new elite variety. This value was substantially higher than that of the next closest line at 69.17% (Table 3). The heatmap for Chr01 (Figure 1) visually corroborated these quantitative results; TianLong1 is represented by a nearly uniform high-identity (red) bar, while other candidates display large regions of genetic divergence (blue).

Table 3.
Identity analysis results of the elite query variety against the top reference lines.
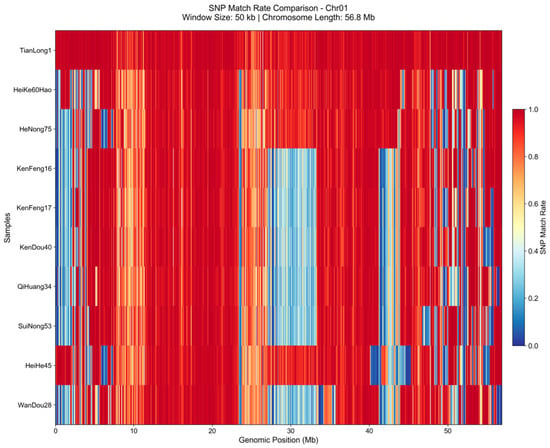
Figure 1.
GIPA-generated heatmap comparing the SNP match rate of the top 10 reference lines against the query variety on Chr01. Each row represents a reference line. The x-axis shows the genomic position along the chromosome, divided into 50 kb windows. The color within each window indicates the SNP match rate, where dark red signifies high genetic identity (1.0) and dark blue signifies genetic divergence (0.0).
The high identity score indicates that TianLong1 can be considered a near-isogenic line of the target variety. Utilizing this donor allows breeders to bypass the typical 5–6 generations of backcrossing, potentially reducing the process to 1–2 crosses for validation. This application of GIPA can therefore substantially reduce the breeding cycle duration, saving considerable time and resources.
3.2. Case Study 2: Parentage Analysis of Commercial Maize Hybrids
Accurate parentage information is vital for hybrid seed quality control and intellectual property protection. We evaluated GIPA’s Parentage module by identifying the parental inbred lines for three widely grown commercial maize hybrids (JK968, YF303, and ZD958) from a panel of elite inbred lines.
The analysis produced a ranked list of potential parental combinations for each hybrid, with clear and decisive results (Table 4). For all three hybrids, the top-ranking combination achieved a match score exceeding 97%, while the second-best combination scored significantly lower (by at least 10 percentage points). The identified parental pairs matched the known pedigrees for these commercial hybrids. Specifically, GIPA identified Jing724 × Jing92 as the parents for JK968 (98.46% match), CT1669 × CT3354 for YF303 (97.60% match), and Chang7-2 × Zheng58 for ZD958 (97.32% match).

Table 4.
Top 5 parental combination results for three commercial maize hybrids.
To visually validate these high scores, GIPA generated genome-wide heatmaps for the top-ranking parental combination of each hybrid, using JK968 as an example (Figure 2). The heatmaps for JK968 display a consistent pattern of high genetic identity, represented by the overwhelming prevalence of dark red coloration across all chromosomes. The near absence of divergent regions (blue) provides strong visual corroboration for the quantitative match scores.

Figure 2.
Genome-wide SNP match rate heatmaps for the identified parental combinations of JK968 vs. (Jing724 × Jing92). Each horizontal bar represents a chromosome, colored by SNP match rate in 50 kb windows. The consistent red color indicates a high match score across the entire genome.
The combination of unambiguous quantitative ranking and comprehensive visual confirmation demonstrates GIPA’s reliability and precision for applications in variety authentication and seed purity testing.
4. Discussion
GIPA was developed to fill a software gap in modern plant breeding. While powerful and complex tools for quantitative genetics exist, such as PLINK [19] and TASSEL [20], they often require multi-step command sequences and significant bioinformatics expertise to perform the routine tasks of identity and parentage verification. GIPA’s primary advantage is not necessarily raw computational speed but a dramatic reduction in operational complexity. It consolidates error correction, parentage-specific logic, and direct visualization into a single, intuitive command, making these analyses accessible to non-specialists.
GIPA’s main advantage is its focus on practical applications. It is not intended to replace comprehensive population genetics suits for estimating quantitative relatedness coefficients. Instead, it is highly optimized to provide rapid, definitive answers to the discrete logistical questions breeders face daily: ‘Is this sample what I think it is?’ and ‘Who are the parents of this hybrid?’ The sliding window correction algorithm is a practical feature that makes the results more reliable by correcting for the random genotyping errors that are common in high-throughput sequencing data [29]. Similarly, the automated classification of inbred and hybrid lines simplifies and speeds up parentage analysis.
The comparison with phylogenetic trees highlights a key advantage of GIPA. While phylogenetics is the gold standard for inferring evolutionary history [30], it can be an indirect tool for identifying the most genetically similar individual. A complex dendrogram may obscure the simple, quantitative answer a breeder needs. In contrast, GIPA’s ranked list and heatmaps provide a more direct, quantitative, and visually intuitive answer. This output is precisely tailored for rapid decision-making, such as selecting the best backcross parent from a panel or verifying the identity of a seed lot, tasks where clarity and speed are paramount.
The case studies show GIPA’s practical value in different, important breeding situations. The soybean analysis demonstrated its usefulness for strategic donor selection, a key step in efficient marker-assisted backcrossing (MABC) [15,31]. By identifying a transgenic line (TianLong1) that was highly similar to the target variety, GIPA showed a direct path to reducing a multi-year backcrossing program to a simple validation cross [32]. In the maize study, the tool accurately identified the correct parental pairs, and the results matched the known pedigrees. This proves it is effective for important tasks like authenticating commercial varieties and controlling seed quality [33].
Beyond these applications, GIPA can be useful in other ways. The clear results from the maize study suggest it has strong potential to resolve sample mix-ups: a common and expensive problem in large-scale breeding and germplasm management [34]. If a tray of seedlings loses its labels, GIPA can reliably screen them against a database of potential parents to rescue valuable genetic material. Furthermore, the whole-genome identity score calculated by GIPA serves as a direct and quantitative estimate of the recurrent parent genome (RPG) recovery. This allows breeders to precisely track the progress of backcrossing, verify the genetic purity of advanced lines, and make informed decisions on which individuals to advance to the next generation, ensuring breeding records are accurate and program goals are met efficiently [15,35].
Despite its strengths, GIPA has some limitations. Its analysis is based on SNPs and does not account for larger structural variations (SVs). The tool’s accuracy depends heavily on the quality and density of the input SNP data. Future development will focus on including SV data and creating a graphical user interface (GUI) to make the tool easier for non-specialists to use. Furthermore, the current implementation is tailored for diploid species, and its parentage analysis module cannot be directly applied to polyploid crops. Expanding the tool’s logic to accommodate various ploidy levels is a primary goal for future work.
5. Conclusions
GIPA is a practical software tool for identity and parentage analysis in crop breeding. Its key advantage lies in its integration of robust quantitative analysis with clear, visual heatmaps, providing more direct and actionable answers than traditional methods. We have shown this innovative approach can dramatically shorten breeding cycles by optimizing donor selection and reliably authenticate commercial hybrids for quality control. By transforming complex genomic data into easy-to-understand results, GIPA is a valuable tool that helps breeders make faster, data-driven decisions.
Author Contributions
Conceptualization, Y.-X.Y.; software, Y.-X.Y.; validation, Y.-F.Y.; data curation, X.-Y.M.; visualization, Y.W.; writing—original draft preparation, Y.-X.Y.; supervision, Z.-C.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Key Research, and Development Program of Zhejiang Province (2023C02033).
Data Availability Statement
The whole-genome resequencing data supporting the findings of this study are publicly available in the NCBI Sequence Read Archive (SRA) under the BioProject accession numbers PRJNA681974, PRJNA1202942, and PRJNA1170466. The GIPA software, including its source code and documentation, is freely available for academic and non-commercial use on GitHub at: https://github.com/nhyyx37/GIPA (accessed on 19 October 2025).
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| GIPA | Genomic Identity and Parentage Analysis |
| SNP | Single Nucleotide Polymorphisms |
| VCF | Variant Call Format |
References
- He, J.; Zhao, X.; Laroche, A.; Lu, Z.-X.; Liu, H.; Li, Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front. Plant Sci. 2014, 5, 484. [Google Scholar] [CrossRef]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genomics-assisted breeding for crop improvement. Trends Plant Sci. 2005, 10, 621–630. [Google Scholar] [CrossRef]
- Bohra, A.; Chand Jha, U.; Godwin, I.D.; Kumar Varshney, R. Genomic interventions for sustainable agriculture. Plant Biotechnol. J. 2020, 18, 2388–2405. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, A.; Hao, Y.; Xia, X.; Khan, A.; Xu, Y.; Varshney, R.K.; He, Z. Crop breeding chips and genotyping platforms: Progress, challenges, and perspectives. Mol. Plant 2017, 10, 1047–1064. [Google Scholar] [CrossRef] [PubMed]
- Gill, T.; Gill, S.K.; Saini, D.K.; Chopra, Y.; de Koff, J.P.; Sandhu, K.S. A comprehensive review of high throughput phenotyping and machine learning for plant stress phenotyping. Phenomics 2022, 2, 156–183. [Google Scholar] [CrossRef]
- Collard, B.C.; Jahufer, M.; Brouwer, J.; Pang, E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef]
- Varshney, R.K.; Bohra, A.; Yu, J.; Graner, A.; Zhang, Q.; Sorrells, M.E. Designing future crops: Genomics-assisted breeding comes of age. Trends Plant Sci. 2021, 26, 631–649. [Google Scholar] [CrossRef]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef]
- Liu, Y.; Du, H.; Li, P.; Shen, Y.; Peng, H.; Liu, S.; Zhou, G.-A.; Zhang, H.; Liu, Z.; Shi, M. Pan-genome of wild and cultivated soybeans. Cell 2020, 182, 162–176.e13. [Google Scholar] [CrossRef]
- Jones, A.G.; Ardren, W.R. Methods of parentage analysis in natural populations. Mol. Ecol. 2003, 12, 2511–2523. [Google Scholar] [CrossRef]
- Frisch, M.; Melchinger, A.E. Selection theory for marker-assisted backcrossing. Genetics 2005, 170, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Hospital, F. Selection in backcross programmes. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1503–1511. [Google Scholar] [CrossRef]
- Fraiture, M.-A.; Herman, P.; Taverniers, I.; De Loose, M.; Deforce, D.; Roosens, N.H. Current and new approaches in GMO detection: Challenges and solutions. BioMed Res. Int. 2015, 2015, 392872. [Google Scholar] [CrossRef]
- Josia, C.; Mashingaidze, K.; Amelework, A.B.; Kondwakwenda, A.; Musvosvi, C.; Sibiya, J. SNP-based assessment of genetic purity and diversity in maize hybrid breeding. PLoS ONE 2021, 16, e0249505. [Google Scholar] [CrossRef]
- Myles, S.; Boyko, A.R.; Owens, C.L.; Brown, P.J.; Grassi, F.; Aradhya, M.K.; Prins, B.; Reynolds, A.; Chia, J.-M.; Ware, D. Genetic structure and domestication history of the grape. Proc. Natl. Acad. Sci. USA 2011, 108, 3530–3535. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Jones, O.R.; Wang, J. COLONY: A program for parentage and sibship inference from multilocus genotype data. Mol. Ecol. Resour. 2010, 10, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Huisman, J. Pedigree reconstruction from SNP data: Parentage assignment, sibship clustering and beyond. Mol. Ecol. Resour. 2017, 17, 1009–1024. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Harris, C.R.; Millman, K.J.; Van Der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- McKinney, W. Data structures for statistical computing in Python. Scipy 2010, 445, 51–56. [Google Scholar]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Huang, X.; Feng, Q.; Qian, Q.; Zhao, Q.; Wang, L.; Wang, A.; Guan, J.; Fan, D.; Weng, Q.; Huang, T. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009, 19, 1068–1076. [Google Scholar] [CrossRef]
- Pompanon, F.; Bonin, A.; Bellemain, E.; Taberlet, P. Genotyping errors: Causes, consequences and solutions. Nat. Rev. Genet. 2005, 6, 847–859. [Google Scholar] [CrossRef] [PubMed]
- Naruya Saitou, M.N. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Qi, Y.; Sun, G.; Zhang, S.; Li, W.; Wang, Y. Improving Soybean Breeding Efficiency Using Marker-Assisted Selection. Mol. Plant Breed. 2024, 15, 259–268. [Google Scholar] [CrossRef]
- Bhat, J.A.; Feng, X.; Mir, Z.A.; Raina, A.; Siddique, K.H. Recent advances in artificial intelligence, mechanistic models, and speed breeding offer exciting opportunities for precise and accelerated genomics-assisted breeding. Physiol. Plant. 2023, 175, e13969. [Google Scholar] [CrossRef] [PubMed]
- Mumm, R.H. A look at product development with genetically modified crops: Examples from maize. J. Agric. Food Chem. 2013, 61, 8254–8259. [Google Scholar] [CrossRef]
- Gowda, M.; Worku, M.; Nair, S.K.; Palacios-Rojas, N.; Huestis, G.; Prasanna, B. Quality Assurance/Quality Control (QA/QC) in Maize Breeding and Seed Production: Theory and Practice; CIMMYT: Nairobi, Kenya, 2017; Volume 13. [Google Scholar]
- Sundaram, R.M.; Vishnupriya, M.; Laha, G.S.; Rani, N.S.; Rao, P.S.; Balachandran, S.M.; Reddy, G.A.; Sarma, N.P.; Sonti, R.V. Introduction of bacterial blight resistance into Triguna, a high yielding, mid-early duration rice variety. Biotechnol. J. Healthc. Nutr. Technol. 2009, 4, 400–407. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).