Abstract
Anaerobic digestate from manure, a byproduct of biogas production, is increasingly used as an organic fertilizer in circular agriculture systems. This study assessed the microbiological impact of maize fertigation with anaerobic digestate, focusing on fecal indicators (Escherichia coli, Salmonella), antibiotic resistance genes (ARGs), and integrons. The trial was conducted in a commercial maize field, where on-site manure-based anaerobic digestate was applied via center-pivot irrigation. Leaf samples were collected two days (2 dai) and four weeks (4 wai) after the last fertigation. E. coli and Salmonella were assessed by culturable methods, while ARGs and integrons were analyzed by qPCR. Results showed that E. coli (3 MPN/g) and Salmonella were detected at 2 dai but were undetectable at 4 wai and in the control condition, suggesting transient contamination. The abundance of tetW was approximately tenfold higher in digestate-treated plants than in the control, while no consistent changes were observed for the other genes. Overall, fertigation with anaerobic digestate appears to pose minimal microbiological impact within the specific conditions of this study, although it may act as a source of specific resistance determinants. Although limited by the use of single treated and control plots, this study offers preliminary insight into microbial and resistance gene dynamics in the phyllosphere, providing a basis for future replicated hypothesis-driven studies.
Keywords:
feedlot manure; anaerobic digestion; fecal indicators; antibiotic resistance genes; tetW; ermB; blaCTX-M; intI1; a-int1 1. Introduction
The “One Health” concept emphasizes the link between human, animal, and environmental health, proposing a unified approach to global health challenges [1]. Antimicrobial resistance (AMR) is a priority concern under this framework, as the misuse and overuse of antibiotics in recent decades in both human medicine and livestock production is accelerating its emergence and dissemination [2,3]. Globally, antimicrobials used in food animals represent approximately 73% of total antibiotic consumption, with a projected increase of 11.5% by 2030 relative to 2017 [3]. In livestock farming, a significant portion of these antibiotics is used for non-therapeutic purposes, including growth promotion and disease prophylaxis, contributing to the spread of AMR [3].
Unmetabolized antibiotics in manure promote an increase in antimicrobial resistance genes (ARGs) levels, recognized by the WHO as contaminants of emerging concern, by applying selective pressure on microbial communities [2,4]. These genes can be transferred between bacteria in animal populations and from animals to humans, increasing the risk of treatment failures in both veterinary and human healthcare [5]. There is strong scientific evidence indicating that antimicrobial use on farms imposes a public health impact due to infections with resistant pathogens [6]. As many of the microbes that are pathogenic in livestock animals are also pathogenic in humans, there is a potential for resistant strains to develop in animals and subsequently infect humans [1].
The dissemination of ARGs represents one of the main challenges to global public health [2,3,7]. On the other hand, mobile genetic elements (MGEs), such as integrons, play a crucial role in the spread and assembly of ARGs by facilitating horizontal gene transfer both within and between bacterial populations. This makes them a significant reservoir of resistance determinants, which should be regarded as a threat from the “One Health” perspective [3].
Without effective measures to stop the spread of AMR, it is projected to result in 10 million deaths per year by 2050. To address this urgent issue, a “One Health” approach would be necessary, involving multidisciplinary strategies and the collaboration of various national and international agencies to combat this problem [1].
A Circular Economy is a systemic framework intended to address global challenges such as waste, climate change, pollution, and biodiversity loss. As a sustainable alternative to the traditional linear economic model, it represents a new paradigm focused on minimizing waste and pollution, optimizing resource efficiency by keeping products and materials in use, and restoring natural systems [8]. Within this framework, waste is redefined as a resource that can be reintegrated into the production process [9]. Agricultural operations generate substantial volumes of agro-waste, generally accompanied by limited disposal options. As a result, it contributes to exacerbating various key problems like greenhouse gas emissions, soil degradation, and biodiversity loss [10]. Taking this into account, it is urgent to consider innovative solutions to transform agro-waste into valuable resources. One critical aspect of this challenge is the improper disposal of animal manure, which, despite its potential to cause ecological damage, can be a valuable resource if integrated into a circular concept [11]. In this regard, anaerobic digestion of manure emerges as an attractive and cost-effective treatment. This process consists of the microbial decomposition of organic material in a bioreactor under anaerobic conditions, generating biogas. The biogas is then converted into electrical and thermal energy or, more recently, upgraded to biomethane, while anaerobic digestate, a byproduct, holds significant potential as a sustainable fertilizer in agricultural systems [12]. Animal manure anaerobic digestate contributes to nutrient recycling, reduces reliance on chemical fertilizers, and ultimately supports environmental sustainability [13]. However, its use presents certain risks that need to be carefully considered.
The influence of the anaerobic digestion process on the survival of pathogenic or multidrug-resistant bacteria is a topic that has received considerable attention. In many cases, the persistence of antibiotics, ARGs to commonly used antibiotics in the livestock sector, as well as MGEs involved in the spread of resistance in agroecosystems, has been reported [14,15]. These compounds can be exchanged between bacteria in the microbiota of animals, plants, soils, and humans through horizontal gene transfer, thus contributing to the global problem of AMR and multidrug resistance [7]. This horizontal gene transfer also extends to plant-associated environments. Specifically, microorganisms establish a diverse range of intimate associations with plants, inhabiting the phyllosphere (the above-ground plant surface), the plant interior as endophytes, and the rhizosphere (the surrounding soil and roots) [16]. In fact, the presence of ARGs has been detected in the phyllosphere of many plants, among them maize (Zea mays L.), even though little is known about the composition of the antibiotic resistome and the factors influencing the diversity and abundance of ARGs [17]. Maize is a versatile cereal crop that plays a central role in global agriculture. Approximately 60% of its production is dedicated to livestock feed. It is a primary energy source in diets for poultry, swine, and cattle due to its high energy value, palatability, and digestibility. In beef and dairy cattle, maize forages, including silage, are extensively utilized due to its unique nutritional and functional properties, including their starch content and presence of essential fatty acids, with feedlot diets often comprising around 75–80% of maize [18].
The use of anaerobic digestate as a fertilizer may introduce complex interactions that could influence the composition and dynamics of microbial communities, potentially allowing the persistence of pathogens and ARGs within the phyllosphere and, consequently, threatening the health of cattle fed with maize silage. In agricultural practice, fertigation—the application of liquid fertilizers through irrigation systems—is a common approach to efficiently deliver nutrients to crops. Therefore, it is crucial to investigate how the application of anaerobic digestates by fertigation could affect the spread of ARGs, providing a deeper understanding of the potential risks and benefits associated with using anaerobic digestate as a fertilizer in sustainable agricultural practices. This study aimed to evaluate microbiological impacts of fertigation with anaerobic digestate within a privately managed, commercial maize operation practicing circular agriculture. Anaerobic digestate generated on-site from feedlot manure was applied through the system’s only available center pivot. While replication was limited by the realities of large-scale farming infrastructure, this case study offers valuable, context-specific data on pathogen and ARG dynamics, providing insight for both scientific advancement and sustainable practice.
2. Materials and Methods
2.1. Study Site and Climate Context
This study was carried out in collaboration with a privately owned commercial maize operation located in Coronel Suárez, Buenos Aires Province, Argentina. The farm is actively implementing a circular agriculture model, whereby anaerobic digestate produced on-site from cattle manure is reused as fertigation input for maize. The climate in Coronel Suárez is temperate, with a mean annual temperature of approximately 15 °C and annual rainfall averaging 800 to 1000 mm, relatively evenly distributed. In 2023, the site received approximately 870.7 mm of precipitation, with a typical dry period in late winter and occasional semi-dry conditions in mid-summer (January–February). The soils in both fields are classified as clay loam Petrocalcic Hapludols [19].
2.2. Field Design and Agronomic Practices
The study was conducted in two maize fields. The treated field (37°39′30″ S, 61°52′19″ W) is equipped with the operation’s only center-pivot irrigation system, delivering overhead sprinkler application that fully wets the canopy. This field received liquid anaerobic digestate through fertigation. The control field (37°32′48.39″ S, 61°56’25.92″ W) was located 14 km northwest of the treated site to avoid cross-contamination, while maintaining similar environmental and soil conditions. Neither field had a history of animal grazing, and both are exclusively dedicated to crop production. Maize was sown in December 2023 using DUO 30 (Corteva Agriscience, Buenos Aires, Argentina) at a density of 65,000 plants/ha in the anaerobic digestate-treated field (60 ha), and NK 897 (Syngenta, Buenos Aires, Argentina) at 40,000 plants/ha in the control field (26 ha). Diammonium phosphate was applied prior to sowing at 60 kg/ha (treated) and 80 kg/ha (control), and light harrowing was conducted only in the treated field to prepare the seedbed. Due to logistical and operational constraints on this commercial farm, only one digestate-treated field and one control field were available for study. Therefore, differences in hybrid, plant density, and base fertilizer rates may act as potential confounding factors in the interpretation of results.
2.3. Anaerobic Digestate Source and Application
The liquid anaerobic digestate used in this study was produced on-site at the BiodeS plant operated by Agro De Souza S.A. (Coronel Suárez, Argentina), using two complete mix anaerobic reactors fueled entirely by feedlot cattle manure. The system features a European-style design and includes key components from Vogelsang® GmbH & Co. KG, (Essen, Oldenburg, Germany). Anaerobic digestion is carried out under mesophilic conditions (39 °C), with a hydraulic retention time of 40 days. Starting December 12th, a total of 100,842 L per hectare of anaerobic digestate was applied to 50 ha of maize via the pivot system. Anaerobic digestate was pumped directly from the plant’s settling lagoon to the center pivot for fertigation. Due to the operational realities of working within a commercial circular system with a single pivot, only one field per treatment was available. Although this limits field-level replication, the study design reflects applied sustainability practices already in use, offering a unique opportunity to assess microbial and antimicrobial resistance dynamics under real-world conditions. Photographic documentation is provided in the Supplementary Material (Figure S1).
2.4. Use of Antibiotics in Cattle
Throughout the duration of the trial, the animals in the feedlot were administered the following antibiotics: Maxibiotic™ (a tetracycline-based antibiotic, Biogenesis Bagó, Garín, Argentina), and Maxytil Platinum™ (Biogenesis Bagó, Garín, Argentina), and Biomacrotil™ (Calier, Buenos Aires, Argentina) (all of which are macrolides containing tylosin). Macrolides and tetracyclines are two classes of antibiotics commonly used in veterinary medicine. Macrolides, such as erythromycin and tylosin, are often used to control respiratory diseases and gastrointestinal infections in animals, while tetracyclines, like oxytetracycline and chlortetracycline, are frequently employed for a broad range of bacterial infections, including those affecting the respiratory tract, digestive system, and reproductive health of animals [20,21].
2.5. Characterization of Anaerobic Digestate
The following parameters were determined: NH4+-N, NO3−-N and total Kjeldahl nitrogen (TKN) by the semi-micro-Kjeldahl method. Total carbon (C) was determined by dry combustion (Elemental Analyser Exeter CE 440, Exeter Analytical UK Ltd., Coventry, England). Total solid (TS) was determined by drying samples at 105 °C to constant weight. The ash content was determined by burning samples at 550 °C for 2 h. The volatile solid (VS) was calculated by subtracting the ash mass from the TS mass. The determinations for total and available phosphorus (TP and AP, respectively) and heavy metals were carried out with an Inductively Coupled Plasma Optical Emission Spectrometer (ICP-OES), Shimadzu Simultaneous 9000 (Shimadzu, Canby, OR, USA) (Table 1). In Argentina, digestate application is regulated by Resolution 19/2019 [22]. In compliance with this normative, stability measurements were performed using Residual Biogas Potential (RB) and Volatile Fatty Acids (VFA). The obtained values (RB = 0.107 L/g organic matter and VFA = 0.042 chemical oxygen demand/g organic matter) were within the regulatory limits (<0.25 and <0.43, respectively), confirming that the digestate was stable. Total coliforms, E. coli, and Salmonella were determined prior to the field sampling, following the standard methodology specified by Resolution 19/2019 [22]. In the initial anaerobic digestate sample, the total coliform count was 43 MPN/g fresh weight, with fecal coliforms and E. coli present at levels below detection (<3 MPN/g). However, the Salmonella count was 113 MPN/4 g fresh weight, which exceeded the limit set by the regulation for this pathogen (<3 MPN/4 g).

Table 1.
Chemical and spectroscopy characterization of digestate.
2.6. Plant Sampling
The first sampling was conducted 2 days after the final fertigation event with anaerobic digestate (2 dai, 16 March 2024), allowing time for plant surfaces to dry. The second sampling occurred 4 weeks later (4 wai, 12 April 2024), just before the scheduled maize harvest for silage. In the control field (Figure 1a), three 100 m transects (T1, T2, T3) were established in a North–South direction, spaced 20 m apart. At each transect, leaf samples were collected from the top to bottom of three maize plants at three different points (0, 50, and 100 m), using nitrile gloves to prevent contamination. These nine leaflets were composited into one sample per transect (N = 3). In the digestate-treated field (Figure 1b), a similar approach was used. Three transects (T1, T2, T3), spaced 20 m apart and aligned North–South, were sampled at 0 m (upslope), 50 m (mid-slope), and 250 m (downslope). Again, leaf samples were collected from three plants at each point and combined into a composite sample per transect (N = 3). Each transect thus represented a composite sampling unit within its respective field. These transects serve as subsamples rather than independent replicates, consistent with field-based ecological studies where logistical or landscape constraints precluded full replication. While the experimental design lacks field-level replication, the sampling structure allows meaningful comparison between treatment conditions and provides valuable insight into microbial and ARG dynamics under real-world conditions.
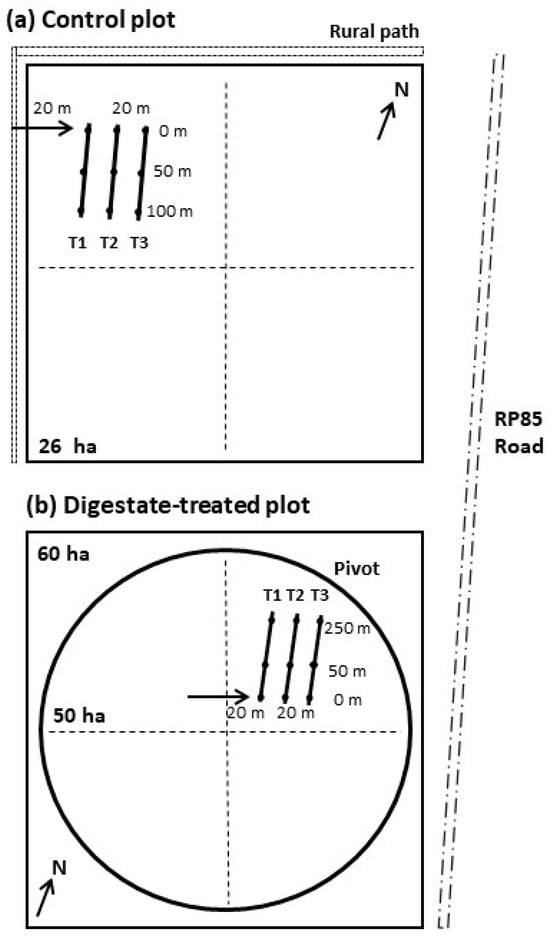
Figure 1.
Spatial representation of the control and digestate-irrigated fields. The control field is located 14 km north of the digestate–irrigated plot, along Provincial Road N°85. Three transects (T1–T3) were established in each plot, proportional to the area of each one. Plots and their elements are not drawn to scale. (a) In the control plot, the transects were 100 m long, separated by approximately 20 m. Samples were taken at 0 m, 50 m, and 100 m. (b) In the digestate-treated plot, the transects were 250 m long, separated by approximately 20 m. Samples were taken at the Upslope (0 m), Mid-slope (50 m), and Downslope (250 m) locations. The 50-ha circle represents the area irrigated by the pivot.
2.7. Screening for Indicators of Fecal Contamination and Pathogens
Total coliforms, E. coli, and Salmonella were determined in the phyllosphere of maize plants. Since there is no specific regulation, the standard established in the Argentine Food Code for the analysis of fresh leafy vegetables [23] was used as a reference. To estimate the concentration of coliforms in each sample, the Most Probable Number (MPN) method was used. Briefly, 20 g of leaf were added to a flask with 180 mL of peptone water (PW, Britania Laboratories, Buenos Aires, Argentina) and shaken for 2 min to obtain the first dilution 1:10 (10−1). Decimal dilutions were prepared by adding 1 mL of the 1:10 dilution to a tube containing 9 mL of peptone water until reaching the 10−3 dilution. Then, 1 mL of each prepared dilution was inoculated in triplicate into tubes with 9 mL of Lauryl sulfate medium (Britania Laboratories, Buenos Aires, Argentina) with a Durham tube inside. Tubes were incubated at 37 °C ± 2 °C for 24 h. After incubation, the tubes were examined for gas production and turbidity. For positive tubes, the procedure continued with inoculation in MacConkey broth and agar plates (Britania Laboratories, Buenos Aires, Argentina). The detection limit was 3 MPN/g. For screening of Salmonella spp., 20 g of leaf was added to a flask with 180 mL of PW, shaken for 2 min and incubated at 35 °C for 24 ± 2 h. Following, 0.1 mL of the culture was transferred into a tube with 10 mL of Rappaport Vassiliadis broth (RV, Britania Laboratories, Buenos Aires, Argentina) and 1 mL into a tube with 10 mL of Tetrathionate Broth (TT, Britania Laboratories, Buenos Aires, Argentina). The RV tubes were incubated for 24 ± 2 h at 42 ± 0.2 °C, and the TT tubes for 24 ± 2 h at 35 ± 2 °C. Then, 10 µL of each tube was streaked onto three different media plates: brilliant green agar (Britania Laboratories, Buenos Aires, Argentina), Xylose Lysine Deoxycholate agar (Britania Laboratories, Buenos Aires, Argentina) and Hektoen Enteric agar (Britania Laboratories, Buenos Aires, Argentina) and incubated at 35 °C for 24 ± 2 h.
2.8. Extraction of Total DNA from the Microbial Community
The extraction and purification of metagenomic DNA were performed using the DNeasy PowerSoil Pro Kits® (Qiagen, Hilden, Germany). For the anaerobic digestate, extraction was performed using 2 mL of homogenized fresh sample, which was centrifuged at 10,000× g for 10 min to obtain a pellet. The supernatant was discarded, and the pellet was stored at −20 °C until extraction. For DNA extraction from the phyllosphere, the procedure described by Zhu et al. (2017) was followed with minor modifications [24]. Briefly, 10 g of leaf (cut into 2 cm pieces) were soaked in 40 mL of Phosphate Buffer (60 mM pH 7.6) with 10% Tween, left for 1 h under reciprocal shaking, filtered and then centrifuged for 15 min at 10,000 rpm (Heraeus Biofuge pico centrifuge, Kendro, Germany). The pellets were recovered, and the DNeasy PowerSoil Pro Kits® (Qiagen, Hilden, Germany) was followed. The obtained DNA was subsequently quantified using the QuantiFluor® dsDNA System (Promega Corporation, Madison, WI, USA) in a Quantus fluorometer (Promega Corporation, Madison, WI, USA). The quality and integrity of the DNA were assessed from 1% agarose gel and absorbance ratios (260:230 and 260:280 nm ratio) in a DS-11 FX spectrophotometer (DeNovix Inc., Wilmington, DE, USA).
2.9. Analysis of Total Bacteria, Resistance Genes and Integrons
To study the treatment effect on the total number of bacteria, a quantitative real-time PCR (qPCR) of the 16S rRNA gene was conducted. Antibiotic resistance in the samples was determined using previously optimized qPCR protocols evaluated in different environmental samples [3]. Specifically, the genes ermB, tetW, blaCTX-M group 1, associated with resistance to different antibiotics (Table 2 and Table 3), and the integrons intI1 and a-int1 (Table 2), associated with horizontal gene transfer, were assessed. All amplifications, baseline corrections, melting curve analysis, and standard curve assessments were conducted in ABI 7500 Real—Time System and its associated software (7500 Software v2.0.3, Applied Biosystems, Foster City, CA, USA). In all cases, the SYBR Green system was used. SYBR Green reactions were conducted using SsoAdvanced™ Universal SYBR® Green Supermix (BioRad, Hercules, CA, USA), employing two different thermocycler programs. For 16S rRNA, the protocol was as follows: 5 min of initial denaturation at 95 °C, and 35 cycles of 15 s denaturation at 95 °C, 30 s annealing (see temperature in Table 2) and 45 s extension at 72 °C. For resistance genes and integrons, 10 min of initial denaturation at 95 °C, and 40 cycles of 15 s denaturation at 95 °C, 20 s annealing (see temperatures in Table 2) and 30 s extension at 72 °C. In all cases, the melt curve analysis was at 65–95 °C and the detection limit was approximately 2 × 101 gene copies per reaction.

Table 2.
Primers used in this study to quantify total bacteria (16s rRNA), resistance genes (tetW, ermB, blaCTX-M), integrons (intI1) and anthropogenic class 1 integrons (a-int1).

Table 3.
Antibiotic resistance mechanisms of the ARGs tetW, ermB and blaCTX-M.
2.10. Statistical Analysis
MPN of bacteria was reported as the median and 95% confidence interval of three replicates. Statistical analyses of the resistance genes data were performed as repeated measures analysis of variance (RM-ANOVA) using R studio version 4.4.2 [31]. Data were analyzed using a linear mixed-effects model (lme procedure fit with REML) in the nlme package (version 3.1-90) in R. The analysis tested the effects of fertilization treatment and time elapsed since fertilization on the abundance of resistance genes. Statistical analyses were conducted using linear mixed-effects models, accounting for repeated measures. Due to the logistical limitations of conducting research within a production-scale circular agriculture system, our statistical approach permits exploratory conclusions, appropriate for this real-world case study context [32]. Transect was included as a random effect, and fertilization treatment, time, and their interaction as fixed effects. The Compound Symmetry (CS) model was selected, using Aikake’s Information criterion (AIC), Bayesian information criterion (BIC) and log of restricted maximum likelihood (logLik) criterion. Comparisons were calculated using the emmeans function from the emmeans package version 1.10.5. This function provides adjusted means for each factor level and was used to conduct pairwise comparisons to assess the differences between group means. Given the absence of true replication at the field level, statistical comparisons should be interpreted as indicative of trends rather than definitive inferences.
3. Results
3.1. Detection of Indicators of Fecal Contamination and Pathogens
As shown in Table 4, in the 2 dai sampling, the control situation showed low levels of total coliforms and fecal coliforms, with no detection of E. coli or Salmonella, consistent with the fact that maize was not irrigated with anaerobic digestate. In contrast, the phyllosphere of maize that was fertigated with anaerobic digestate (AD) exhibited higher levels of total coliforms, with both E. coli and Salmonella present. In the 4 wai sampling, the control had no detectable total coliforms or fecal coliforms and neither E. coli nor Salmonella were detected, as observed in 2 dai sampling. The AD phyllosphere sampled 4 wai showed a lower concentration of total coliforms than in the first sampling date, and no detectable fecal coliforms, E. coli, or Salmonella (Table 4).

Table 4.
Detection of total coliforms, fecal coliforms, E. coli, and Salmonella in maize leaves in both sampling dates (2 days after last irrigation with digestate, 2 dai, and 4 weeks after last irrigation with digestate, 4 wai) for treated (AD) and control samples. Results are presented as the median of MPN/g (Most Probable Number per gram) along with the corresponding 95% confidence intervals (CI). The presence or absence of Salmonella/20 g of leaves is noted for each sample.
3.2. Analysis of Antibiotic Resistance Genes
Due to the absence of field-level replication, statistical tests cannot be interpreted as confirmatory. For transparency, descriptive p-values are provided in the Supplementary Material (Tables S1 and S2) but are not shown in the main text. Differences in 16S rRNA gene abundance were examined across treatments (control and AD) and sampling times (2 dai and 4 wai) (Table 5 and Table S1). At 2 dai, levels were generally lower in the control than in AD, with a similar pattern observed in the control across the sampling times. The abundances of the ARGs tetW, ermB, blaCTX-M, and integrons intI1 and a-int1 were also analyzed. Most genes showed no apparent differences among treatments or sampling times, except for two cases (Table 5 and Table S1). Levels of tetW were generally higher in AD than in the control (Table 5, Figure S2), with values in AD around tenfold greater. Patterns for a-int1, varied across treatments and sampling times. At the second sampling, levels tended to be lower in AD than in the control, and in AD they were lower at 4 wai compared with 2 dai. No notable differences between treatments were observed at 2 dai (Table 5 and Table S1). No clear differences were observed in the other ARGs tested (ermB, blaCTX-M, and intI1). Similar patterns were observed for relative abundances (i.e., resistance gene abundance normalized to 16S rRNA gene abundance). Specifically, tetW relative abundance tended to be higher in digestate-treated plants, whereas a-int1 relative abundance tended to be lower in the AD treatment at 4 wai compared with the earlier sampling (2 dai) and with the control at both time points (Tables S2 and S3).

Table 5.
Mean values of absolute abundance [log10 (gene copies µg−1 DNA)]. The mean values, standard errors of the mean (S.E.) and number of observations (n) are indicated. T represents the effect of fertilization treatment (Control vs. AD), S refers to the sampling time (2 dai vs. 4 wai), and T × S indicates the interaction between fertilization treatment and sampling time. Boldface denotes the lowest mean values among treatments, indicating comparative trends.
4. Discussion
This study offers valuable preliminary data from a commercial circular agriculture system, where on-site anaerobic digestate was applied to maize through the farm’s existing irrigation infrastructure. While the field-level design was constrained by the realities of production-scale farming, the findings represent a rare insight into pathogen and ARG behavior under real-world, sustainability-oriented conditions. Our approach is consistent with other field studies conducted under similar constraints [32], where data were still recognized as valuable contributions to the broader understanding of environmental change. Although field-level replication was not feasible, our findings highlight potential short-term microbial impacts and set the stage for future research involving replicated experimental designs. In this research, we focused on exploring the microbial impacts associated with the use of anaerobic digestate from manure for the fertigation of maize using a center-pivot system that results in full wetting of the foliage. Emphasis was placed on the potential presence of pathogenic microorganisms and the persistence and spread of ARGs. One of the key findings was the detection of E. coli and Salmonella in maize plants that were irrigated with anaerobic digestate. In addition, we identified the presence of ARGs in these plants, suggesting that anaerobic digestate could serve as a vector for the transmission of ARGs into the agricultural environment.
In anaerobic digestates, the survival of bacteria is influenced by various factors, such as oxygen concentration, temperature, pH, and moisture content. Moreover, the risk of pathogen presence is higher when manure is used as a feedstock, as it commonly contains coliforms and pathogens such as Salmonella, Campylobacter, and some protozoa, which can persist in anaerobic digestates [33,34]. Their inactivation during the anaerobic process is highly variable and depends on many factors [35]. For example, while anaerobic digestion is generally effective at reducing microbial load, the process tends to be more efficient in laboratory-scale digesters than in full-scale systems [35]. Temperature also plays a crucial role in pathogen inactivation, with digesters operating at thermophilic temperatures typically achieving more effective microbial reduction than mesophilic ones [36]. In our research, the initial anaerobic digestate sample had undetectable levels of E. coli, but a count of Salmonella which exceeded the limit set by the regulation for this pathogen (Table 4). This could be influenced by the conditions during anaerobic digestion, as observed in other studies, as well as by the storage conditions [35,37]. The presence of pathogens in the anaerobic digestate emphasizes the importance of conducting thorough microbiological evaluations to assess potential health risks for humans and animals [35]. Regarding the effects of anaerobic digestate applied to soil or plants, it is known that microbial populations associated with manure vary significantly depending on the animal source, its feeding regime, and waste management practices, among others [38,39]. The application of manure and anaerobic digestate can enrich the soil with pathogens, which poses a potential risk to crops, livestock, and humans, especially in areas with intensive agricultural practices [40]. In our study, E. coli and Salmonella were detected in the phyllosphere of maize 2 days after the last irrigation with anaerobic digestate (2 dai), but not in the second sampling (4 wai) nor in the non-irrigated crops (Table 4). The results of our research may suggest that the presence of these pathogens is transient and potentially linked to the immediate effects of fertigation with anaerobic digestate, with their prevalence decreasing over time. Nevertheless, even though the prevalence of E. coli and Salmonella appears to decline over time, their short-term detection may still pose a temporary risk that requires careful consideration in terms of management and food safety. Notably, the absence of these pathogens at four weeks should not be interpreted as a lack of risk, since harvest in commercial systems may occur earlier, during the period when pathogens were still detectable.
The antibiotics administered to livestock are often excreted in their feces, and traces of these antibiotics can be detected in the fecal anaerobic fermentation byproducts and in the soil, promoting the increment of ARGs [4,14]. The tetW gene has frequently been reported as persistent in anaerobic digestates derived from different feedstocks, showing undesirable removal efficiencies or even increments in its abundance compared to raw manure [41,42]. Our observations suggested an enrichment of tetW in AD maize samples. This may be linked to the fact that, during the trial, the animals in the feedlot were administered a tetracycline-based antibiotic. Since the increase in the abundance of this gene in maize phyllosphere was only observed at the first sampling (2 dai, Table 5 and Table S1, Figure S2), this transient rise may be attributed to short-term enrichment from the anaerobic digestate. While tetW has been detected in soils treated with organic amendments [43,44,45], the risk of its transfer to the phyllosphere has received little attention. To our knowledge, no previous studies have reported the presence of tetW in the phyllosphere.
The gene ermB, together with the blaCTX-M gene, is among the recommended targets for monitoring resistance [46]. In addition, they have been classified as posing the highest risk for human health based on several factors, including their presence in human-associated environments, their ability to spread through MGEs, and their prevalence in many pathogens [3,47]. Regarding ermB, our observations did not indicate notable differences between treated and untreated crops or across sampling dates (Table 5). Although previous studies have reported the environmental persistence of ermB in soils [48,49], the phyllosphere may not sustain comparable microbial dynamics. Consequently, the addition of anaerobic digestate might have little influence on the occurrence of this gene in plant tissues. Even though macrolides were administered to the animals in the feedlot, potentially introducing ermB-carrying bacteria through manure, no evident changes were detected in our study. This could be related to low concentrations of antibiotic residues in the manure or digestate, insufficient to affect ermB in the phyllosphere, or to a rapid degradation of the gene in this environment [45]. Similarly, no notable differences in blaCTX-M were observed between treatments or across sampling dates (Table 5). Previous studies have reported contrasting results regarding blaCTX-M responses to the application of animal-derived fertilizers. While some studies observed increases in its relative abundance in treated soils [15,50], others found no clear effects. For instance, Allegrini et al. (2024) reported stable blaCTX-M levels in the rhizosphere of ryegrass following anaerobic digestate application even after 190 days of cultivation [45]. Additionally, it is important to note that blaCTX-M represents a family of resistance genes, with M being just one of its many subtypes. Other blaCTX variants were not assessed; therefore, our observations do not exclude the presence or persistence of other blaCTX subtypes, and, given the lack of field replication, should be interpreted as preliminary trends rather than generalizable patterns.
Among the three documented classes of integrons, class 1 (intI1) has been found mostly among clinical and environmental isolates. Class 1 integrons have been detected in soils treated with both raw and digested manures, as well as in the rhizosphere of plants fertilized with these materials [14,45,49]. We evaluated the presence and persistence of intI1 and a-int1, a specific type of class 1 integron that is primarily associated with antibiotic resistance in environments impacted by human activities [30]. While no differences were apparent between irrigated and non-irrigated crops, nor across sampling dates for intI1, a-int1 displayed a different pattern (Table 5 and Table S1). In particular, a trend toward lower abundance of this integron was observed in the AD phyllosphere at 4 wai (Table 5 and Table S1). Although somewhat unexpected, this result could be explained by considering the changes in the environmental conditions within the phyllosphere of the treated plants, which may differ from those of the control crop. In plants treated with anaerobic digestate, which involves the addition of organic matter and microbial load, there is potential for increased microbial competition [51]. The introduction of a complex mixture of microorganisms could alter the composition of the microbial community, potentially influencing the behavior of specific microorganisms associated with the integron. Furthermore, the effects of anaerobic digestate on the phyllosphere are likely to vary over time [45], as natural processes, including microbial degradation and plant physiological responses, may progressively reduce the abundance of the a-int1 integron.
The benefits of using anaerobic digestates as organic fertilizers have been widely recognized [12,13,37]. However, while they could improve soil health and crop productivity, their use also presents potential risks related to the persistence and spread of pathogens and ARGs, which should not be overlooked. Our study highlights the importance of evaluating the microbiological quality of anaerobic digestate both before and after its application to soil or crops to minimize the risks of pathogen and ARGs transmission. As agricultural systems increasingly rely on organic amendments like anaerobic digestates, it is crucial to consider better practices for managing and reducing the risks associated with their use. Further research is needed to find a balance between the benefits of the obtention and use of anaerobic digestates and the potential environmental and public health concerns, ensuring their responsible and sustainable use.
Limitations and Scope of the Study
This study was conducted under real-world conditions on a working commercial farm, which provides practical relevance but also entails design constraints. Only one digestate-treated field and one control field were available, with inherent differences in maize variety, planting density, and soil preparation. Maize leaves from three composite transects were sampled per field, but these represent subsamples rather than true replicates. Consequently, the study design does not permit statistical inference or broad generalization beyond the specific site and timeframe examined. Observed patterns in microbial markers may also reflect site-specific variables not captured in the design, and therefore, p-values should be viewed as exploratory.
Our intent was not to present a replicated field trial, but to document trends under commercial conditions and generate hypotheses for future research. Given that the work was restricted to a single farm, crop, climate, and type of digestate, the findings should be regarded as context-dependent and hypothesis-generating rather than conclusive.
5. Conclusions
Despite the aforementioned constraints, the study provides preliminary insights into short-term microbial dynamics of maize phyllosphere following digestate fertigation and highlights key parameters that warrant further investigation in future trials. The main observations include the transient detection of E. coli and Salmonella shortly after digestate application (2 dai) and a temporary increase in the tetW resistance gene in the maize phyllosphere, while other ARGs and integrons showed no notable changes. Overall, under the conditions of this study, fertigation with anaerobic digestate appears to pose minimal short-term impact on the phyllosphere, though these conclusions are based on limited data and should be interpreted with caution.
From an environmental and economic perspective, the use of on-site manure-based digestate contributes to circular agriculture and nutrient recycling, with potential benefits for soil health and crop productivity. Nevertheless, careful management is recommended to reduce potential microbial risks. Suggested best practices include microbiological assessment of the anaerobic digestate before application, monitoring of indicators of fecal contamination, pathogens, and ARGs, and careful consideration of the timing and method of fertigation to minimize transient contamination. In this regard, guidelines developed under the Nitrates Directive (91/676/EEC) recommend composting of solids and pasteurization of the liquid fraction to ensure regulatory compliance while preserving agronomic benefits [52].
Future research should therefore focus on replicated field trials to confirm the observations reported here, assess the fate of a wider spectrum of ARGs and mobile genetic elements, and evaluate their persistence through subsequent steps of the agricultural cycle, including various time points throughout the crop cycle and in silage fed to livestock. Integrating agronomic, microbiological, and regulatory perspectives will support the development of improved management strategies and promote the safe, sustainable use of digestates. This will align with One Health principles, balancing agricultural productivity with environmental and public health considerations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy15102398/s1, Figure S1. Photographic overview of the experimental site: (a) General view of the plant and the feedlot area, with the digestate lagoon visible in the lower left, (b) Irrigation area showing the circular pattern traced by the pivot (seen from a distance), and (c) Pivot in operation. Table S1. Differences in gene absolute abundance. p-values and degrees of freedom associated with the main effects (T, S and T × S). Due to the absence of true replication at the field level, statistical comparisons should be interpreted as indicative trends rather than confirmed differences. Table S2. Differences in gene absolute abundance. p-values and degrees of freedom associated with the main effects (T, S and T × S). Due to the absence of true replication at the field level, statistical comparisons should be interpreted as indicative trends rather than confirmed differences. Table S3. Mean values of relative abundance [log10 (gene copies µg−1 DNA)]. The mean values, standard errors of the mean (S.E.) and number of observations (n) are indicated. T represents the effect of fertilization treatment (Control vs. AD), S refers to the sampling time (2 dai vs. 4 wai), and T × S indicates the interaction between fertilization treatment and sampling. Boldface denotes the lowest mean values among treatments, indicating comparative trends. Figure S2. Changes in the abundance of the tetW gene between control and AD samples. Values are expressed as the mean of absolute abundances [log10 (gene copies µg−1 DNA)], with error bars representing the S.E. Because true field replication was not performed, statistical comparisons should be interpreted as indicative of trends rather than confirmed differences.
Author Contributions
Conceptualization, M.C.Z.; methodology, C.F., M.V.V., J.B. and G.A.I.; resources, M.C.Z. and G.A.I.; formal analysis and data curation, C.F., M.C.Z. and M.B.V.; writing—original draft preparation, C.F.; writing—review and editing, M.B.V., G.A.I., M.C.Z. and M.A.; supervision, project administration, and funding acquisition, M.C.Z. and M.B.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Agencia Nacional de Promoción Científica (ANPCyT, PICT 2020-02289), the Secretaría de Ciencia y Tecnología—UNS (PGI 24/A276), and the United States Department of Agriculture, USDA-NIFA (HATCH Grant, Nº. ILLU-802-978).
Data Availability Statement
Dataset is available on request from the authors.
Acknowledgments
C.F and M.V.V acknowledge CONICET for the fellowships granted. Artificial intelligence tools (ChatGPT, OpenAI, GPT-5) were used solely to improve the English language and readability of the manuscript. No AI tools were used for the generation of scientific content, data analysis, or interpretation.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ARG | Antibiotic Resistance Genes |
| AMR | Antimicrobial Resistance |
| WHO | World Health Organization |
| MGEs | Mobile genetic elements |
| MPN | Most Probable Number |
| AD | Sample fertigated with Digestate |
| dai | Days After Fertigation |
| wai | Weeks After Fertigation |
References
- Pitt, S.J.; Gunn, A. The One Health Concept. Br. J. Biomed. Sci. 2024, 81, 12366. [Google Scholar] [CrossRef]
- World Health Organization. Antimicrobial Resistance; Fact Sheet. WHO. 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 22 January 2025).
- Allegrini, M.; Zabaloy, M.C. Anaerobic digestates in agricultural soils: A systematic review of their effects on antibiotic resistance genes. Rev. Argent. Microbiol. 2024, 56, 394–401. [Google Scholar] [CrossRef]
- Huang, Z.; Hu, L.X.; Yang, J.B.; Liu, Y.S.; He, L.Y.; Zhao, J.L.; Ying, G.G. Comprehensive discovery and migration evaluation of antimicrobial drugs and their transformation products in a swine farm by target, suspect, and nontarget screening. Environ. Int. 2023, 181, 108304. [Google Scholar] [CrossRef]
- Samreen, A.I.; Malak, H.A.; Abulreesh, H.H. Environmental antimicrobial resistance and its drivers: A potential threat to public health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Hoelzer, K.; Wong, N.; Thomas, J.; Talkington, K.; Jungman, E.; Coukell, A. Antimicrobial drug use in food-producing animals and associated human health risks: What, and how strong, is the evidence? BMC Vet. Res. 2017, 13, 211. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.; Dantas, G. The shared antibiotic resistome of soil bacteria and human pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef] [PubMed]
- Ellen MacArthur Foundation. It’s Time for a Circular Economy. 2021. Available online: https://www.ellenmacarthurfoundation.org/topics/circular-economy-introduction/overview (accessed on 3 December 2024).
- Abad-Segura, E.; González-Zamar, M.D.; Belmonte-Ureña, L.J. Effects of circular economy policies on the environment and sustainable growth: Worldwide research. Sustainability 2020, 12, 5792. [Google Scholar] [CrossRef]
- Sathish Kumar, R.K.; Sasikumar, R.; Dhilipkumar, T. Exploiting agro-waste for cleaner production: A review focusing on biofuel generation, bio-composite production, and environmental considerations. J. Clean. Prod. 2024, 435, 140536. [Google Scholar] [CrossRef]
- Hollas, C.E.; Guedes Cubas do Amaral, K.; Valles Lange, M.; Mayumi Higarashi, M.; Radis Steinmetz, R.L.; Barros, E.C.; Ferronato Mariani, L.; Nakano, V.; Kunz, A.; Sanches-Pereira, A.; et al. Life cycle assessment of waste management from the Brazilian pig chain residues in two perspectives: Electricity and biomethane production. J. Clean. Prod. 2022, 354, 131654. [Google Scholar] [CrossRef]
- Iocoli, G.A.; Zabaloy, M.C.; Pasdevicelli, G.; Gómez, M.A. Use of biogas digestates obtained by anaerobic digestion and codigestion as fertilizers: Characterization, soil biological activity and growth dynamic of Lactuca sativa L. Sci. Total Environ. 2019, 647, 11–19. [Google Scholar] [CrossRef]
- Zoui, O.; Baroudi, M.; Drissi, S.; Abouabdillah, A.; Abd-Elkader, O.H.; Plavan, G.; Bourioug, M. Utilization of digestate as an organic manure in corn silage culture: An in-depth investigation of its profound influence on soil’s physicochemical properties, crop growth parameters, and agronomic performance. Agronomy 2023, 13, 1715. [Google Scholar] [CrossRef]
- Cao, H.; Jiao, Q.; Cheng, L.; Song, L.; Xun, M.; Yang, H. Occurrence and prevalence of antibiotic resistance genes in apple orchard after continual application of anaerobic fermentation residues of pig manure. Environ. Sci. Pollut. Res. Int. 2023, 30, 29229–29242. [Google Scholar] [CrossRef] [PubMed]
- Nõlvak, H.; Truu, M.; Kanger, K.; Tampere, M.; Espenberg, M.; Loit, E.; Raave, H.; Truu, J. Inorganic and organic fertilizers impact the abundance and proportion of antibiotic resistance and integron integrase genes in agricultural grassland soil. Sci. Total Environ. 2016, 562, 678–689. [Google Scholar] [CrossRef]
- Methe, B.A.; Hiltbrand, D.; Roach, J.; Xu, W.; Gordon, S.G.; Goodner, B.W.; Stapleton, A.E. Functional gene categories differentiate maize leaf drought-related microbial epiphytic communities. PLoS ONE 2020, 15, e0237493. [Google Scholar] [CrossRef]
- Chen, Q.L.; An, X.L.; Zheng, B.X.; Ma, Y.B.; Su, J.Q. Long-term organic fertilization increased antibiotic resistome in phyllosphere of maize. Sci. Total Environ. 2018, 645, 1230–1237. [Google Scholar] [CrossRef]
- Kaul, J.; Jain, K.; Olakh, O. An overview on role of yellow maize in food, feed and nutrition security. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 3037–3048. [Google Scholar] [CrossRef]
- USDA Natural Resources Conservation Service. Soil Survey Staff. In Keys to Soil Taxonomy, 13th ed.; USDA Natural Resources Conservation Service: Washington, DC, USA, 2022. [Google Scholar]
- Cameron, A.; McAllister, T.A. Antimicrobial usage and resistance in beef production. J. Anim. Sci. Biotechnol. 2016, 7, 68. [Google Scholar] [CrossRef]
- Dinos, G.P. The macrolide antibiotic renaissance. Br. J. Pharmacol. 2017, 174, 2967–2983. [Google Scholar] [CrossRef] [PubMed]
- Norma Técnica Para la Aplicación Agrícola de Digerido Proveniente de Plantas de Digestión Anaeróbica. Available online: https://servicios.infoleg.gob.ar/infolegInternet/anexos/315000-319999/319167/res19.pdf (accessed on 27 September 2025).
- Código Alimentario Argentino, Capítulo XI. Alimentos Vegetales. Available online: https://www.argentina.gob.ar/sites/default/files/capitulo_xi_vegetales_actualiz_2025-071.pdf (accessed on 27 September 2025).
- Zhu, B.; Chen, Q.; Chen, S.; Zhu, Y.G. Does organically produced lettuce harbor higher abundance of antibiotic resistance genes than conventionally produced? Environ. Int. 2017, 98, 152–159. [Google Scholar] [CrossRef]
- Pollock, J.; Muwonge, A.; Hutchings, M.R.; Mainda, G.; Bronsvoort, B.M.; Gally, D.; Corbishley, A. Resistance to change: AMR gene dynamics on a commercial pig farm with high antimicrobial usage. Sci. Rep. 2020, 10, 1708. [Google Scholar] [CrossRef]
- Aminov, R.I.; Chee-Sanford, J.C.; Garrigues, N.; Mehboob, A.; Mackie, R.I. Detection of tetracycline resistance genes by PCR methods. Methods Mol. Biol. 2004, 268, 3–13. [Google Scholar] [CrossRef]
- Szczepanowski, R.; Linke, B.; Krahn, I.; Gartemann, K.H.; Gützkow, T.; Eichler, W.; Pühler, A.; Schlüter, A. Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology 2009, 155, 2306–2319. [Google Scholar] [CrossRef] [PubMed]
- Marti, E.; Jofre, J.; Balcazar, J.L. Prevalence of antibiotic resistance genes and bacterial community composition in a river influenced by a wastewater treatment plant. PLoS ONE 2013, 8, e78906. [Google Scholar] [CrossRef]
- Barraud, O.; Baclet, M.C.; Denis, F.; Ploy, M.C. Quantitative multiplex real-time PCR for detecting class 1, 2 and 3 integrons. J. Antimicrob. Chemother. 2010, 65, 1642–1645. [Google Scholar] [CrossRef] [PubMed]
- Quintela-Baluja, M.; Frigon, D.; Abouelnaga, M.; Jobling, K.; Romalde, J.L.; Gomez Lopez, M.; Graham, D.W. Dynamics of integron structures across a wastewater network—Implications to resistance gene transfer. Water Res. 2021, 206, 117720. [Google Scholar] [CrossRef]
- R—“Pile of Leaves”, version 4.4.2; The R Foundation for Statistical Computing: Vienna, Austria, 2024.
- Reemts, C.M.; Hansen, L.L. Short-term effects of repeated wildfires in oak-juniper woodlands. Fire Ecol. 2013, 9, 64–79. [Google Scholar] [CrossRef]
- Bicudo, J.R.; Goyal, S.M. Pathogens and manure management systems: A review. Environ. Technol. 2003, 24, 115–130. [Google Scholar] [CrossRef]
- Spencer, J.L.; Guan, J. Public health implications related to spread of pathogens in manure from livestock and poultry operations. Methods Mol. Biol. 2004, 268, 503–515. [Google Scholar] [CrossRef]
- Burch, T.R.; Spencer, S.K.; Borchardt, S.S.; Larson, R.A.; Borchardt, M.A. Fate of manure-borne pathogens during anaerobic digestion and solids separation. J. Environ. Qual. 2018, 47, 336–344. [Google Scholar] [CrossRef]
- Gantzer, C.; Gaspard, P.; Galvez, L.; Huyard, A.; Dumouthier, N.; Schwartzbrod, J. Monitoring of bacterial and parasitological contamination during various treatments of sludge. Water Res. 2001, 35, 3763–3770. [Google Scholar] [CrossRef]
- Ndubuisi-Nnaji, U.; Ofon, U.; Asira, A.E.; Dickson, N. Anaerobic digestion of untreated manure: Environmental risk assessment of resultant digestates. World J. Appl. Sci. Technol. 2023, 14, 73–79. [Google Scholar] [CrossRef]
- Albihn, A.; Vinnerås, B. Biosecurity and arable use of manure and biowaste—Treatment alternatives. Livest. Sci. 2007, 112, 232–239. [Google Scholar] [CrossRef]
- Lopatto, E.; Choi, J.; Colina, A.; Ma, L.; Howe, A.; Hinsa-Leasure, S. Characterizing the soil microbiome and quantifying antibiotic resistance gene dynamics in agricultural soil following swine CAFO manure application. PLoS ONE 2019, 14, e0220770. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Lv, T.; Shi, M.; Wu, S.; Carvalho, P.N.; Dong, R. Stabilization of preliminary anaerobically digested slurry in post-storage: Dynamics of chemical characteristics and hygienic quality. Water Air Soil Pollut. 2017, 306, 3493. [Google Scholar] [CrossRef]
- Ben, W.; Wang, J.; Cao, R.; Yang, M.; Zhang, Y.; Qiang, Z. Distribution of antibiotic resistance in the effluents of ten municipal wastewater treatment plants in China and the effect of treatment processes. Chemosphere 2017, 172, 392–398. [Google Scholar] [CrossRef]
- Chen, X.; Tang, R.; Wang, Y.; Yuan, S.; Wang, W.; Ali, I.M.; Hu, Z.H. Effect of ultrasonic and ozone pretreatment on the fate of enteric indicator bacteria and antibiotic resistance genes, and anaerobic digestion of dairy wastewater. Bioresour. Technol. 2021, 320, 124356. [Google Scholar] [CrossRef] [PubMed]
- Pu, C.; Liu, H.; Ding, G.; Sun, Y.; Yu, X.; Chen, J.; Ren, J.; Gong, X. Impact of direct application of biogas slurry and residue in fields: In situ analysis of antibiotic resistance genes from pig manure to fields. J. Hazard. Mater. 2018, 344, 441–449. [Google Scholar] [CrossRef]
- Tran, T.T.; Scott, A.; Tien, Y.C.; Murray, R.; Boerlin, P.; Pearl, D.L.; Liu, K.; Robertson, J.; Nash, J.H.E.; Topp, E. On-farm anaerobic digestion of dairy manure reduces the abundance of antibiotic resistance associated gene targets and the potential for plasmid transfer. Appl. Environ. Microbiol. 2021, 87, e0298020. [Google Scholar] [CrossRef]
- Allegrini, M.; Iocoli, G.A.; Zabaloy, M.C. Combined use of digestate and inorganic fertilizer alleviates the burden of class 1 integrons in perennial ryegrass rhizosphere without compromising aerial biomass production. Environ. Sci. Pollut. Res. Int. 2024, 31, 47132–47143. [Google Scholar] [CrossRef]
- Luby, E.; Ibekwe, A.M.; Zilles, J.; Pruden, A. Molecular methods for assessment of antibiotic resistance in agricultural ecosystems: Prospects and challenges. J. Environ. Qual. 2016, 45, 441–453. [Google Scholar] [CrossRef]
- Zhang, A.N.; Gaston, J.M.; Dai, C.L.; Zhao, S.; Poyet, M.; Groussin, M.; Yin, X.; Li, L.G.; van Loosdrecht, M.C.M.; Topp, E.; et al. An omics-based framework for assessing the health risk of antimicrobial resistance genes. Nat. Commun. 2021, 12, 4765. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.; Tien, Y.C.; Drury, C.F.; Reynolds, W.D.; Topp, E. Enrichment of antibiotic resistance genes in soil receiving composts derived from swine manure, yard wastes, or food wastes, and evidence for multiyear persistence of swine Clostridium spp. Can. J. Microbiol. 2018, 64, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.Y.; Meng, M.; Qi, L.; Li, Y.; Yao, H. Environmental risks in swine biogas slurry-irrigated soils: A comprehensive analysis of antibiotic residues, resistome, and bacterial pathogens. Environ. Int. 2024, 191, 108954. [Google Scholar] [CrossRef]
- Nüesch-Inderbinen, M.; Stephan, R. Epidemiology of extended-spectrum β-lactamase producing Escherichia coli in the human-livestock environment. Curr. Clin. Microbiol. Rep. 2016, 3, 1–9. [Google Scholar] [CrossRef]
- De Mandal, S.; Jeon, J. Phyllosphere Microbiome in Plant Health and Disease. Plants 2023, 12, 3481. [Google Scholar] [CrossRef] [PubMed]
- Huygens, D.; Orveillon, G.; Lugato, E.; Tavazzi, S.; Comero, S.; Jones, A.; Gawlik, B.; Saveyn, H.G.M. Technical Proposals for the Safe Use of Processed Manure Above the Threshold Established for Nitrate Vulnerable Zones by the Nitrates Directive (91/676/EEC); Publications Office of the European Union: Luxembourg, 2020. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).