Genome-Wide Analysis of NAC Gene Family and Its Cold-Responsive Transcriptional Dynamics in Coffea arabica
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of NAC Genes in C. arabica
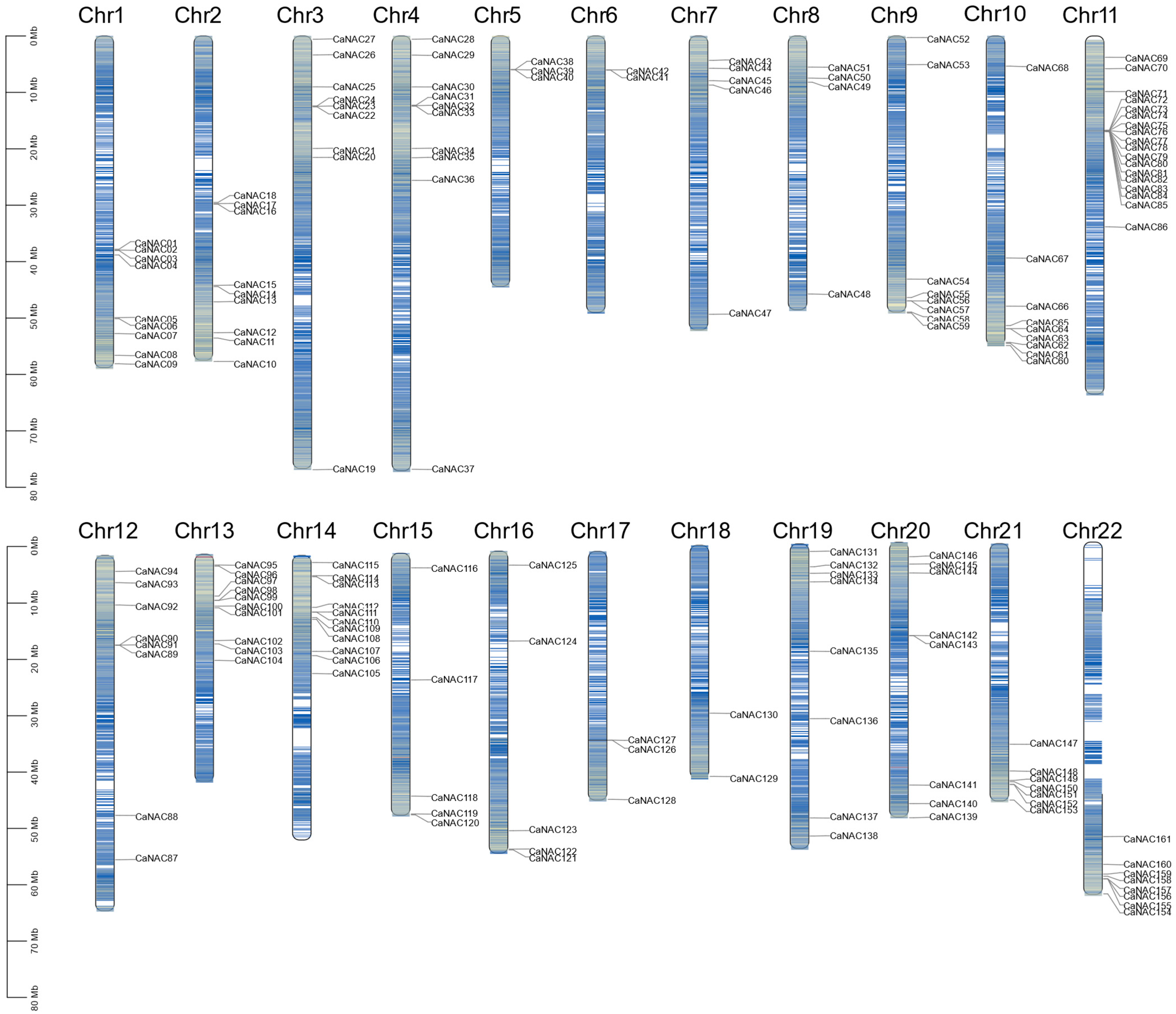
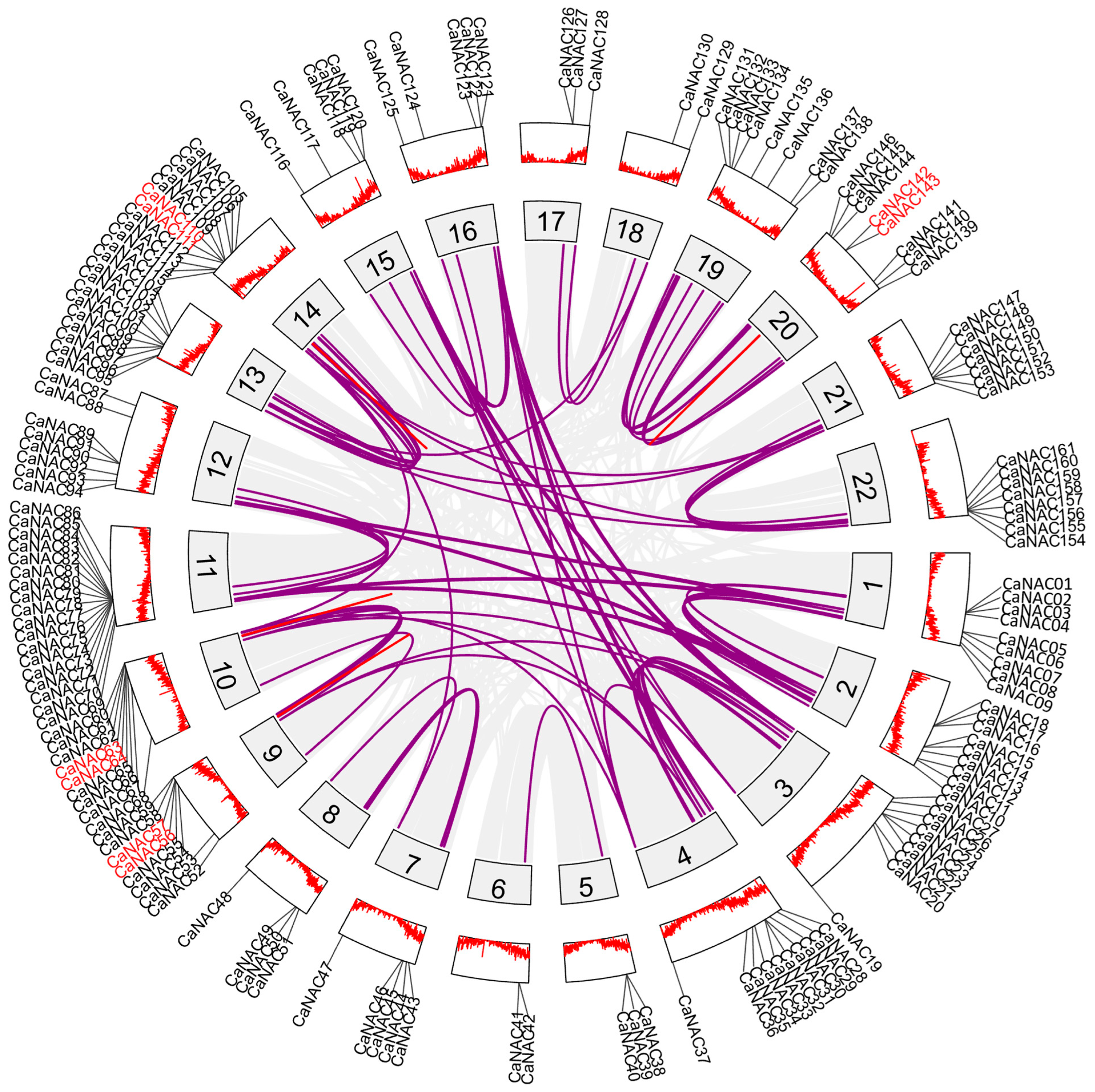
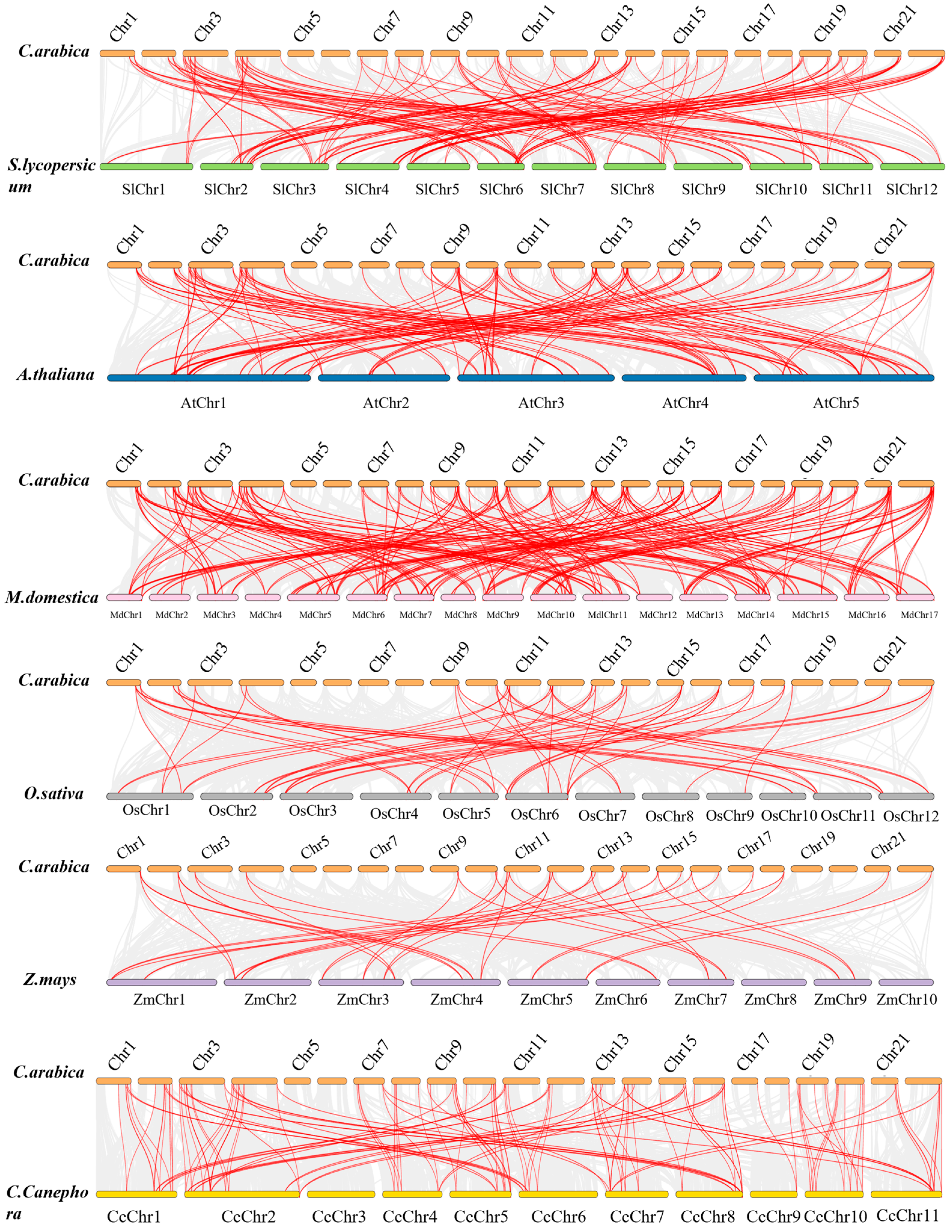
2.2. Chromosomal Distribution, Gene Duplication and Syntenic Analysis
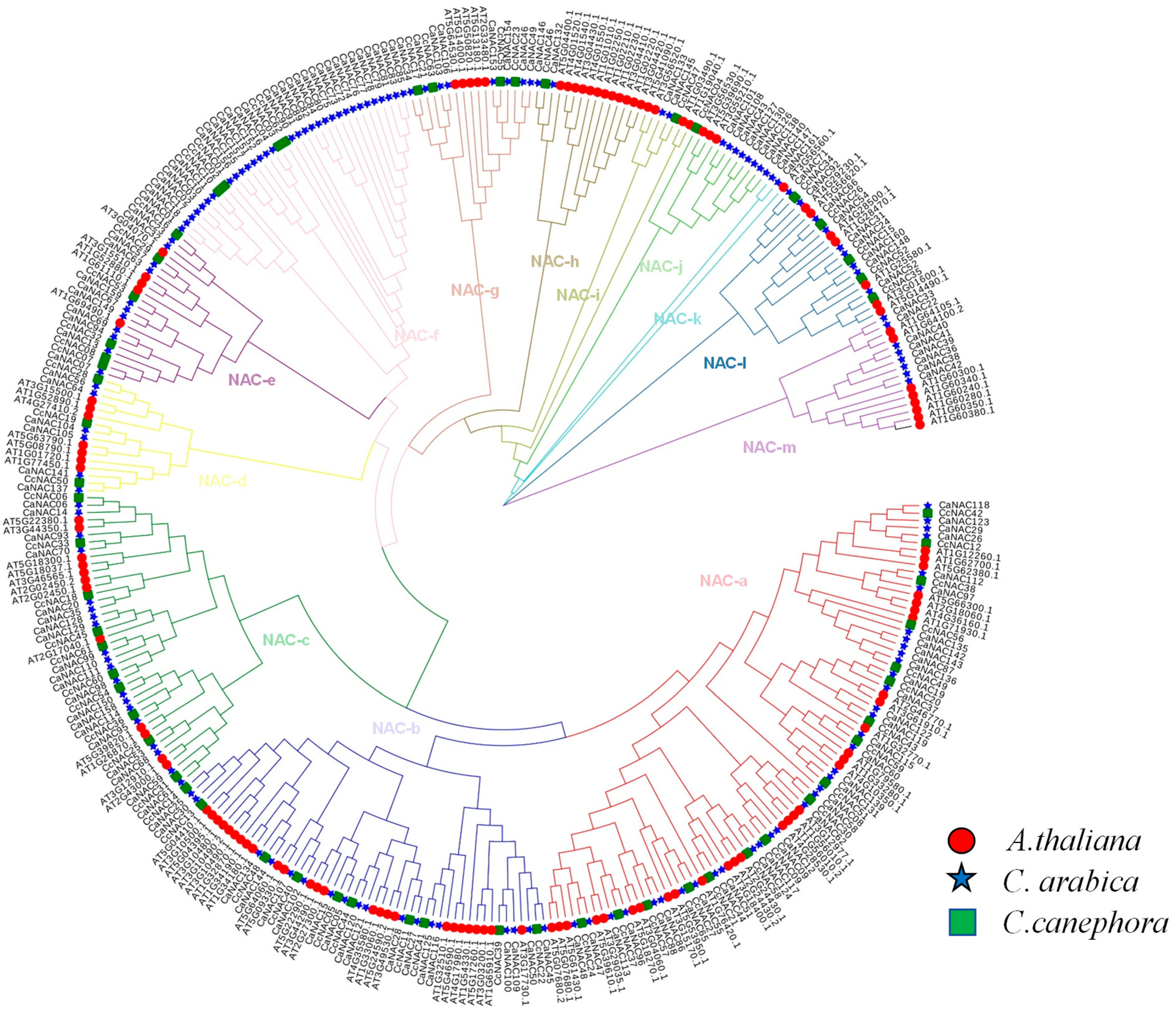
2.3. Sequence Alignment and Phylogenetic Analysis of CaNACs
2.4. Gene Structure, Conserved Motif, and Cis-Element Analyses of CaNACs
2.5. Plant Materials and RNA-Seq
2.6. Physiological Parameter Assays
2.7. cDNA Synthesis and qRT-PCR
3. Results
3.1. Identification and Characterization of NAC Genes in C. arabica
3.2. Chromosomal Locations, Gene Duplication, and Syntenic Analysis
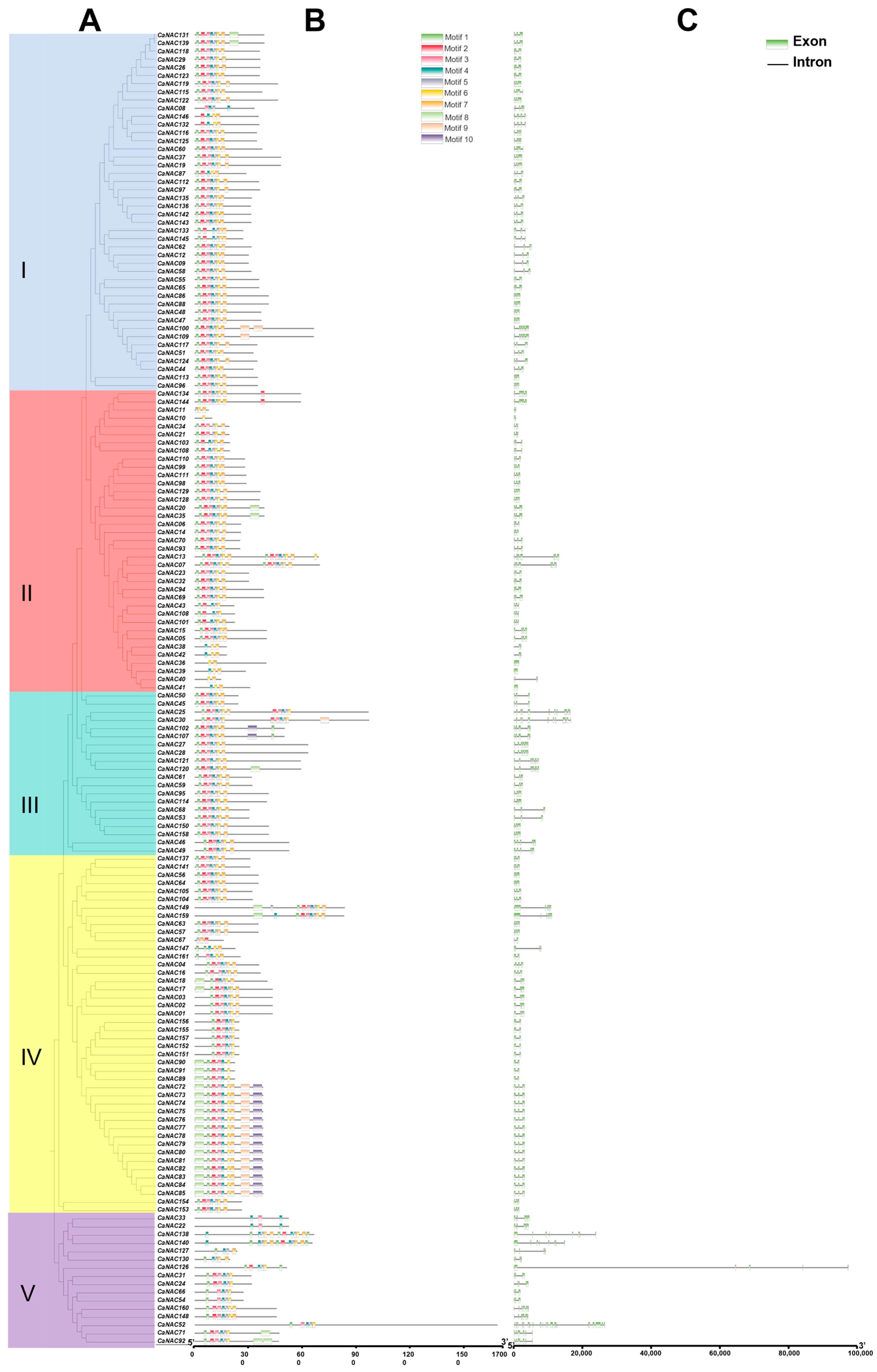
3.3. Phylogenetic Analysis of CaNACs
3.4. Analysis of CaNAC Gene Structure and Protein Domains
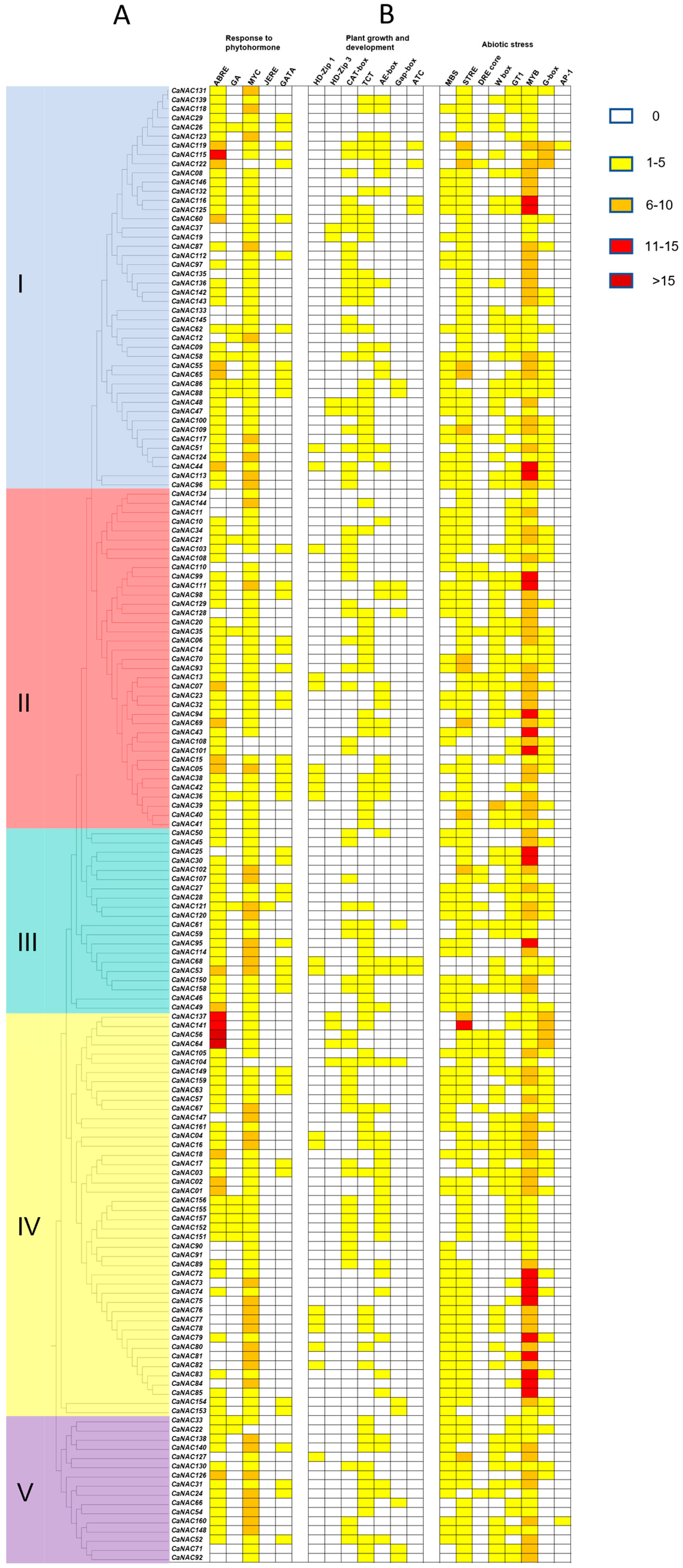
3.5. Cis-Element Analysis of CaNAC Genes
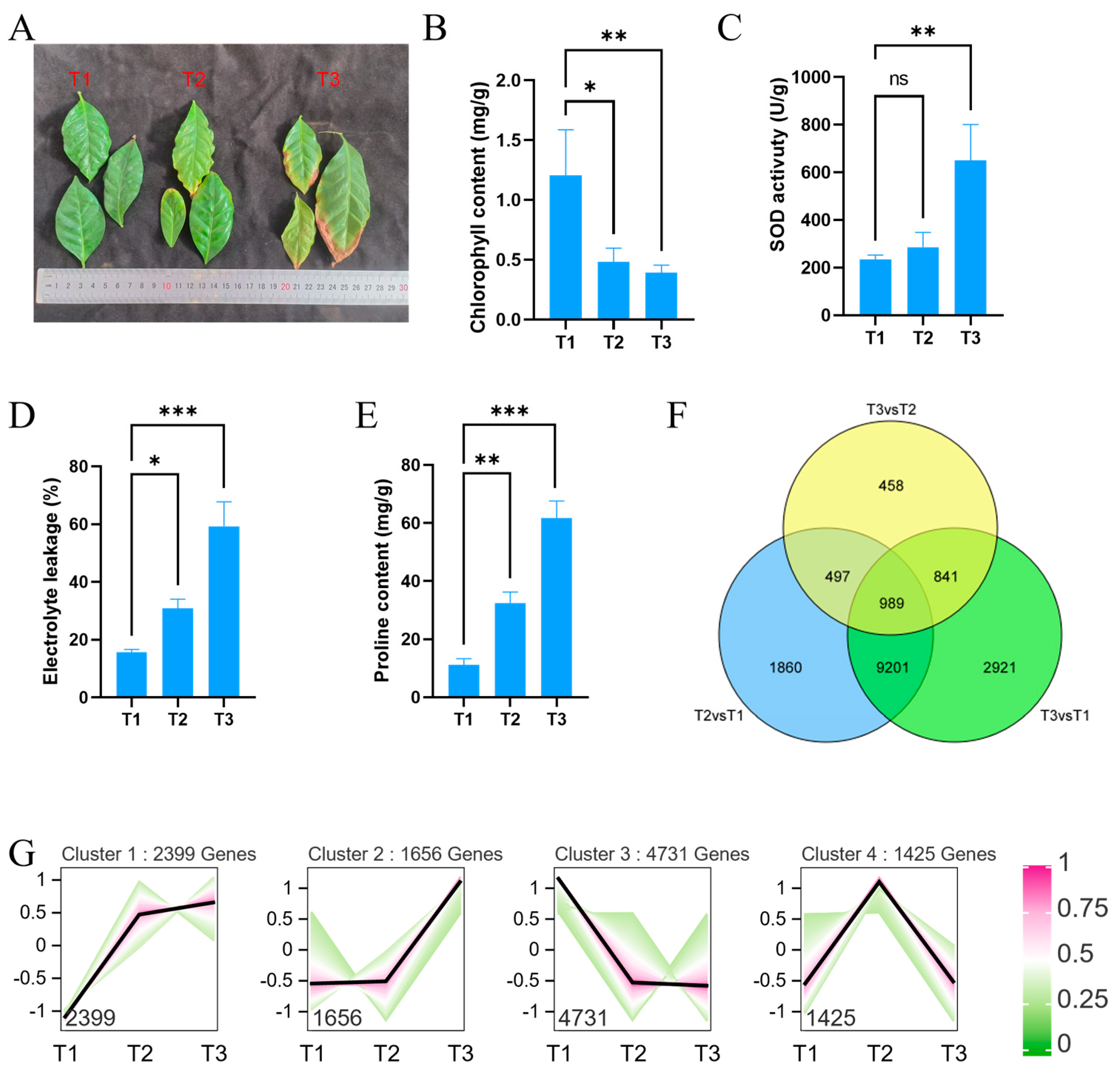
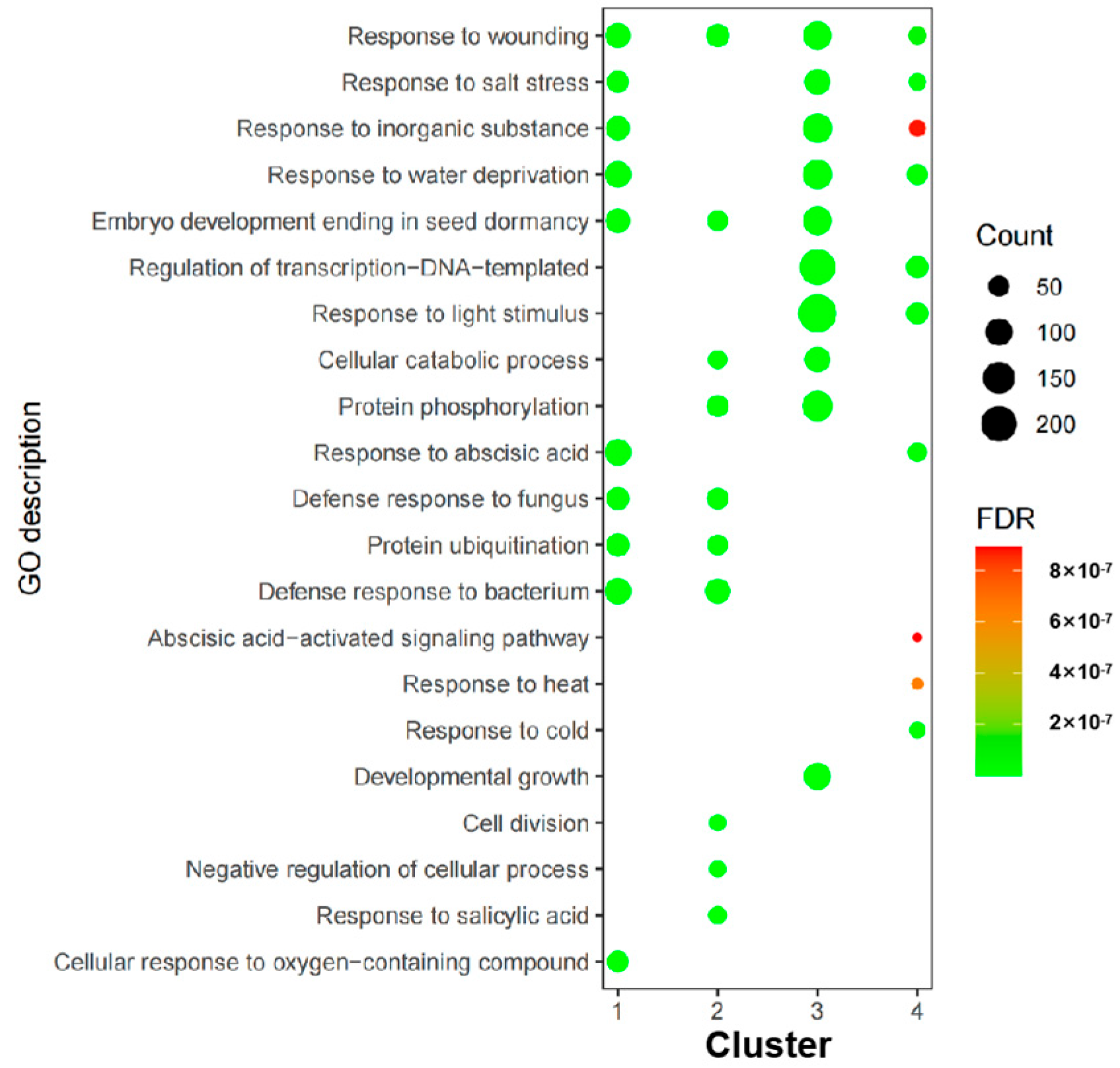
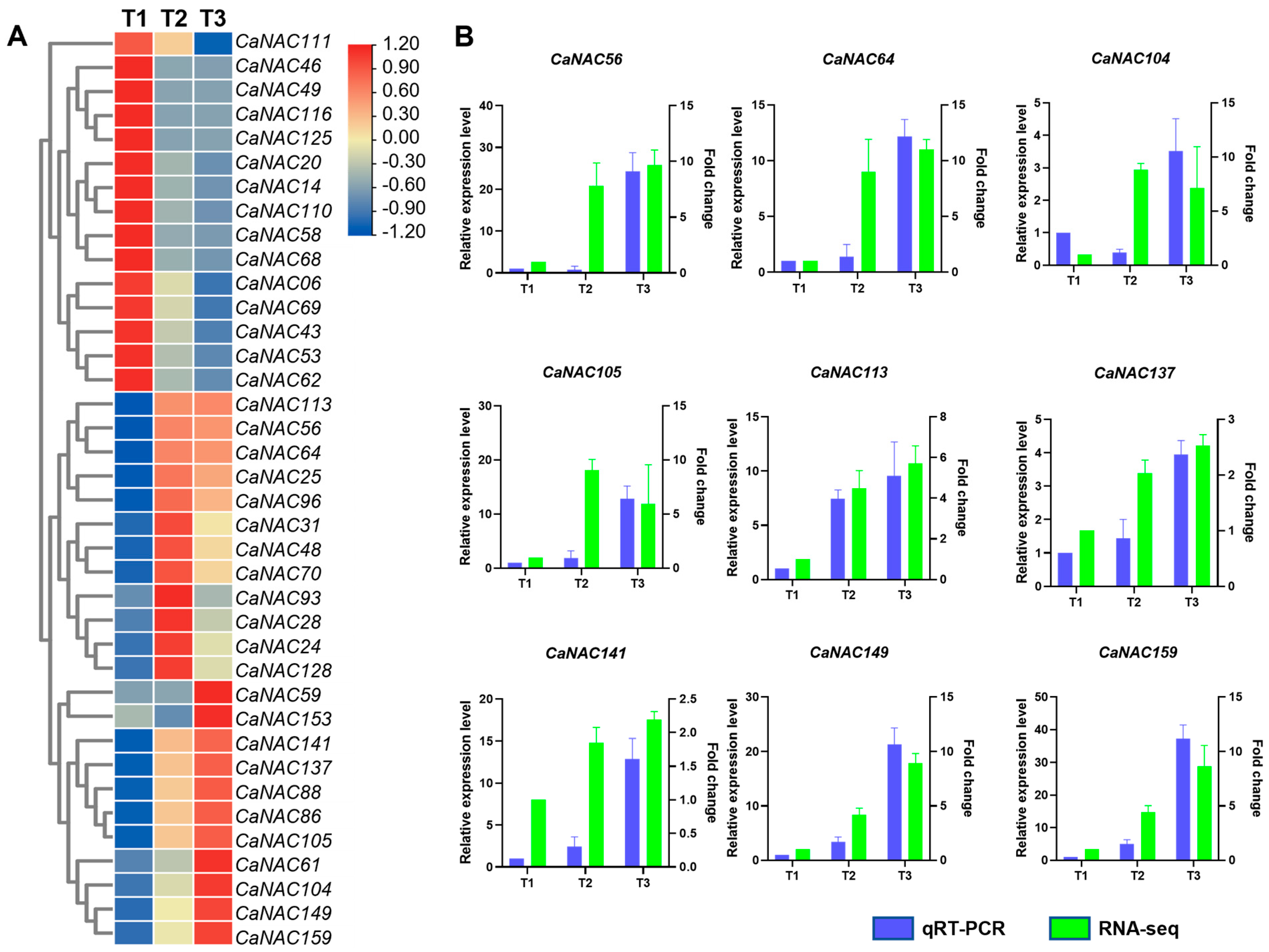
3.6. RNA-Seq Analysis of CaNAC Genes Under Low Temperature Conditions
4. Discussion
4.1. CaNAC Gene Identification and Evolutionary Analysis in C. arabica
4.2. Cis-Element Analysis Reveals the Roles of CaNACs
4.3. Identification of Cold-Responsive CaNAC Genes in C. arabica
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kadonaga, J.T. Eukaryotic Transcription: An Interlaced Network of Transcription Factors and Chromatin-Modifying Machines. Cell 1998, 92, 307–313. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis among Eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC Transcription Factors: Structurally Distinct, Functionally Diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC Proteins: Regulation and Role in Stress Tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Ernst, H.A.; Olsen, A.N.; Larsen, S.; Lo Leggio, L. Structure of the Conserved Domain of ANAC, a Member of the NAC Family of Transcription Factors. EMBO Rep. 2004, 5, 297–303. [Google Scholar] [CrossRef]
- Huang, G.; Dong, B.; Jiang, J.; Chen, S.; Fang, W.; Liu, Y.; Huang, G.; Dong, B.; Jiang, J.; Chen, S.; et al. CmNAC083 Regulates Resistance to Alternaria alternata via Reactive Oxygen Species and Jasmonic Acid Signaling Pathways in Chrysanthemum morifolium. Ornam. Plant Res. 2023, 3, 16. [Google Scholar] [CrossRef]
- Zhang, H.; Cui, X.; Guo, Y.; Luo, C.; Zhang, L. Picea Wilsonii Transcription Factor NAC2 Enhanced Plant Tolerance to Abiotic Stress and Participated in RFCP1-Regulated Flowering Time. Plant Mol. Biol. 2018, 98, 471–493. [Google Scholar] [CrossRef] [PubMed]
- Pimenta, M.R.; Silva, P.A.; Mendes, G.C.; Alves, J.R.; Caetano, H.D.N.; Machado, J.P.B.; Brustolini, O.J.B.; Carpinetti, P.A.; Melo, B.P.; Silva, J.C.F.; et al. The Stress-Induced Soybean NAC Transcription Factor GmNAC81 Plays a Positive Role in Developmentally Programmed Leaf Senescence. Plant Cell Physiol. 2016, 57, 1098–1114. [Google Scholar] [CrossRef]
- Tran, L.-S.P.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and Functional Analysis of Arabidopsis Stress-Inducible NAC Transcription Factors That Bind to a Drought-Responsive Cis-Element in the Early Responsive to Dehydration Stress 1 Promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Ying, S.; Zhang, D.-F.; Shi, Y.-S.; Song, Y.-C.; Wang, T.-Y.; Li, Y. A Maize Stress-Responsive NAC Transcription Factor, ZmSNAC1, Confers Enhanced Tolerance to Dehydration in Transgenic Arabidopsis. Plant Cell Rep. 2012, 31, 1701–1711. [Google Scholar] [CrossRef]
- Ma, N.-N.; Zuo, Y.-Q.; Liang, X.-Q.; Yin, B.; Wang, G.-D.; Meng, Q.-W. The Multiple Stress-Responsive Transcription Factor SlNAC1 Improves the Chilling Tolerance of Tomato. Physiol. Plant. 2013, 149, 474–486. [Google Scholar] [CrossRef] [PubMed]
- Song, S.-Y.; Chen, Y.; Chen, J.; Dai, X.-Y.; Zhang, W.-H. Physiological Mechanisms Underlying OsNAC5-Dependent Tolerance of Rice Plants to Abiotic Stress. Planta 2011, 234, 331–345. [Google Scholar] [CrossRef]
- Nakashima, K.; Tran, L.-S.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional Analysis of a NAC-Type Transcription Factor OsNAC6 Involved in Abiotic and Biotic Stress-Responsive Gene Expression in Rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef]
- Redillas, M.C.F.R.; Jeong, J.S.; Kim, Y.S.; Jung, H.; Bang, S.W.; Choi, Y.D.; Ha, S.H.; Reuzeau, C.; Kim, J.K. The Overexpression of OsNAC9 Alters the Root Architecture of Rice Plants Enhancing Drought Resistance and Grain Yield under Field Conditions. Plant Biotechnol. J. 2012, 10, 792–805. [Google Scholar] [CrossRef]
- Jeong, J.S.; Kim, Y.S.; Baek, K.H.; Jung, H.; Ha, S.H.; Do Choi, Y.; Kim, M.; Reuzeau, C.; Kim, J.-K. Root-Specific Expression of OsNAC10 Improves Drought Tolerance and Grain Yield in Rice under Field Drought Conditions. Plant Physiol. 2010, 153, 185–197. [Google Scholar] [CrossRef]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Lang, Z.; Zhu, J.-K. Cold Responsive Gene Transcription Becomes More Complex. Trends Plant Sci. 2015, 20, 466–468. [Google Scholar] [CrossRef]
- Li, S.; He, L.; Yang, Y.; Zhang, Y.; Han, X.; Hu, Y.; Jiang, Y. INDUCER OF CBF EXPRESSION 1 Promotes Cold-Enhanced Immunity by Directly Activating Salicylic Acid Signaling. Plant Cell 2024, 36, 2587–2606. [Google Scholar] [CrossRef]
- Hwarari, D.; Guan, Y.; Ahmad, B.; Movahedi, A.; Min, T.; Hao, Z.; Lu, Y.; Chen, J.; Yang, L. ICE-CBF-COR Signaling Cascade and Its Regulation in Plants Responding to Cold Stress. Int. J. Mol. Sci. 2022, 23, 1549. [Google Scholar] [CrossRef] [PubMed]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 Overexpression Induces COR Genes and Enhances Freezing Tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef]
- Zhang, L.; Xing, L.; Dai, J.; Li, Z.; Zhang, A.; Wang, T.; Liu, W.; Li, X.; Han, D. Overexpression of a Grape WRKY Transcription Factor VhWRKY44 Improves the Resistance to Cold and Salt of Arabidopsis Thaliana. Int. J. Mol. Sci. 2024, 25, 7437. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Tan, Y.; Wei, R.; Wang, X.; Mubeen, S.; Chen, C.; Cao, S.; Wang, C.; Chen, P. Genome-Wide Identification of bHLH Transcription Factors in Kenaf (Hibiscus cannabinus L.) and Gene Function Analysis of HcbHLH88. Physiol. Mol. Biol. Plants 2024, 30, 1517–1532. [Google Scholar] [CrossRef]
- An, J.-P.; Wang, X.-F.; Zhang, X.-W.; Xu, H.-F.; Bi, S.-Q.; You, C.-X.; Hao, Y.-J. An Apple MYB Transcription Factor Regulates Cold Tolerance and Anthocyanin Accumulation and Undergoes MIEL1-Mediated Degradation. Plant Biotechnol. J. 2020, 18, 337–353. [Google Scholar] [CrossRef]
- Shan, W.; Kuang, J.-F.; Lu, W.-J.; Chen, J.-Y. Banana Fruit NAC Transcription Factor MaNAC1 Is a Direct Target of MaICE1 and Involved in Cold Stress through Interacting with MaCBF1. Plant Cell Environ. 2014, 37, 2116–2127. [Google Scholar] [CrossRef]
- Hou, X.-M.; Zhang, H.-F.; Liu, S.-Y.; Wang, X.-K.; Zhang, Y.-M.; Meng, Y.-C.; Luo, D.; Chen, R.-G. The NAC Transcription Factor CaNAC064 Is a Regulator of Cold Stress Tolerance in Peppers. Plant Sci. 2020, 291, 110346. [Google Scholar] [CrossRef]
- An, J.-P.; Li, R.; Qu, F.-J.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. An Apple NAC Transcription Factor Negatively Regulates Cold Tolerance via CBF-Dependent Pathway. J. Plant Physiol. 2018, 221, 74–80. [Google Scholar] [CrossRef]
- Jin, C.; Li, K.-Q.; Xu, X.-Y.; Zhang, H.-P.; Chen, H.-X.; Chen, Y.-H.; Hao, J.; Wang, Y.; Huang, X.-S.; Zhang, S.-L. A Novel NAC Transcription Factor, PbeNAC1, of Pyrus Betulifolia Confers Cold and Drought Tolerance via Interacting with PbeDREBs and Activating the Expression of Stress-Responsive Genes. Front. Plant Sci. 2017, 8, 1049. [Google Scholar] [CrossRef]
- Herrera, J.C.; Combes, M.C.; Anthony, F.; Charrier, A.; Lashermes, P. Introgression into the Allotetraploid Coffee (Coffea arabica L.): Segregation and Recombination of the C. Canephora Genome in the Tetraploid Interspecific Hybrid (C. arabicax C. canephora). Theor. Appl. Genet. 2002, 104, 661–668. [Google Scholar] [CrossRef] [PubMed]
- DaMatta, F.M.; Ramalho, J.D.C. Impacts of Drought and Temperature Stress on Coffee Physiology and Production: A Review. Braz. J. Plant Physiol. 2006, 18, 55–81. [Google Scholar] [CrossRef]
- de Sousa, G.F.; Silva, M.A.; de Morais, E.G.; Van Opbergen, G.A.Z.; Van Opbergen, G.G.A.Z.; de Oliveira, R.R.; Amaral, D.; Brown, P.; Chalfun-Junior, A.; Guilherme, L.R.G. Selenium Enhances Chilling Stress Tolerance in Coffee Species by Modulating Nutrient, Carbohydrates, and Amino Acids Content. Front. Plant Sci. 2022, 13, 1000430. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ye, T.; Fan, L.; Liu, X.; Zhang, M.; Zhu, Y.; Gole, T.W. Chill Topped Historical Arabica Coffee Yield Loss among Climate Stressors in Yunnan, China, Followed by Drought. npj Nat. Hazards 2025, 2, 32. [Google Scholar] [CrossRef]
- Salojärvi, J.; Rambani, A.; Yu, Z.; Guyot, R.; Strickler, S.; Lepelley, M.; Wang, C.; Rajaraman, S.; Rastas, P.; Zheng, C.; et al. The Genome and Population Genomics of Allopolyploid Coffea Arabica Reveal the Diversification History of Modern Coffee Cultivars. Nat. Genet. 2024, 56, 721–731. [Google Scholar] [CrossRef]
- Prakash, A.; Jeffryes, M.; Bateman, A.; Finn, R.D. The HMMER Web Server for Protein Sequence Similarity Search. Curr. Protoc. Bioinform. 2017, 60, 3.15.1–3.15.23. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc: A Package of Web Servers for Predicting Subcellular Localization of Proteins in Various Organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, T.; Chen, X.; Zhang, S.; Zhu, J.; Tang, B.; Wang, A.; Dong, L.; Zhang, Z.; Yu, C.; Sun, Y.; et al. The Genome Sequence Archive Family: Toward Explosive Data Growth and Diverse Data Types. Genom. Proteom. Bioinform. 2021, 19, 578–583. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce Acceleration-Supported Software for Integrated Quality Control and Preprocessing of High-Throughput Sequencing Data. Gigascience 2018, 7, 1–6. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide Dismutases: I. Occurrence in Higher Plants. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef]
- Maehly, A.C.; Chance, B. The Assay of Catalases and Peroxidases. Methods Biochem. Anal. 1954, 1, 357–424. [Google Scholar]
- Lichtenthaler, H.K. Chlorophylls and Carotenoids: Pigments of Photosynthetic Biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar]
- Dionisio-Sese, M.L.; Tobita, S. Antioxidant Responses of Rice Seedlings to Salinity Stress. Plant Sci. 1998, 135, 1–9. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid Determination of Free Proline for Water-Stress Studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-Wide Analysis of NAC Transcription Factor Family in Rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef]
- Singh, A.K.; Sharma, V.; Pal, A.K.; Acharya, V.; Ahuja, P.S. Genome-Wide Organization and Expression Profiling of the NAC Transcription Factor Family in Potato (Solanum tuberosum L.). DNA Res. 2013, 20, 403–423. [Google Scholar] [CrossRef]
- Li, W.; Li, X.; Chao, J.; Zhang, Z.; Wang, W.; Guo, Y. NAC Family Transcription Factors in Tobacco and Their Potential Role in Regulating Leaf Senescence. Front. Plant Sci. 2018, 9, 1900. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, H.; Yang, Y.; Tang, K.; Yang, Y.; Ouyang, W.; Du, G. Genome-Wide Identification of NAC Family Genes and Their Expression Analyses in Response to Osmotic Stress in Cannabis sativa L. Int. J. Mol. Sci. 2024, 25, 9466. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Mochida, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. Genome-Wide Survey and Expression Analysis of the Plant-Specific NAC Transcription Factor Family in Soybean During Development and Dehydration Stress. DNA Res. 2011, 18, 263–276. [Google Scholar] [CrossRef]
- Jin, J.F.; Wang, Z.Q.; He, Q.Y.; Wang, J.Y.; Li, P.F.; Xu, J.M.; Zheng, S.J.; Fan, W.; Yang, J.L. Genome-Wide Identification and Expression Analysis of the NAC Transcription Factor Family in Tomato (Solanum lycopersicum) during Aluminum Stress. BMC Genom. 2020, 21, 288. [Google Scholar] [CrossRef]
- Blanc, G.; Hokamp, K.; Wolfe, K.H. A Recent Polyploidy Superimposed on Older Large-Scale Duplications in the Arabidopsis Genome. Genome Res. 2003, 13, 137–144. [Google Scholar] [CrossRef]
- Denoeud, F.; Carretero-Paulet, L.; Dereeper, A.; Droc, G.; Guyot, R.; Pietrella, M.; Zheng, C.; Alberti, A.; Anthony, F.; Aprea, G.; et al. The Coffee Genome Provides Insight into the Convergent Evolution of Caffeine Biosynthesis. Science 2014, 345, 1181–1184. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Wang, H.; Gao, H.; Yu, S.; Liu, C.; Wang, Y.; Sun, Y.; Zhang, D. Alternative Splicing of Functional Genes in Plant Growth, Development, and Stress Responses. Int. J. Mol. Sci. 2025, 26, 5864. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Cao, Y.; Ma, L. Alternative Splicing in Plant Genes: A Means of Regulating the Environmental Fitness of Plants. Int. J. Mol. Sci. 2017, 18, 432. [Google Scholar] [CrossRef]
- Wang, Q.; Zhou, L.; Yuan, M.; Peng, F.; Zhu, X.; Wang, Y. Genome-Wide Identification of NAC Gene Family Members of Tree Peony (Paeonia suffruticosa Andrews) and Their Expression under Heat and Waterlogging Stress. Int. J. Mol. Sci. 2024, 25, 9312. [Google Scholar] [CrossRef]
- Yang, Q.; Li, Z.; Wang, X.; Jiang, C.; Liu, F.; Nian, Y.; Fu, X.; Zhou, G.; Liu, L.; Wang, H. Genome-Wide Identification and Characterization of the NAC Gene Family and Its Involvement in Cold Response in Dendrobium Officinale. Plants 2023, 12, 3626. [Google Scholar] [CrossRef]
- Liu, S.; Guan, Y.; Weng, Y.; Liao, B.; Tong, L.; Hao, Z.; Chen, J.; Shi, J.; Cheng, T. Genome-Wide Identification of the NAC Gene Family and Its Functional Analysis in Liriodendron. BMC Plant Biol. 2023, 23, 415. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and Validation of Promoters and Cis-Acting Regulatory Elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef]
- Peleke, F.F.; Zumkeller, S.M.; Gültas, M.; Schmitt, A.; Szymański, J. Deep Learning the Cis-Regulatory Code for Gene Expression in Selected Model Plants. Nat. Commun. 2024, 15, 3488. [Google Scholar] [CrossRef]
- Jin, M.; Gan, S.; Jiao, J.; He, Y.; Liu, H.; Yin, X.; Zhu, Q.; Rao, J. Genome-Wide Analysis of the bZIP Gene Family and the Role of AchnABF1 from Postharvest Kiwifruit (Actinidia chinensis Cv. Hongyang) in Osmotic and Freezing Stress Adaptations. Plant Sci. 2021, 308, 110927. [Google Scholar] [CrossRef]
- Li, Z.; Huang, Y.; Shen, Z.; Wu, M.; Huang, M.; Hong, S.-B.; Xu, L.; Zang, Y. Advances in Functional Studies of Plant MYC Transcription Factors. Theor. Appl. Genet. 2024, 137, 195. [Google Scholar] [CrossRef]
- Ohta, M.; Sato, A.; Renhu, N.; Yamamoto, T.; Oka, N.; Zhu, J.-K.; Tada, Y.; Suzaki, T.; Miura, K. MYC-Type Transcription Factors, MYC67 and MYC70, Interact with ICE1 and Negatively Regulate Cold Tolerance in Arabidopsis. Sci. Rep. 2018, 8, 11622. [Google Scholar] [CrossRef]
- Zhang, L.; Song, J.; Lin, R.; Tang, M.; Shao, S.; Yu, J.; Zhou, Y. Tomato SlMYB15 Transcription Factor Targeted by Sly-miR156e-3p Positively Regulates ABA-Mediated Cold Tolerance. J. Exp. Bot. 2022, 73, 7538–7551. [Google Scholar] [CrossRef]
- Acidri, R.; Sawai, Y.; Sugimoto, Y.; Handa, T.; Sasagawa, D.; Masunaga, T.; Yamamoto, S.; Nishihara, E. Exogenous Kinetin Promotes the Nonenzymatic Antioxidant System and Photosynthetic Activity of Coffee (Coffea arabica L.) Plants Under Cold Stress Conditions. Plants 2020, 9, 281. [Google Scholar] [CrossRef]
- Xiong, H.; He, H.; Chang, Y.; Miao, B.; Liu, Z.; Wang, Q.; Dong, F.; Xiong, L. Multiple Roles of NAC Transcription Factors in Plant Development and Stress Responses. J. Integr. Plant Biol. 2025, 67, 510–538. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Jiang, Y.; Yang, Y.; Xiao, Z.; Bai, X.; Gao, J.; Tan, S.; Hur, Y.; Hao, S.; He, F. Identification and Expression Analysis of the NAC Gene Family in Coffea canephora. Agronomy 2019, 9, 670. [Google Scholar] [CrossRef]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Tran, L.-S.P.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A Dehydration-Induced NAC Protein, RD26, Is Involved in a Novel ABA-Dependent Stress-Signaling Pathway. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef] [PubMed]
- Hickman, R.; Hill, C.; Penfold, C.A.; Breeze, E.; Bowden, L.; Moore, J.D.; Zhang, P.; Jackson, A.; Cooke, E.; Bewicke-Copley, F.; et al. A Local Regulatory Network around Three NAC Transcription Factors in Stress Responses and Senescence in A Rabidopsis Leaves. Plant J. 2013, 75, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Duan, A.-Q.; Yang, X.-L.; Feng, K.; Liu, J.-X.; Xu, Z.-S.; Xiong, A.-S. Genome-Wide Analysis of NAC Transcription Factors and Their Response to Abiotic Stress in Celery (Apium graveolens L.). Comput. Biol. Chem. 2020, 84, 107186. [Google Scholar] [CrossRef]
- Zhang, X.M.; Yu, H.J.; Sun, C.; Deng, J.; Zhang, X.; Liu, P.; Li, Y.Y.; Li, Q.; Jiang, W.J. Genome-Wide Characterization and Expression Profiling of the NAC Genes under Abiotic Stresses in Cucumis sativus. Plant Physiol. Biochem. 2017, 113, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-X.; Liu, Z.-W.; Wu, Z.-J.; Li, H.; Zhuang, J. Transcriptome-Wide Identification and Expression Analysis of the NAC Gene Family in Tea Plant [Camellia sinensis (L.) O. Kuntze]. PLoS ONE 2016, 11, e0166727. [Google Scholar] [CrossRef]
- Wu, Y.; Deng, Z.; Lai, J.; Zhang, Y.; Yang, C.; Yin, B.; Zhao, Q.; Zhang, L.; Li, Y.; Yang, C.; et al. Dual Function of Arabidopsis ATAF1 in Abiotic and Biotic Stress Responses. Cell Res. 2009, 19, 1279–1290. [Google Scholar] [CrossRef]
- Yang, X.; Wang, X.; Ji, L.; Yi, Z.; Fu, C.; Ran, J.; Hu, R.; Zhou, G. Overexpression of a Miscanthus lutarioriparius NAC Gene MlNAC5 Confers Enhanced Drought and Cold Tolerance in Arabidopsis. Plant Cell Rep. 2015, 34, 943–958. [Google Scholar] [CrossRef]
- Sharma, R.; Singh, G.; Bhattacharya, S.; Singh, A. Comparative Transcriptome Meta-Analysis of Arabidopsis thaliana under Drought and Cold Stress. PLoS ONE 2018, 13, e0203266. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Ma, W.; Feng, H.; Cai, W. NAC056 Transcription Factor Confers Freezing Tolerance by Positively Regulating the Expression of CBFs and NIA1 in Arabidopsis. Plant Commun. 2024, 5, 100923. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; Zhu, J.; Lan, Z.; He, F.; Dong, X. Genome-Wide Analysis of NAC Gene Family and Its Cold-Responsive Transcriptional Dynamics in Coffea arabica. Agronomy 2025, 15, 2394. https://doi.org/10.3390/agronomy15102394
Gao J, Zhu J, Lan Z, He F, Dong X. Genome-Wide Analysis of NAC Gene Family and Its Cold-Responsive Transcriptional Dynamics in Coffea arabica. Agronomy. 2025; 15(10):2394. https://doi.org/10.3390/agronomy15102394
Chicago/Turabian StyleGao, Jing, Junjie Zhu, Zenan Lan, Feifei He, and Xiangshu Dong. 2025. "Genome-Wide Analysis of NAC Gene Family and Its Cold-Responsive Transcriptional Dynamics in Coffea arabica" Agronomy 15, no. 10: 2394. https://doi.org/10.3390/agronomy15102394
APA StyleGao, J., Zhu, J., Lan, Z., He, F., & Dong, X. (2025). Genome-Wide Analysis of NAC Gene Family and Its Cold-Responsive Transcriptional Dynamics in Coffea arabica. Agronomy, 15(10), 2394. https://doi.org/10.3390/agronomy15102394





