Advances of QTL Localization and GWAS Application in Crop Resistances Against Plant-Parasitic Nematodes
Abstract
1. Introduction
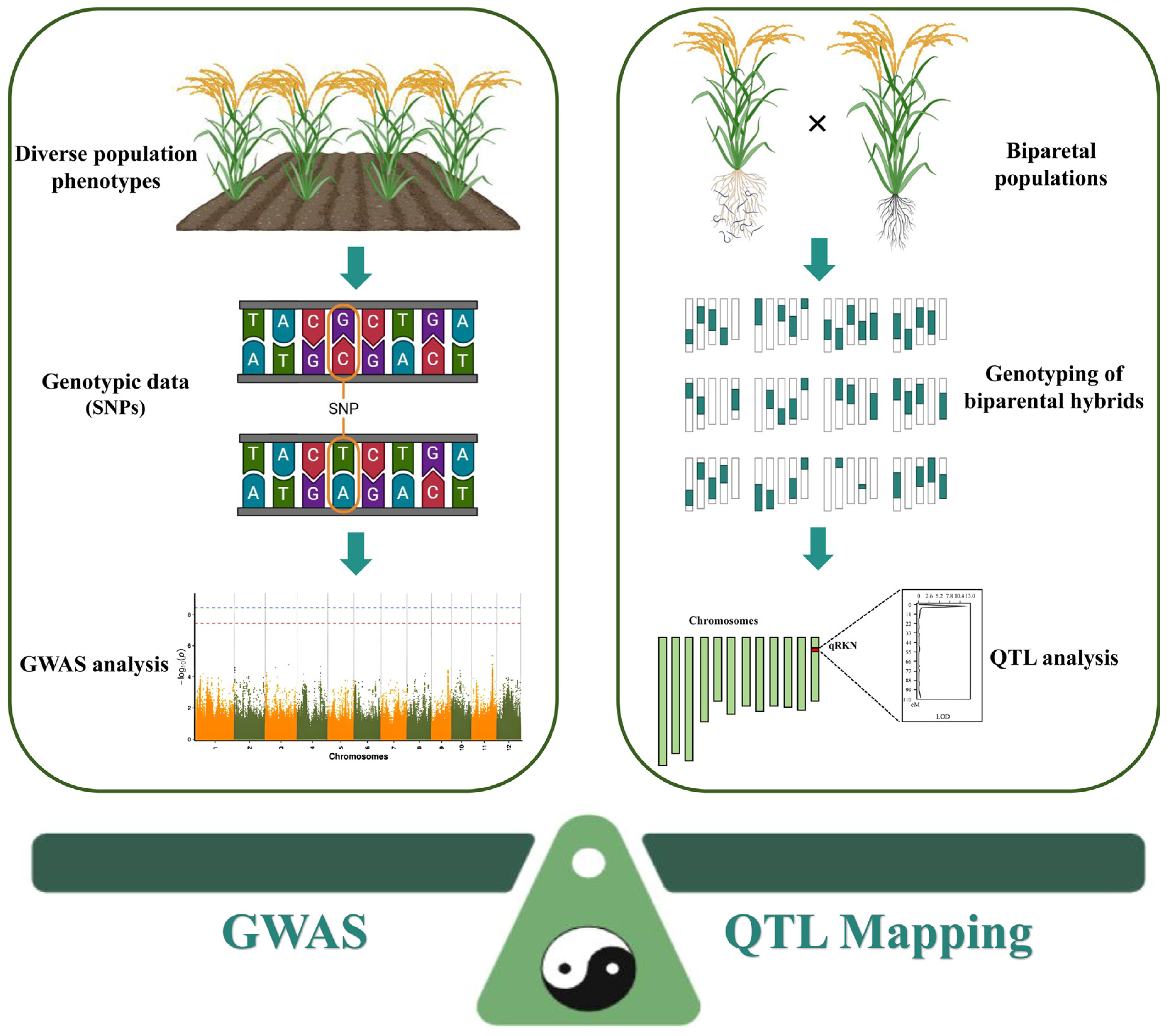
2. Comparing QTL Mapping and GWAS in PPNs Resistance
2.1. QTL Mapping in PPNs Resistance: Methodology and Constraints
2.2. GWAS in PPNs Resistance: Opportunities and Challenges
3. QTL Mapping for Crop Resistance to PPNs
3.1. QTL Mapping for Soybean Cyst Nematode
3.2. QTL Mapping for Cereal Cyst Nematode and Root-Lesion Nematode
3.3. QTL Mapping for Rice Root-Knot Nematode
3.4. QTL Mapping for Southern Root-Knot Nematode
4. Application of GWAS for Crop Resistance to PPNs
4.1. Application of GWAS for Crop Resistance to Soybean Cyst Nematode
4.2. Application of GWAS for Crop Resistance to Cereal Cyst Nematode
4.3. Application of GWAS for Crop Resistance to Rice Root-Knot Nematode
4.4. Application of GWAS for Crop Resistance to Southern Root-Knot Nematode
5. Applications and Future Perspectives
5.1. Application of Resistance Genes in Breeding Programs
5.2. New Strategies in Breeding Crops for Resistance to PPNs
6. Conclusions and Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef]
- Garnett, T.; Appleby, M.C.; Balmford, A.; Bateman, I.J.; Benton, T.G.; Bloomer, P.; Burlingame, B.; Dawkins, M.; Dolan, L.; Fraser, D.; et al. Sustainable intensification in agriculture: Premises and policies. Science 2013, 341, 33–34. [Google Scholar] [CrossRef]
- Kantor, C.; Eisenback, J.D.; Kantor, M. Biosecurity risks to human food supply associated with plant-parasitic nematodes. Front. Plant Sci. 2024, 15, 1404355. [Google Scholar] [CrossRef]
- Jones, J.T.; Haegeman, A.; Danchin, E.G.J.; Gaur, H.S.; Helder, J.; Jones, M.G.K.; Kikuchi, T.; Manzanilla-López, R.; Palomares-Rius, J.E.; Wesemael, W.M.L.; et al. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Dayi, M. Evolution of parasitism genes in the plant parasitic nematodes. Sci. Rep. 2024, 14, 3733. [Google Scholar] [CrossRef]
- Meresa, B.K.; Matthys, J.; Kyndt, T. Biochemical defence of plants against parasitic nematodes. Plants 2024, 13, 2813. [Google Scholar] [CrossRef]
- Mendoza-de Gives, P. Soil-Borne nematodes: Impact in agriculture and livestock and sustainable strategies of prevention and control with special reference to the use of nematode natural enemies. Pathogens 2022, 11, 640. [Google Scholar] [CrossRef] [PubMed]
- Abd-Elgawad, M.M.M. Upgrading strategies for managing nematode pests on profitable crops. Plants 2024, 13, 1558. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Liu, J.; Liu, Q.; Sun, J.B.; Zhao, Y.X.; Liu, J.; Gao, W.S.; Chen, Y.Q.; Sui, P. Knowledge domain and research progress in the field of crop rotation from 2000 to 2020: A scientometric review. Environ. Sci. Pollut. Res. 2023, 30, 86598–86617. [Google Scholar] [CrossRef]
- Ayaz, M.; Zhao, J.T.; Zhao, W.; Chi, Y.K.; Ali, Q.; Ali, F.; Khan, A.R.; Yu, Q.; Yu, J.W.; Wu, W.C.; et al. Biocontrol of plant parasitic nematodes by bacteria and fungi: A multi-omics approach for the exploration of novel nematicides in sustainable agriculture. Front. Microbiol. 2024, 15, 1433716. [Google Scholar] [CrossRef]
- Opdensteinen, P.; Charudattan, R.; Hong, J.C.; Rosskopf, E.N.; Steinmetz, N.F. Biochemical and nanotechnological approaches to combat phytoparasitic nematodes. Plant Biotechnol. J. 2024, 22, 2444–2460. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.X.; Li, Q.X.; Song, B.A. Chemical nematicides: Recent research progress and outlook. J. Agric. Food Chem. 2020, 68, 12175–12188. [Google Scholar] [CrossRef] [PubMed]
- Vasantha-Srinivasan, P.; Park, K.B.; Kim, K.Y.; Jung, W.J.; Han, Y.S. The role of Bacillus species in the management of plant-parasitic nematodes. Front. Microbiol. 2025, 15, 1510036. [Google Scholar] [CrossRef] [PubMed]
- Han, S.J.; Schliemann, W.; Liu, S.M. Editorial: Resistance of plants to parasitic nematodes and its application in breeding. Front. Plant Sci. 2024, 15, 1439535. [Google Scholar] [CrossRef]
- Afzal, A.; Mukhtar, T. Revolutionizing nematode management to achieve global food security goals—An overview. Heliyon 2024, 10, e25325. [Google Scholar] [CrossRef]
- Sun, L.X.; Lai, M.Y.; Ghouri, F.; Nawaz, M.A.; Ali, F.; Baloch, F.S.; Nadeem, M.A.; Aasim, M.; Shahid, M.Q. Modern plant breeding techniques in crop improvement and genetic diversity: From molecular markers and gene editing to artificial intelligence-a critical review. Plants 2024, 13, 2676. [Google Scholar] [CrossRef]
- Abd-Elgawad, M.M.M. Understanding Molecular plant-nematode interactions to develop alternative approaches for nematode control. Plants 2022, 11, 2141. [Google Scholar] [CrossRef]
- Ali, M.A.; Azeem, F.; Abbas, A.; Joyia, F.A.; Li, H.J.; Dababat, A.A. Transgenic strategies for enhancement of nematode resistance in plants. Front. Plant Sci. 2017, 8, 750. [Google Scholar] [CrossRef]
- Dababat, A.A.; Paulitz, T.; Laasli, S.-E.; Lahlali, R.; Li, H.; Mokrini, F.; Dreisigacker, S.J.B. From genes to fields: Marker-Assisted Selection for nematode resistance in crops. Integr. Plant Biotechnol. 2025, 3, 1–18. [Google Scholar] [CrossRef]
- Shen, C. Analysis of complex traits and molecular selection in annual crops. Agronomy 2024, 14, 948. [Google Scholar] [CrossRef]
- Altaf, M.T.; Tatar, M.; Ali, A.; Liaqat, W.; Mortazvi, P.; Kayihan, C.; Ölmez, F.; Nadeem, M.A.; Javed, J.; Gou, J.Y.; et al. Advancements in QTL mapping and GWAS application in plant improvement. Turk. J. Bot. 2024, 48, 948. [Google Scholar] [CrossRef]
- Nasim, A.; Hao, J.W.; Tawab, F.; Jin, C.; Zhu, J.M.; Luo, S.; Nie, X.J. Micronutrient biofortification in wheat: QTLs, candidate genes and molecular mechanism. Int. J. Mol. Sci. 2025, 26, 2178. [Google Scholar] [CrossRef]
- Susmitha, P.; Kumar, P.; Yadav, P.; Sahoo, S.; Kaur, G.; Pandey, M.K.; Singh, V.; Tseng, T.; Gangurde, S.S. Genome-wide association study as a powerful tool for dissecting competitive traits in legumes. Front. Plant Sci. 2023, 14, 1123631. [Google Scholar] [CrossRef] [PubMed]
- Snehi, S.; Choudhary, M.; Kumar, S.; Jayaswal, D.; Kumar, S.; Prakash, N.R. Mapping of quantitative traits loci: Harnessing genomics revolution for dissecting complex traits. In Genomics Data Analysis for Crop Improvement; Anjoy, P., Kumar, K., Chandra, G., Gaikwad, K., Eds.; Springer Nature: Singapore, 2024; pp. 125–157. [Google Scholar]
- Cook, D.E.; Lee, T.G.; Guo, X.L.; Melito, S.; Wang, K.; Bayless, A.M.; Wang, J.P.; Hughes, T.J.; Willis, D.K.; Clemente, T.E.; et al. Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 2012, 338, 1206–1209. [Google Scholar] [CrossRef]
- Liu, S.M.; Kandoth, P.K.; Warren, S.D.; Yeckel, G.; Heinz, R.; Alden, J.; Yang, C.L.; Jamai, A.; El-Mellouki, T.; Juvale, P.S.; et al. A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature 2012, 492, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, C.; Taguchi-Shiobara, F.; Ikeda, C.; Iwahashi, M.; Matsui, T.; Yamashita, Y.; Ogura, R. Mapping soybean rhg2 locus, which confers resistance to soybean cyst nematode race 1 in combination with rhg1 and Rhg4 derived from PI 84751. Breed. Sci. 2020, 70, 474–480. [Google Scholar] [CrossRef]
- Caldwell, B.E.; Brim, C.A.; Ross, J.P. Inheritance of resistance of soybeans to the cyst nematode, Heterodera glycines. Agron. J. 1960, 52, 635–636. [Google Scholar] [CrossRef]
- Glover, K.D.; Wang, D.; Arelli, P.R.; Carlson, S.R.; Cianzio, S.R.; Diers, B.W. Near isogenic lines confirm a soybean cyst nematode resistance gene from PI 88788 on linkage group J. Crop Sci. 2004, 44, 936–941. [Google Scholar] [CrossRef]
- Kazi, S.; Shultz, J.; Afzal, J.; Hashmi, R.; Jasim, M.; Bond, J.; Arelli, P.R.; Lightfoot, D.A. Iso-lines and inbred-lines confirmed loci that underlie resistance from cultivar ‘Hartwig’ to three soybean cyst nematode populations. Theor. Appl. Genet. 2010, 120, 633–644. [Google Scholar] [CrossRef]
- Yu, N.; Diers, B.W. Fine mapping of the SCN resistance QTL cqSCN-006 and cqSCN-007 from Glycine soja PI 468916. Euphytica 2017, 213, 54. [Google Scholar] [CrossRef]
- Arelli, P.R.; Concibido, V.C.; Young, L.D. QTLs associated with resistance in soybean PI567516C to synthetic nematode population infecting cv. Hartwig. J. Crop Sci. Biotechnol. 2010, 13, 163–167. [Google Scholar] [CrossRef]
- Guo, B.; Sleper, D.A.; Arelli, P.R.; Shannon, J.G.; Nguyen, H.T. Identification of QTLs associated with resistance to soybean cyst nematode races 2, 3 and 5 in soybean PI 90763. Theor. Appl. Genet. 2005, 111, 965–971. [Google Scholar] [CrossRef]
- Kretschmer, J.M.; Chalmers, K.J.; Manning, S.; Karakousis, A.; Barr, A.R.; Islam, A.; Logue, S.J.; Choe, Y.W.; Barker, S.J.; Lance, R.C.M.; et al. RFLP mapping of the Ha2 cereal cyst nematode resistance gene in barley. Theor. Appl. Genet. 1997, 94, 1060–1064. [Google Scholar] [CrossRef]
- Barr, A.R.; Chalmers, K.J.; Karakousis, A.; Kretschmer, J.M.; Manning, S.; Lance, R.C.M.; Lewis, J.; Jeffries, S.P.; Langridge, P. RFLP mapping of a new cereal cyst nematode resistance locus in barley. Plant Breed. 1998, 117, 185–187. [Google Scholar] [CrossRef]
- Van Gansbeke, B.; Khoo, K.H.P.; Lewis, J.G.; Chalmers, K.J.; Mather, D.E. Fine mapping of Rha2 in barley reveals candidate genes for resistance against cereal cyst nematode. Theor. Appl. Genet. 2019, 132, 1309–1320. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Qiu, D.; Sun, L.; Sun, Y.; Ren, Y.K.; Zhang, H.J.; Li, J.T.; Zou, J.W.; Wu, P.P.; Hu, J.H.; et al. Resistance to Heterodera filipjevi and H. avenae in winter wheat is conferred by different QTL. Phytopathology 2020, 110, 472–482. [Google Scholar] [CrossRef]
- Dababat, A.; Arif, M.A.R.; Toktay, H.; Atiya, O.; Shokat, S.; E-Orakci, G.; Imren, M.; Singh, S. A GWAS to identify the cereal cyst nematode (Heterodera filipjevi) resistance loci in diverse wheat prebreeding lines. J. Appl. Genet. 2021, 62, 93–98. [Google Scholar] [CrossRef]
- Zwart, R.S.; Thompson, J.P.; Godwin, I.D. Identification of quantitative trait loci for resistance to two species of root-lesion nematode (Pratylenchus thornei and P-neglectus) in wheat. Aust. J. Agric. Res. 2005, 56, 345–352. [Google Scholar] [CrossRef]
- Rahman, M.S.; Linsell, K.J.; Taylor, J.D.; Hayden, M.J.; Collins, N.C.; Oldach, K.H. Fine mapping of root lesion nematode (Pratylenchus thornei) resistance loci on chromosomes 6D and 2B of wheat. Theor. Appl. Genet. 2020, 133, 635–652. [Google Scholar] [CrossRef] [PubMed]
- Mhatre, P.H.; Pankaj; Sirohi, A.; Singh, A.K.; Ellur, R.K.; Bhowmick, P.K.; Singh, V.K. Molecular mapping of rice root-knot nematode (Meloidogyne graminicola) resistance gene in Asian rice (Oryza sativa L.) using STMS markers. Indian J. Genet. Plant Breed. 2017, 77, 163–165. [Google Scholar] [CrossRef]
- Galeng-Lawilao, J.; Kumar, A.; De Waele, D. QTL mapping for resistance to and tolerance for the rice root-knot nematode, Meloidogyne graminicola. BMC Genet. 2018, 19, 53. [Google Scholar] [CrossRef]
- Lahari, Z.; Ribeiro, A.; Talukdar, P.; Martin, B.; Heidari, Z.; Gheysen, G.; Price, A.H.; Shrestha, R. QTL-seq reveals a major root-knot nematode resistance locus on chromosome 11 in rice (Oryza sativa L.). Euphytica 2019, 215, 117. [Google Scholar] [CrossRef]
- Wang, C.L.; Ulloa, M.; Mullens, T.R.; Yu, J.Z.; Roberts, P.A. QTL Analysis for transgressive resistance to root-knot nematode in interspecific cotton (Gossypium spp.) progeny derived from susceptible parents. PLoS ONE 2012, 7, e34874. [Google Scholar] [CrossRef]
- Giordani, W.; Gama, H.C.; Chiorato, A.F.; Marques, J.P.R.; Huo, H.Q.; Benchimol-Reis, L.L.; Camargo, L.E.A.; Garcia, A.A.F.; Vieira, M.L.C. Genetic mapping reveals complex architecture and candidate genes involved in common bean response to Meloidogyne incognita infection. Plant Genome 2022, 15, e20161. [Google Scholar] [CrossRef]
- Mistanoglu, I.; Özalp, T.; Devran, Z. The efficacy of molecular markers associated with virulence in root-knot nematodes. Nematology 2020, 22, 147–154. [Google Scholar] [CrossRef]
- Du, H.P.; Fang, C.; Li, Y.R.; Kong, F.J.; Liu, B.H. Understandings and future challenges in soybean functional genomics and molecular breeding. J. Integr. Plant Biol. 2023, 65, 468–495. [Google Scholar] [CrossRef] [PubMed]
- Concibido, V.C.; Denny, R.L.; Boutin, S.R.; Hautea, R.; Orf, J.H.; Young, N.D. DNA marker analysis of loci underlying resistance to soybean cyst nematode (Heterodera glycines Ichinohe). Crop Sci. 1994, 34, 240–246. [Google Scholar] [CrossRef]
- Matsye, P.D.; Kumar, R.; Hosseini, P.; Jones, C.M.; Tremblay, A.; Alkharouf, N.W.; Matthews, B.F.; Klink, V.P. Mapping cell fate decisions that occur during soybean defense responses. Plant Mol. Biol. 2011, 77, 513–528. [Google Scholar] [CrossRef]
- Bayless, A.M.; Smith, J.M.; Song, J.Q.; McMinn, P.H.; Teillet, A.; August, B.K.; Bent, A.F. Disease resistance through impairment of α-SNAP-NSF interaction and vesicular trafficking by soybean Rhg1. Proc. Natl. Acad. Sci. USA 2016, 113, E7375–E7382. [Google Scholar] [CrossRef] [PubMed]
- Cook, S.D.; Nichols, D.S.; Smith, J.; Chourey, P.S.; McAdam, E.L.; Quittenden, L.; Ross, J.J. Auxin Biosynthesis: Are the indole-3-acetic acid and phenylacetic acid biosynthesis pathways mirror images? Plant Physiol. 2016, 171, 1230–1241. [Google Scholar] [CrossRef]
- Concibido, V.C.; Diers, B.W.; Arelli, P.R. A decade of QTL mapping for cyst nematode resistance in soybean. Crop Sci. 2004, 44, 1121–1131. [Google Scholar] [CrossRef]
- Meksem, K.; Pantazopoulos, P.; Njiti, V.N.; Hyten, L.D.; Arelli, P.R.; Lightfoot, D.A. ‘Forrest’ resistance to the soybean cyst nematode is bigenic: Saturation mapping of the Rhg1 and Rhg4 loci. Theor. Appl. Genet. 2001, 103, 710–717. [Google Scholar] [CrossRef]
- Chang, S.J.C.; Doubler, T.W.; Kilo, V.Y.; AbuThredeih, J.; Prabhu, R.; Freire, V.; Suttner, R.; Klein, J.; Schmidt, M.E.; Gibson, P.T.; et al. Association of loci underlying field resistance to soybean sudden death syndrome (SDS) and cyst nematode (SCN) race 3. Crop Sci. 1997, 37, 965–971. [Google Scholar] [CrossRef]
- Ros, R.; Muñoz-Bertomeu, J.; Krueger, S. Serine in plants: Biosynthesis, metabolism, and functions. Trends Plant Sci. 2014, 19, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Kandoth, P.K.; Liu, S.M.; Prenger, E.; Ludwig, A.; Lakhssassi, N.; Heinz, R.; Zhou, Z.; Howland, A.; Gunther, J.; Eidson, S.; et al. Systematic Mutagenesis of serine hydroxymethyltransferase reveals an essential role in nematode resistance. Plant Physiol. 2017, 175, 1370–1380. [Google Scholar] [CrossRef]
- Wu, X.Y.; Zhou, G.C.; Chen, Y.X.; Wu, P.; Liu, L.W.; Ma, F.F.; Wu, M.; Liu, C.C.; Zeng, Y.J.; Chu, A.E.; et al. Soybean cyst nematode resistance emerged via artificial selection of duplicated serine hydroxymethyltransferase genes. Front. Plant Sci. 2016, 7, 998. [Google Scholar] [CrossRef]
- Barloy, D.; Lemoine, J.; Abelard, P.; Tanguy, A.M.; Rivoal, R.; Jahier, J. Marker-assisted pyramiding of two cereal cyst nematode resistance genes from Aegilops variabilis in wheat. Mol. Breed. 2007, 20, 31–40. [Google Scholar] [CrossRef]
- Zhang, R.Q.; Feng, Y.G.; Li, H.F.; Yuan, H.X.; Dai, J.L.; Cao, A.Z.; Xing, L.P.; Li, H.L. Cereal cyst nematode resistance gene CreV effective against Heterodera filipjevi transferred from chromosome 6VL of Dasypyrum villosum to bread wheat. Mol. Breed. 2016, 36, 122. [Google Scholar] [CrossRef]
- Safari, E.; Gororo, N.N.; Eastwood, R.F.; Lewis, J.; Eagles, H.A.; Ogbonnaya, F.C. Impact of Cre1, Cre8 and Cre3 genes on cereal cyst nematode resistance in wheat. Theor. Appl. Genet. 2005, 110, 567–572. [Google Scholar] [CrossRef]
- Wang, X.M.; Cheng, R.; Xu, D.C.; Huang, R.L.; Li, H.X.; Jin, L.; Wu, Y.F.; Tang, J.Y.; Sun, C.H.; Peng, D.L.; et al. MG1 interacts with a protease inhibitor and confers resistance to rice root-knot nematode. Nat. Commun. 2023, 14, 3354. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.M.; Kandoth, P.K.; Lakhssassi, N.; Kang, J.W.; Colantonio, V.; Heinz, R.; Yeckel, G.; Zhou, Z.; Bekal, S.; Dapprich, J.; et al. The soybean GmSNAP18 gene underlies two types of resistance to soybean cyst nematode. Nat. Commun. 2017, 8, 14822. [Google Scholar] [CrossRef]
- Shaibu, A.S.; Zhang, S.R.; Ma, J.K.; Feng, Y.; Huai, Y.Y.; Qi, J.; Li, J.; Abdelghany, A.M.; Azam, M.; Htway, H.T.P.; et al. The GmSNAP11 contributes to resistance to soybean cyst nematode race 4 in Glycine max. Front. Plant Sci. 2022, 13, 939763. [Google Scholar] [CrossRef]
- Usovsky, M.; Gamage, V.A.; Meinhardt, C.G.; Dietz, N.; Triller, M.; Basnet, P.; Gillman, J.D.; Bilyeu, K.D.; Song, Q.J.; Dhital, B.; et al. Loss-of-function of an α-SNAP gene confers resistance to soybean cyst nematode. Nat. Commun. 2023, 14, 7629. [Google Scholar] [CrossRef]
- Ogbonnaya, F.C.; Seah, S.; Delibes, A.; Jahier, J.; López-Braña, I.; Eastwood, R.F.; Lagudah, E.S. Molecular-genetic characterisation of a new nematode resistance gene in wheat. Theor. Appl. Genet. 2001, 102, 623–629. [Google Scholar] [CrossRef]
- Eastwood, R.F.; Lagudah, E.S.; Appels, R.; Hannah, M.; Kollmorgen, J.F. Triticum tauschii: A novel source of resistance to cereal cyst nematode (Heterodera avenae). Aust. J. Agric. Res. 1991, 42, 69–77. [Google Scholar] [CrossRef]
- Eastwood, R.F.; Lagudah, E.S.; Appels, R. A directed search for DNA sequences tightly linked to cereal cyst nematode resistance genes in Triticum tauschii. Genome 1994, 37, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Jahier, J.; Abelard, P.; Tanguy, A.M.; Dedryver, F.; Rivoal, R.; Khatkar, S.; Bariana, H.S. The Aegilops ventricosa segment on chromosome 2AS of the wheat cultivar ‘VPM1’ carries the cereal cyst nematode resistance gene Cre5. Plant Breed. 2001, 120, 125–128. [Google Scholar] [CrossRef]
- Jahier, J.; Rivoal, R.; Yu, M.Q.; Abelard, P.; Tanguy, A.M.; Barloy, D. Transfer of genes for resistance to cereal cyst nematode from Aegilops variabilis Eig to wheat. J. Genet. Breed. 1998, 52, 253–257. [Google Scholar]
- Rivoal, R.; Bekal, S.; Valette, S.; Gauthier, J.P.; Fradj, M.B.H.; Mokabli, A.; Jahier, J.; Nicol, J.; Yahyaoui, A. Variation in reproductive capacity and virulence on different genotypes and resistance genes of Triticeae, in the cereal cyst nematode species complex. Nematology 2001, 3, 581–592. [Google Scholar] [CrossRef]
- Williams, K.J.; Taylor, S.P.; Bogacki, P.; Pallotta, M.; Bariana, H.S.; Wallwork, H. Mapping of the root lesion nematode (Pratylenchus neglectus) resistance gene Rlnn1 in wheat. Theor. Appl. Genet. 2002, 104, 874–879. [Google Scholar] [CrossRef]
- Hwang, C.F.; Bhakta, A.V.; Truesdell, G.M.; Pudlo, W.M.; Williamson, V.M. Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell 2000, 12, 1319–1329. [Google Scholar] [CrossRef]
- Ammiraju, J.S.S.; Veremis, J.C.; Huang, X.; Roberts, P.A.; Kaloshian, I. The heat-stable root-knot nematode resistance gene Mi-9 from Lycopersicon peruvianum is localized on the short arm of chromosome 6. Theor. Appl. Genet. 2003, 106, 478–484. [Google Scholar] [CrossRef]
- Shihab, K.M.; Abood, I.D. Genetic segregation of tomato trihybrid, double cross and detection of Mi1. 2, Mi-3 resistance genes against root-knot nematode (Meloidogyne spp.). Int. J. Agric. Stat. Sci. 2019, 15, 153–162. [Google Scholar]
- Fukino, N.; Ohara, T.; Monforte, A.; Sugiyama, M.; Sakata, Y.; Kunihisa, M.; Matsumoto, S. Identification of QTLs for resistance to powdery mildew and SSR markers diagnostic for powdery mildew resistance genes in melon (Cucumis melo L.). Theor. Appl. Genet. 2008, 118, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Boissot, N.; Thomas, S.; Sauvion, N.; Marchal, C.; Pavis, C.; Dogimont, C. Mapping and validation of QTLs for resistance to aphids and whiteflies in melon. Theor. Appl. Genet. 2010, 121, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cao, H.Y.; Ling, J.; Yang, Y.H.; Li, Y.; Xie, B.Y.; Zhao, J.L.; Mao, Z.C. Molecular cloning and functional analysis of the pepper resistance gene Me3 to root-knot nematode. Hortic. Plant J. 2023, 9, 133–144. [Google Scholar] [CrossRef]
- Nicol, J.M.; Rivoal, R. Global knowledge and its application for the integrated control and management of nematodes on wheat. In Integrated management and Biocontrol of Vegetable and Grain Crops Nematodes; Springer: Berlin/Heidelberg, Germany, 2008; pp. 251–294. [Google Scholar]
- Dababat, A.A.; Imren, M.; Erginbas-Orakci, G.; Ashrafi, S.; Yavuzaslanoglu, E.; Toktay, H.; Pariyar, S.R.; Elekcioglu, H.I.; Morgounov, A.; Mekete, T. The importance and management strategies of cereal cyst nematodes, Heterodera spp., in Turkey. Euphytica 2015, 202, 173–188. [Google Scholar] [CrossRef]
- Ali, M.A.; Shahzadi, M.; Zahoor, A.; Dababat, A.A.; Toktay, H.; Bakhsh, A.; Nawaz, M.A.; Li, H.J. Resistance to cereal cyst nematodes in wheat and barley: An emphasis on classical and modern approaches. Int. J. Mol. Sci. 2019, 20, 432. [Google Scholar] [CrossRef]
- Jayatilake, D.V.; Tucker, E.J.; Brueggemann, J.; Lewis, J.; Garcia, M.; Dreisigacker, S.; Hayden, M.J.; Chalmers, K.; Mather, D.E. Genetic mapping of the Cre8 locus for resistance against cereal cyst nematode (Heterodera avenae Woll.) in wheat. Mol. Breed. 2015, 35, 66. [Google Scholar] [CrossRef]
- Mokabli, A.; Valette, S.; Gauthier, J.P.; Rivoal, R. Variation in virulence of cereal cyst nematode populations from North Africa and Asia. Nematology 2002, 4, 521–525. [Google Scholar] [CrossRef]
- Vanstone, V.A.; Hollaway, G.J.; Stirling, G.R. Managing nematode pests in the southern and western regions of the Australian cereal industry: Continuing progress in a challenging environment. Australas. Plant Pathol. 2008, 37, 220–234. [Google Scholar] [CrossRef]
- Nicol, J.M.; Rivoal, R.; Trethowan, R.M.; van Ginkel, M.; Mergoum, M.; Singh, R.P. CIMMYT’s approach to identify and use resistance to nematodes and soil-borne fungi, in developing superior wheat germplasm. In Proceedings of the 6th International Wheat Conference, Budapest, Hungary, 5–9 June 2000; pp. 381–389. [Google Scholar]
- Soriano, I.R.; Schmit, V.; Brar, D.S.; Prot, J.C.; Reversat, G. Resistance to rice root-knot nematode Meloidogyne graminicola identified in Oryza longistaminata and O. glaberrima. Nematology 1999, 1, 395–398. [Google Scholar] [CrossRef]
- Bimpong, I.K.; Carpena, A.L.; Mendioro, M.S.; Fernandez, L.; Ramos, J.; Reversat, G.; Brar, D.S. Evaluation of Oryza sativa x O. glaberrima derived progenies for resistance to rootknot nematode and identification of introgressed alien chromosome segments using SSR markers. Afr. J. Biotechnol. 2010, 9, 3988–3997. [Google Scholar]
- Shrestha, R.; Uzzo, F.; Wilson, M.J.; Price, A.H. Physiological and genetic mapping study of tolerance to root-knot nematode in rice. New Phytol. 2007, 176, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Jena, M.; Mohapatra, S.L.; Panda, R.S.; Mohanty, S.K.; Thatoi, H.N.; Sahu, S.C. Genetic loci associated with root-knot nematode resistance in rice cv. Ramakrishna. ORYZA-Int. J. Rice 2013, 50, 132–139. [Google Scholar]
- Li, Z.; Jakkula, L.; Hussey, R.S.; Tamulonis, J.P.; Boerma, H.R. SSR mapping and confirmation of the QTL from PI96354 conditioning soybean resistance to southern root-knot nematode. Theor. Appl. Genet. 2001, 103, 1167–1173. [Google Scholar] [CrossRef]
- Ha, B.K.; Bennett, J.B.; Hussey, R.S.; Finnerty, S.L.; Boerma, H.R. Pedigree analysis of a major QTL conditioning soybean resistance to southern root-knot nematode. Crop Sci. 2004, 44, 758–763. [Google Scholar] [CrossRef]
- Thomas, T.; Sakure, A.A.; Kumar, S.; Mishra, A.; Ahmad, S.; Rojasara, Y.M.; Vaja, M.B.; Patel, D.A. The Mi-1 gene is a key regulator of defence mechanisms and cellular gene dynamics in response to root-knot nematodes. Plant Cell Rep. 2025, 44, 1–44. [Google Scholar] [CrossRef]
- El-Sappah, A.H.; Islam, M.M.; El-awady, H.H.; Yan, S.; Qi, S.M.; Liu, J.Y.; Cheng, G.T.; Liang, Y. Tomato natural resistance genes in controlling the root-knot nematode. Genes 2019, 10, 925. [Google Scholar] [CrossRef]
- Salmeron, J.M.; Oldroyd, G.E.D.; Rommens, C.M.T.; Scofield, S.R.; Kim, H.S.; Lavelle, D.T.; Dahlbeck, D.; Staskawicz, B.J. Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 1996, 86, 123–133. [Google Scholar] [CrossRef]
- Ellis, J.G.; Lawrence, G.J.; Luck, J.E.; Dodds, P.N. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 1999, 11, 495–506. [Google Scholar] [CrossRef]
- Fourie, H.; Mienie, C.M.S.; Mc Donald, A.H.; De Waele, D. Identification and validation of genetic markers associated with Meloidogyne incognita race 2 resistance in soybean, Glycine max (L.) Merr. Nematology 2008, 10, 651–661. [Google Scholar] [CrossRef]
- Xu, X.Y.; Zeng, L.; Tao, Y.; Vuong, T.; Wan, J.R.; Boerma, R.; Noe, J.; Li, Z.L.; Finnerty, S.; Pathan, S.M.; et al. Pinpointing genes underlying the quantitative trait loci for root-knot nematode resistance in palaeopolyploid soybean by whole genome resequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 13469–13474. [Google Scholar] [CrossRef]
- Jiang, H.P.; Lv, S.C.; Zhou, C.J.; Qu, S.; Liu, F.; Sun, H.W.; Zhao, X.; Han, Y.P. Identification of QTL, QTL-by-environment interactions, and their candidate genes for resistance HG Type 0 and HG Type 1.2.3.5.7 in soybean using 3VmrMLM. Front. Plant Sci. 2023, 14, 1177345. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.W.; Chang, H.X.; Brown, P.J.; Domier, L.L.; Hartman, G.L. Genome-wide association and genomic prediction identifies soybean cyst nematode resistance in common bean including a syntenic region to soybean Rhg1 locus. Hortic. Res. 2019, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wen, Z.X.; Li, W.; Zhang, Y.W.; Zhang, L.F.; Dai, H.Y.; Wang, D.C.; Xu, R. Genome-wide association study for soybean cyst nematode resistance in Chinese elite soybean cultivars. Mol. Breed. 2017, 37, 60. [Google Scholar] [CrossRef]
- Tian, Y.; Li, D.L.; Wang, X.Q.; Zhang, H.; Wang, J.J.; Yu, L.J.; Guo, C.H.; Luan, X.Y.; Liu, X.L.; Li, H.J.; et al. Deciphering the genetic basis of resistance to soybean cyst nematode combining IBD and association mapping. Theor. Appl. Genet. 2023, 136, 50. [Google Scholar] [CrossRef]
- Pariyar, S.R.; Dababat, A.A.; Sannemann, W.; Erginbas-Orakci, G.; Elashry, A.; Siddique, S.; Morgounov, A.; Leon, J.; Grundler, F.M.W. Genome-Wide Association Study in wheat identifies resistance to the cereal cyst nematode Heterodera filipjevi. Phytopathology 2016, 106, 1128–1138. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, D.; Pundir, S.; Singh, V.K.; Kumar, D.; Sharma, R.; Röder, M.S.; Sharma, S.; Sharma, S. Identification of genomic regions associated with cereal cyst nematode (Heterodera avenae Woll.) resistance in spring and winter wheat. Sci. Rep. 2023, 13, 5916. [Google Scholar] [CrossRef]
- Singh, V.K.; Chaturvedi, D.; Pundir, S.; Kumar, D.; Sharma, R.; Kumar, S.; Sharma, S.; Sharma, S. GWAS scans of cereal cyst nematode (Heterodera avenae) resistance in Indian wheat germplasm. Mol. Genet. Genom. 2023, 298, 579–601. [Google Scholar] [CrossRef]
- Taheri, Z.M.; Maafi, Z.T.; Nazari, K.; Nezhad, K.Z.; Rakhshandehroo, F.; Dababat, A.A. Genome-wide association mapping revealed SNP alleles associated with resistance to cereal cyst nematode (Heterodera filipjevi) in wheat. J. Agric. Sci. Technol. 2024, 26, 861–872. [Google Scholar] [CrossRef]
- Hada, A.; Dutta, T.K.; Singh, N.; Singh, B.; Rai, V.; Singh, N.K.; Rao, U. A genome-wide association study in Indian wild rice accessions for resistance to the root-knot nematode Meloidogyne graminicola. PLoS ONE 2020, 15, e0239085. [Google Scholar] [CrossRef]
- Dimkpa, S.O.N.; Lahari, Z.; Shrestha, R.; Douglas, A.; Gheysen, G.; Price, A.H. A genome-wide association study of a global rice panel reveals resistance in Oryza sativa to root-knot nematodes. J. Exp. Bot. 2016, 67, 1191–1200. [Google Scholar] [CrossRef]
- Passianotto, A.L.D.; Sonah, H.; Dias, W.P.; Marcelino-Guimaraes, F.C.; Belzile, F.; Abdelnoor, R.V. Genome-wide association study for resistance to the southern root-knot nematode (Meloidogyne incognita) in soybean. Mol. Breed. 2017, 37, 148. [Google Scholar] [CrossRef]
- Tylka, G.L.; Mullaney, M.P. Soybean Cyst Nematode-Resistant Soybean Varieties for Iowa; Iowa State University, University Extension: Ames, IA, USA, 2002. [Google Scholar]
- Cook, D.E.; Bayless, A.M.; Wang, K.; Guo, X.L.; Song, Q.J.; Jiang, J.M.; Bent, A.F. Distinct copy number, coding sequence, and locus methylation patterns underlie Rhg1-mediated soybean resistance to soybean cyst nematode. Plant Physiol. 2014, 165, 630–647. [Google Scholar] [CrossRef]
- Patil, G.B.; Lakhssassi, N.; Wan, J.R.; Song, L.; Zhou, Z.; Klepadlo, M.; Vuong, T.D.; Stec, A.O.; Kahil, S.S.; Colantonio, V.; et al. Whole-genome re-sequencing reveals the impact of the interaction of copy number variants of the rhg1 and Rhg4 genes on broad-based resistance to soybean cyst nematode. Plant Biotechnol. J. 2019, 17, 1595–1611. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, A.M.; Dutta, T.K.; Curtis, R.H.C.; Powers, S.J.; Gaur, H.S.; Kerry, B.R. Chemotaxis can take plant-parasitic nematodes to the source of a chemo-attractant via the shortest possible routes. J. R. Soc. Interface 2011, 8, 568–577. [Google Scholar] [CrossRef]
- Aramov, M.K.; Dzhuraeva, L. Breeding Meloidogyne-Resistant Tomato Varieties with a Jointless Pedicel. 1991. Available online: https://www.cabidigitallibrary.org/doi/full/10.5555/19931697756 (accessed on 19 January 2024).
- Guo, Y.X.; Zhao, G.D.; Gao, X.; Zhang, L.; Zhang, Y.A.; Cai, X.M.; Yuan, X.J.; Guo, X.Q. CRISPR/Cas9 gene editing technology: A precise and efficient tool for crop quality improvement. Planta 2023, 258, 36. [Google Scholar] [CrossRef] [PubMed]
- Zargar, S.M.; Raatz, B.; Sonah, H.; Muslima Nazir, M.N.; Bhat, J.A.; Dar, Z.A.; Agrawal, G.K.; Randeep Rakwal, R.R. Recent advances in molecular marker techniques: Insight into QTL mapping, GWAS and genomic selection in plants. J. Crop Sci. Biotechnol. 2015, 18, 293–308. [Google Scholar] [CrossRef]
- Ibrahim, H.M.M.; Ahmad, E.M.; Martínez-Medina, A.; Aly, M.A.M. Effective approaches to study the plant-root knot nematode interaction. Plant Physiol. Biochem. 2019, 141, 332–342. [Google Scholar] [CrossRef]
- Wu, X.C.; Liu, Y.L.; Xing, M.J.; Yang, C.; Hong, S.Y. Image segmentation for pest detection of crop leaves by improvement of regional convolutional neural network. Sci. Rep. 2024, 14, 24160. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.G.; Go, M.J.; Kang, S.H.; Jeong, S.H.; Lim, K. Revolutionizing CRISPR technology with artificial intelligence. Exp. Mol. Med. 2025, 57, 1419–1431. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.; Mikiciuk, G.; Durlik, I.; Mikiciuk, M.; Lobodzinska, A.; Snieg, M. The IoT and AI in agriculture: The time is now-A systematic review of smart sensing technologies. Sensors 2025, 25, 3583. [Google Scholar] [CrossRef] [PubMed]


| MTAs/QTLs | Chromosome | Plant Species | Nematode Species | Localization Technology | Ref. |
|---|---|---|---|---|---|
| Soybean cyst nematode (SCN) | |||||
| Rhg1 | 18 | Glycine max (L.) Merr. | Heterodera glycines Ichinohe, 1952 | QTL mapping | [25] |
| Rhg4 | 8 | G. max | H. glycines | QTL mapping | [26] |
| Rhg2 | 11 | G. max | H. glycines | QTL mapping | [27] |
| Rhg3 | 14 | G. max | H. glycines | QTL mapping | [28] |
| cqSCN-003 | 16 | G. max | H. glycines | QTL mapping | [29] |
| cqSCN-005 | 17 | G. max | H. glycines | QTL mapping | [30] |
| cqSCN-006 | 15 | G. max | H. glycines | QTL mapping | [31] |
| cqSCN-007 | 18 | G. max | H. glycines | QTL mapping | [31] |
| cqSCN 10 | 10 | G. max | H. glycines | QTL mapping | [32] |
| cqSCN11 | 11 | G. max | H. glycines | QTL mapping | [33] |
| Cereal cyst nematode (CCN) | |||||
| Ha1 | 5D | Hordeum vulgare L. | Heterodera avenae Wollenweber, 1924 | QTL mapping | [34] |
| Ha2 | 5D | H. vulgare | H. avenae | QTL mapping | [34] |
| Ha3 | 1B | H. vulgare | H. avenae | QTL mapping | [35] |
| Ha4 | 7D | H. vulgare | H. avenae | QTL mapping | [35] |
| Rha2 | 1R | H. vulgare | H. avenae | QTL mapping | [36] |
| QCre-ma7D | 7D | Triticum aestivum L. | Heterodera filipjevi (Madzhidov, 1981) Stelter, 1984 | QTL mapping | [37] |
| QCre-ma2A | 2AS | T. aestivum | H. filipjevi | QTL mapping | [37] |
| Q.Cyst.TZARI.1A | 1A | T. aestivum | H. filipjevi | GWAS | [38] |
| Q.Cyst.TZARI.2A | 2A | T. aestivum | H. filipjevi | GWAS | [38] |
| Q.Cyst.TZARI.1B | 2B | T. aestivum | H. filipjevi | GWAS | [38] |
| Q.Cyst.TZARI.2D | 2D | T. aestivum | H. filipjevi | GWAS | [38] |
| Q.Cyst.TZARI.3A | 3A | T. aestivum | H. filipjevi | GWAS | [38] |
| Q.Cyst.TZARI.6B | 6B | T. aestivum | H. filipjevi | GWAS | [38] |
| Q.Cyst.TZARI.6D | 6D | T. aestivum | H. filipjevi | GWAS | [38] |
| Root-lesion nematodes (RLN) | |||||
| QRlnt.lrc | 6DS | T. aestivum | Pratylenchus spp. | QTL mapping | [39] |
| QRlnt.sk-2B | 2B | T. aestivum | Pratylenchus spp. | QTL mapping | [40] |
| QRlnt.sk-6D | 6D | T. aestivum | Pratylenchus spp. | QTL mapping | [40] |
| Rice root-knot nematode (RRKN) | |||||
| Mg1(t) | 10 | Oryza sativa L. | Meloidogyne graminicola Golden & Birchfield, 1965 | QTL mapping | [41] |
| qMGR4.1 | 4 | O. sativa | M. graminicola | QTL mapping | [42] |
| qMGR7.1 | 7 | O. sativa | M. graminicola | QTL mapping | [42] |
| qMGR9.1 | 9 | O. sativa | M. graminicola | QTL mapping | [42] |
| qGR4.1 | 4 | O. sativa | M. graminicola | QTL mapping | [42] |
| qGR8.1 | 8 | O. sativa | M. graminicola | QTL mapping | [42] |
| qYR5.1 | 5 | O. sativa | M. graminicola | QTL mapping | [42] |
| qYR11.1 | 11 | O. sativa | M. graminicola | QTL mapping | [42] |
| qJ2RS2.1 | 2 | O. sativa | M. graminicola | QTL mapping | [42] |
| qJ2RS3.1 | 3 | O. sativa | M. graminicola | QTL mapping | [42] |
| qGR3.1 | 3 | O. sativa | M. graminicola | QTL mapping | [42] |
| qGR5.1 | 5 | O. sativa | M. graminicola | QTL mapping | [42] |
| qMGR11.1 | 11 | O. sativa | M. graminicola | QTL mapping | [43] |
| Southern root-knot nematode (SRKN) | |||||
| qMi-C11 | 11 | Gossypium hirsutum L. | Meloidogyne incognita (Kofoid & White, 1919) Chitwood, 1949 | QTL mapping | [44] |
| S1_257146517 | Pv06 | Phaseolus vulgaris L. | M. incognita | GWAS | [45] |
| S1_320960286 | Pv07 | P. vulgaris | M. incognita | GWAS | [45] |
| S1_380326043 | Pv08 | P. vulgaris | M. incognita | GWAS | [45] |
| S1_510326192 | Pv11 | P. vulgaris | M. incognita | GWAS | [45] |
| S1_46835797 | Pv01 | P. vulgaris | M. incognita | GWAS | [45] |
| S1_98931885 | Pv02 | P. vulgaris | M. incognita | GWAS | [45] |
| S1_234697928 | Pv05 | P. vulgaris | M. incognita | GWAS | [45] |
| S1_425572688 | Pv10 | P. vulgaris | M. incognita | GWAS | [45] |
| Gene | Chromosome | Plant Species | Nematode Species | Ref. |
|---|---|---|---|---|
| GmSHMT08 | 8 | Glycine max (L.) Merr. | Heterodera glycines Ichinohe, 1952 | [26] |
| CreY | 3S V | Aegilops variabilis Eig. | Heterodera avenae Wollenweber, 1924 | [58] |
| Cre8V | 2V | A. variabilis | H. avenae | [58] |
| CreV | 6V | Haynaldia villosa (L.) Schur | Heterodera filipjevi (Madzhidov, 1981) Stelter, 1984 | [59] |
| Cre6 | 5N V | Aegilops ventricosa Tausch | H. avenae | [59] |
| Cre1 | 2BL | Triticum aestivum L. | H. avenae | [60] |
| Cre8 | 6BL | T. aestivum | H. avenae | [60] |
| MG1 | 11 | Oryza sativa L. | Meloidogyne graminicola Golden & Birchfield, 1965 | [61] |
| GmSNAP18 | 18 | G. max | H. glycines | [62] |
| GmSNAP11 | 11 | G. max | H. glycines | [63] |
| GmSNAP02 | 2 | G. max | H. glycines | [64] |
| Cre2 | 6M V | A. ventricosa | H. avenae | [65] |
| Cre3 | 2DL | Triticum tauschii (Coss.) Schmalh. | H. avenae | [66] |
| Cre4 | 2D | T. tauschii | H. avenae | [67] |
| Cre5 | 2AS | A. ventricosa | H. avenae | [68] |
| Cre7 | 2DL | A. ventricosa | H. avenae | [69] |
| CreR | 6RL | Secale cereale L. | H. avenae | [70] |
| Rlnn1 | 7AL | T. aestivum | Pratylenchus spp. | [71] |
| Mi-1 | 6 | Solanum lycopersicum L. | Meloidogyne incognita (Kofoid & White, 1919) Chitwood, 1949 | [72] |
| Mi-9 | 6 | S. lycopersicum | M. incognita | [73] |
| Mi-3 | 12 | S. lycopersicum | M. incognita | [74] |
| Mi-5 | 6 | S. lycopersicum | M. incognita | [74] |
| Me1 | P9 | Piper nigrum L. | M. incognita | [75] |
| Vat | 5 | Cucumis melo L. | M. incognita | [76] |
| Me3 | 9 | Capsicum annuum L. | M. incognita | [77] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.-W.; Wan, L.-W.; Hao, H.-H.; Wu, W.-C.; Liu, Y.-Q.; Yu, X.-Y.; Peng, D.-L.; Peng, H.; Liu, S.-M.; Kong, L.-A.; et al. Advances of QTL Localization and GWAS Application in Crop Resistances Against Plant-Parasitic Nematodes. Agronomy 2025, 15, 2370. https://doi.org/10.3390/agronomy15102370
Yu J-W, Wan L-W, Hao H-H, Wu W-C, Liu Y-Q, Yu X-Y, Peng D-L, Peng H, Liu S-M, Kong L-A, et al. Advances of QTL Localization and GWAS Application in Crop Resistances Against Plant-Parasitic Nematodes. Agronomy. 2025; 15(10):2370. https://doi.org/10.3390/agronomy15102370
Chicago/Turabian StyleYu, Jing-Wen, Ling-Wei Wan, Huan-Huan Hao, Wen-Cui Wu, Ya-Qin Liu, Xi-Yue Yu, De-Liang Peng, Huan Peng, Shi-Ming Liu, Ling-An Kong, and et al. 2025. "Advances of QTL Localization and GWAS Application in Crop Resistances Against Plant-Parasitic Nematodes" Agronomy 15, no. 10: 2370. https://doi.org/10.3390/agronomy15102370
APA StyleYu, J.-W., Wan, L.-W., Hao, H.-H., Wu, W.-C., Liu, Y.-Q., Yu, X.-Y., Peng, D.-L., Peng, H., Liu, S.-M., Kong, L.-A., Kang, H.-X., & Huang, W.-K. (2025). Advances of QTL Localization and GWAS Application in Crop Resistances Against Plant-Parasitic Nematodes. Agronomy, 15(10), 2370. https://doi.org/10.3390/agronomy15102370









