Transcriptome Analysis Revealed the Molecular Mechanism of Cyanogenic Glycoside Synthesis in Flax
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. RNA Extraction, cDNA Library Construction and Transcriptome Sequencing
2.3. Transcriptome Data Quality Control
2.4. Identification and Functional Annotation Analysis of Differentially Expressed Genes (DEGs)
2.5. qRT-PCR Validation
2.6. Statistical Analysis
3. Results
3.1. Sequencing Data Quality Assessment
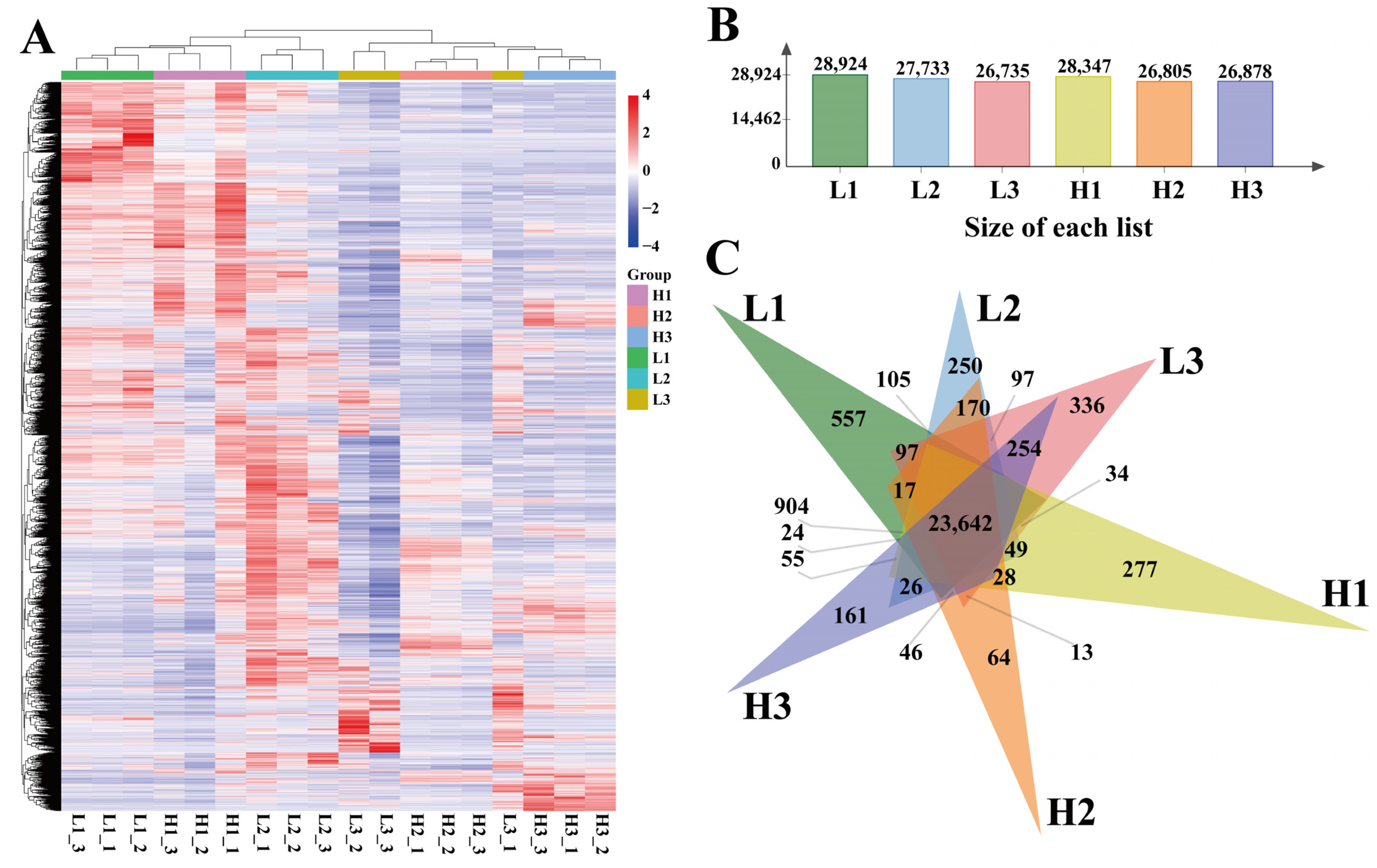
3.2. Correlation Analysis Between Samples
3.3. Genome-Wide Expression Profile Analysis of Different Flax Varieties at Different Developmental Stages
3.4. Identification and Analysis of DEGs in Different Periods of Flax
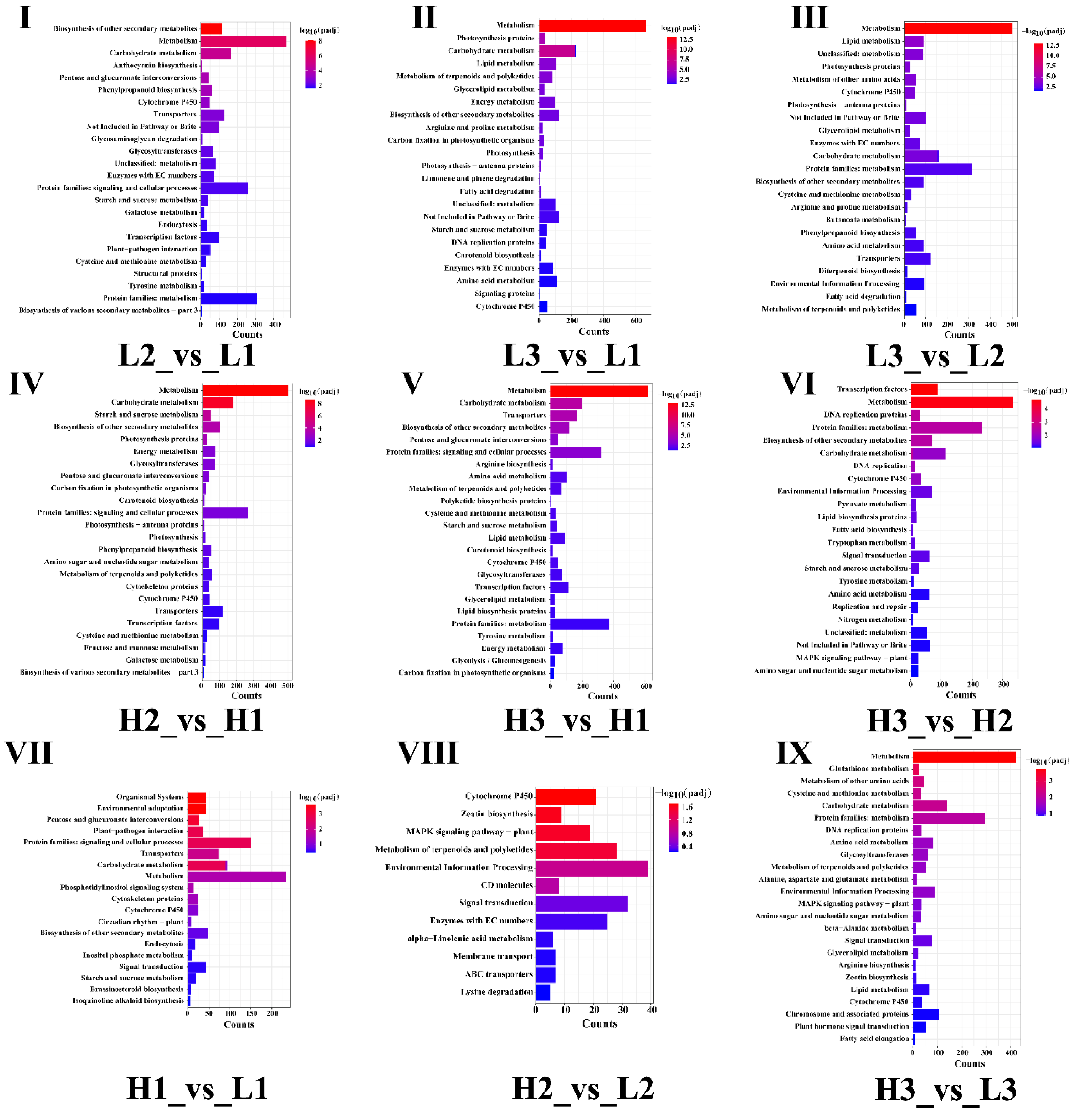
3.5. KEGG Enrichment Analysis of Differentially Expressed Genes
3.6. GO Enrichment Analysis of Differentially Expressed Genes
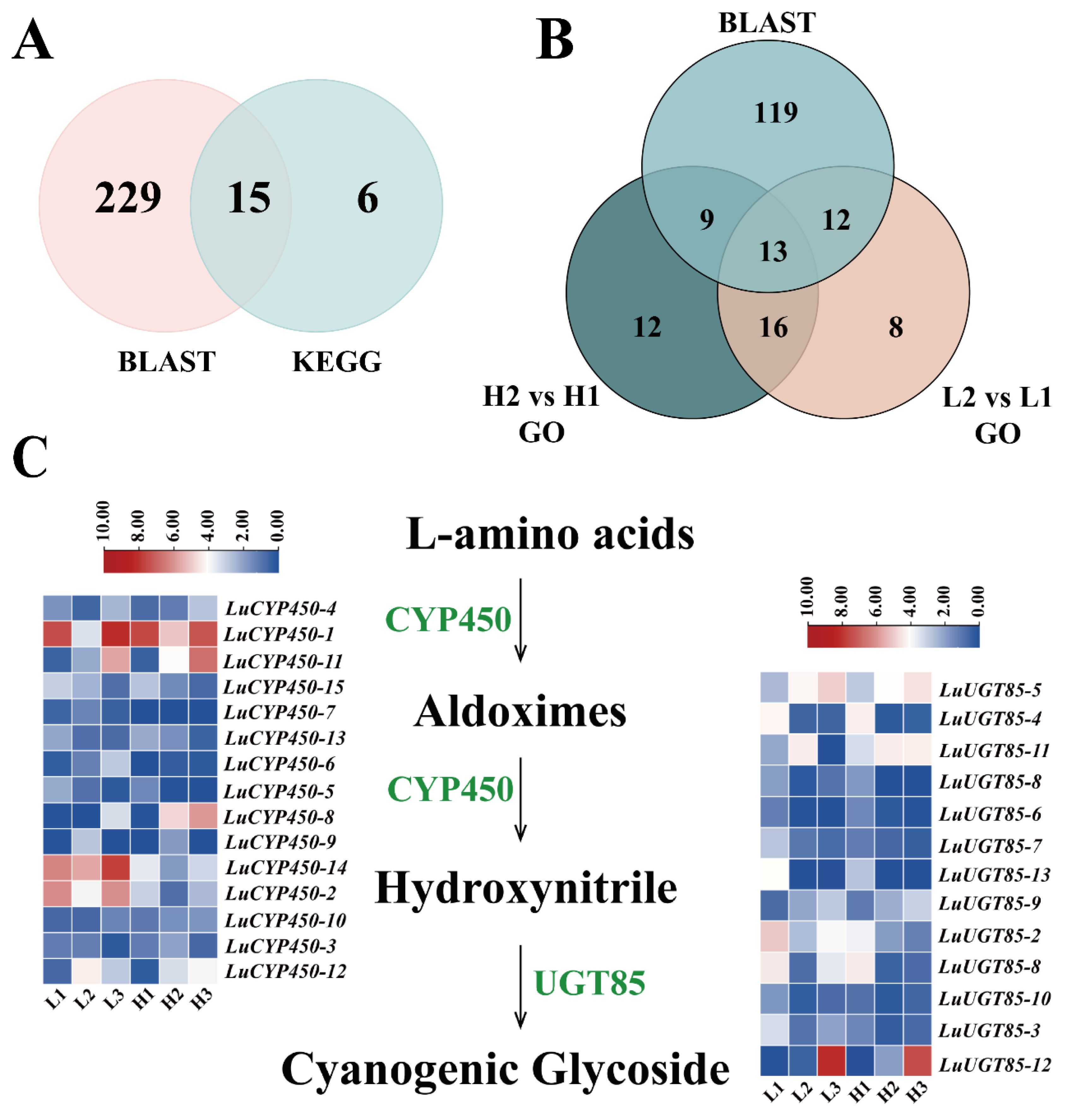
3.7. Screening of Genes Related to Cyanogenic Glycoside Biosynthesis in Flax
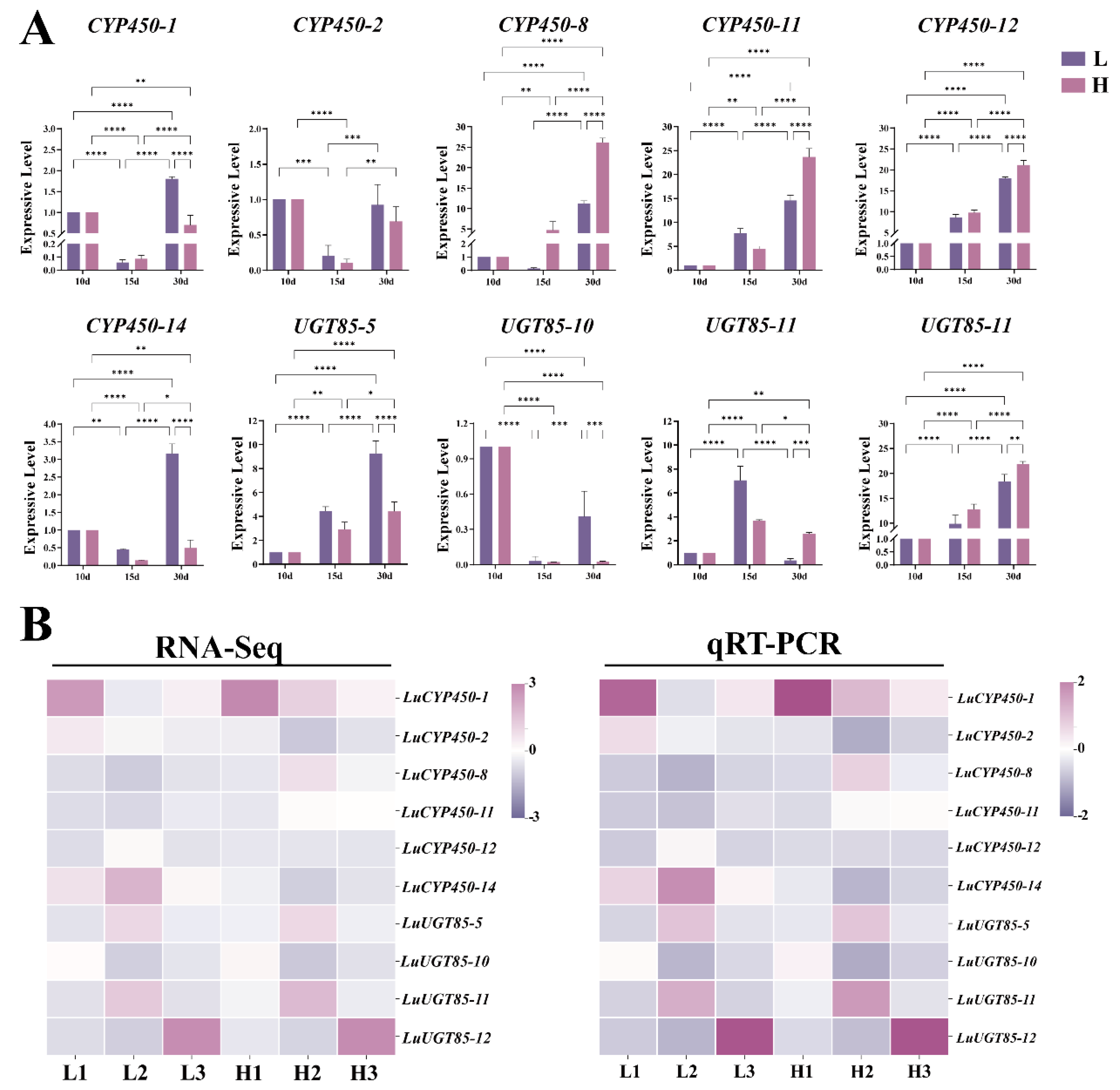
3.8. Cyanogenic Glycoside Synthesis-Related Genes Expression in Flax Seeds
4. Discussion
5. Conclusions
6. Limitations
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xie, D.W.; Dai, Z.G.; Yang, Z.M.; Tang, Q.; Sun, J.; Yang, X.; Song, X.X.; Lu, Y.; Zhao, D.B.; Zhang, L.G.; et al. Genomic variations and association study of agronomic traits in flax. BMC Genom. 2018, 19, e512. [Google Scholar] [CrossRef]
- Yang, J.; Wen, C.T.; Duan, Y.Q.; Deng, Q.C.; Peng, D.F.; Zhang, H.H.; Ma, H.L. The composition, extraction, analysis, bioactivities, bioavailability and applications in food system of flaxseed (Linum usitatissimum L.) oil: A review. Trends Food Sci. Technol. 2021, 118, 252–260. [Google Scholar] [CrossRef]
- Guo, D.L.; Jiang, H.X.; Yan, W.L.; Yang, L.J.; Ye, J.L.; Wang, Y.; Yan, Q.C.; Chen, J.X.; Gao, Y.F.; Duan, L.P.; et al. Resequencing 200 flax cultivated accessions identifies candidate genes related to seed size and weight and reveals signatures of artiffcial selection. Front. Plant Sci. 2020, 10, e1682. [Google Scholar] [CrossRef] [PubMed]
- Sertse, D.; You, F.M.; Ravichandran, S.; Cloutier, S. The genetic structure of flax illustrates environmental and anthropogenic selections that gave rise to its ecogeographical adaptation. Mol. Phylogenet. Evol. 2019, 137, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Kaur, V.; Singh, M.; Wankhede, D.P.; Gupta, K.; Langyan, S.; Aravind, J.; Thangavel, B.; Yadav, S.K.; Kalia, S.; Singh, K.; et al. Diversity of Linum genetic resources in global genebanks: From agro-morphological characterisation to novel genomic technologies—A review. Front. Nutr. 2023, 10, e1165580. [Google Scholar] [CrossRef] [PubMed]
- Mueed, A.; Shibli, S.; Jahangir, M.; Jabbar, S.; Deng, Z.Y. A comprehensive review of flaxseed (Linum usitatissimum L.): Health-affecting compounds, mechanism of toxicity, detoxification, anticancer and potential risk. Crit. Rev. Food Sci. Nutr. 2023, 63, 11081–11104. [Google Scholar] [CrossRef]
- Al-Madhagy, S.; Ashmawy, N.S.; Mamdouh, A.; Eldahshan, O.A.; Farag, M.A. A comprehensive review of the health benefits of flaxseed oil in relation to its chemical composition and comparison with other omega-3-rich oils. Eur. J. Med. Res. 2023, 28, e240. [Google Scholar] [CrossRef]
- Manimurugan, C.; Sujatha, M.; Rathnakumar, A.; Sandhanalakshmi, M.; Zanwar, A. Role of flaxseed (Linum usitatissimum L.) in disease prevention and treatment. Asian Pac. J. Trop. Biomed. 2023, 13, 277–286. [Google Scholar] [CrossRef]
- Kazachkov, M.; Li, Q.; Shen, W.Y.; Wang, L.P.; Gao, P.; Xiang, D.Q.; Datla, R.; Zou, J.T. Molecular identification and functional characterization of a cyanogenic glucosyltransferase from flax (Linum unsitatissimum). PLoS ONE 2020, 15, e0227840. [Google Scholar] [CrossRef]
- Zuk, M.; Pelc, K.; Szperlik, J.; Sawula, A.; Szopa, J. Metabolism of the cyanogenic glucosides in developing flax: Metabolic analysis, and expression pattern of genes. Metabolites 2020, 10, 288. [Google Scholar] [CrossRef]
- Gleadow, R.M.; Møller, B.L. Cyanogenic Glycosides: Synthesis, Physiology, and Phenotypic Plasticity. Annu. Rev. Plant Biol. 2014, 65, 155–185. [Google Scholar] [CrossRef] [PubMed]
- Yulvianti, M.; Zidorn, C. Chemical diversity of plant cyanogenic glycosides: An overview of reported natural products. Molecules 2021, 26, 719. [Google Scholar] [CrossRef] [PubMed]
- Vetter, J. Plant cyanogenic glycosides. Toxicon 2000, 38, 11–36. [Google Scholar] [CrossRef] [PubMed]
- Shim, Y.Y.; Olivia, C.M.; Liu, J.; Boonen, R.; Shen, J.H.; Reaney, M.J.T. Secoisolariciresinol diglucoside and cyanogenic glycosides in gluten free bread fortified with flaxseed meal. J. Agric. Food Chem. 2016, 64, 9551–9558. [Google Scholar] [CrossRef]
- Shahwar, D.; Young, L.W.; Shim, Y.Y.; Reaney, M.J.T. Extractive silylation method for high throughput GC analysis of flaxseed cyanogenic glycosides. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2019, 1132, e121816. [Google Scholar] [CrossRef]
- Oomah, B.D.; Mama, G. Cyanogenic compounds in flaxseed. J. Agric. Food Chem. 1992, 40, 1346–1348. [Google Scholar] [CrossRef]
- Thodberg, S.; Sørensen, M.; Bellucci, M.; Crocoll, C.; Bendtsen, A.K.; Nelson, D.R.; Motawia, M.S.; Møller, B.L.; Neilson, E.H.J. A flavin-dependent monooxygenase catalyzes the initial step in cyanogenic glycoside synthesis in ferns. Commun. Biol. 2020, 3, e507. [Google Scholar] [CrossRef]
- Hansen, K.S.; Kristensen, C.; Tattersall, D.B.; Jones, P.R.; Olsen, C.E.; Bak, S.; Moller, B.L. The in vitro substrate regiospecificity of recombinant UGT85B1, the cyanohydrin glucosyltransferase from Sorghum bicolor. Phytochemistry 2003, 64, 143–151. [Google Scholar] [CrossRef]
- Andersen, M.D.; Busk, P.K.; Svendsen, I.; Moller, B.L. Cytochromes P-450 from cassava (Manihot esculenta crantz) catalyzing the first steps in the biosynthesis of the cyanogenic glucosides linamarin and lotaustralin. J. Biol. Chem. 2000, 275, 1966–1975. [Google Scholar] [CrossRef]
- Ngugi, M.P.; Murugi, N.J.; Oduor, R.O.; Mgutu, A.J.; Ombori, R.O.; Cheriyot, R.C. Determination of cyanogenic compounds content in transgenic acyanogenic kenyan cassava (Manihot esculenta crantz) genotypes: Linking molecular analysis to biochemical analysis. Tech. Univ. Mombasa 2015, 9, 23–29. [Google Scholar]
- Forslund, K.; Morant, M.; Jorgensen, B.; Olsen, C.E.; Asamizu, E.; Sato, S.; Tabata, S.; Bak, S. Biosynthesis of the nitrile glucosides rhodiocyanoside A and D and the cyanogenic glucosides lotaustralin and linamarin in Lotus japonicus. Plant Physiol. 2004, 135, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, K.; Bak, S.; Busk, P.K.; Sorensen, C.; Olsen, C.E.; Puonti-Kaerlas, J.; Moller, B.L. Cassava plants with a depleted cyanogenic glucoside content in leaves and tubers. Distribution of cyanogenic glucosides, their site of synthesis and transport, and blockage of the biosynthesis by RNA interference technology. Plant Physiol. 2005, 139, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Jenrich, R.; Trompetter, I.; Bak, S.; Olsen, C.E.; Moller, B.L.; Piotrowski, M. Evolution of heteromeric nitrilase complexes in Poaceae with new functions in nitrile metabolism. Proc. Natl. Acad. Sci. USA 2007, 104, 18848–18853. [Google Scholar] [CrossRef] [PubMed]
- Soto-Cerda, B.J.; Cloutier, S.; Gajardo, H.A.; Aravena, G.; Quian, R.; You, F.M. Drought response of flax accessions and identification of quantitative trait nucleotides (QTNs) governing agronomic and root traits by genome-wide association analysis. Mol. Breed. 2020, 40, e15. [Google Scholar] [CrossRef]
- Abtahi, M.; Mirlohi, A.; Sharif-Moghaddam, N.; Ataii, E. Revealing seed color variation and their possible association with yield and quality traits in a diversity panel of flax (Linum Usitatissimum L.). Front. Plant Sci. 2022, 13, e1038079. [Google Scholar] [CrossRef]
- Xie, D.W.; Dai, Z.G.; Yang, Z.M.; Sun, J.; Zhao, D.B.; Yang, X.; Zhang, L.G.; Tang, Q.; Su, J.G. Genome-wide association study identifying candidate genes influencing important agronomic traits of flax (Linum usitatissimum L.) using SLAF-seq. Front. Plant Sci. 2018, 8, e2232. [Google Scholar] [CrossRef]
- Yao, Y.; Gu, J.; Luo, Y.; Wang, Y.; Pang, Y.; Shen, G.; Guo, B. Genome-Wide Analysis of UGT Gene Family Identified Key Gene for the Biosynthesis of Bioactive Flavonol Glycosides in Epimedium Pubescens Maxim. Synth. Syst. Biotechnol. 2022, 7, 1095–1107. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Sunbul, M.; Jäschke, A. SRB-2: A promiscuous rainbow aptamer for live-cell RNA imaging. Nucleic Acids Res. 2018, 46, e110. [Google Scholar] [CrossRef]
- Guguchkin, E.; Kasianov, A.; Belenikin, M.; Zobkova, G.; Kosova, E.; Makeev, V.; Karpulevich, E. Enhancing SNV identification in whole-genome sequencing data through the incorporation of known genetic variants into the minimap2 index. BMC Bioinform. 2024, 25, e268. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Spinozzi, G.; Tini, V.; Adorni, A.; Falini, B.; Martelli, M.P. ARPIR: Automatic RNA-Seq pipelines with interactive report. BMC Bioinform. 2020, 21, e574. [Google Scholar] [CrossRef]
- Lachmann, A.; Clarke, D.J.B.; Torre, D.; Xie, Z.R.; Ma’ayan, A. Interoperable RNA-Seq analysis in the cloud. Biochim. Biophys. Acta—Gene Regul. Mech. 2020, 1863, e194521. [Google Scholar] [CrossRef] [PubMed]
- Olney, K.C.; Brotman, S.M.; Andrews, J.P.; Valverde-Vesling, V.A.; Wilson, M.A. Reference genome and transcriptome informed by the sex chromosome complement of the sample increase ability to detect sex differences in gene expression from RNA-Seq data. Biol. Sex Differ. 2020, 11, e42. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Xia, Y.B.; Jiang, G.F.; Wang, X.Q.; Bai, Y.T.; Zhang, P.; Liu, J.N.; Li, L.; Li, H.X.; Huang, L.B.; et al. Identification of quinoa B3 gene family and its expression pattern in response to low temperature stress. Genet. Resour. Crop Evol. 2025, 72, 4345–4360. [Google Scholar] [CrossRef]
- Mitteer, D.R.; Greer, B.D. Using GraphPad Prism’s heat maps for efficient, fine-grained analyses of single-case data. Behav. Anal. Pract. 2022, 15, 505–514. [Google Scholar] [CrossRef]
- Laverty, W.; Kelly, I. Using Excel to Explore the Effects of Assumption Violations on One-Way Analysis of Variance (ANOVA) Statistical Procedures. Open J. Stat. 2019, 9, 458–469. [Google Scholar] [CrossRef]
- Durango, J.M.; Yurango, C.P. Statistical Analysis using traditional technique, Microsoft Excel and Predictive Analytic Software. UIC Res. J. 2016, 19, e1. [Google Scholar] [CrossRef]
- Boulesteix, A.L.; Tutz, G. Identification of interaction patterns and classification with applications to microarray data. Comput. Stat. Data Anal. 2006, 50, 783–802. [Google Scholar] [CrossRef][Green Version]
- O’Connell, T.M. Pathway volcano: An interactive tool for pathway guided visualization of differential expression data. Bioinformatics 2025, 41, ebtaf367. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.H.; Yu, G.C.; Cai, P. ggVennDiagram: An intuitive, easy-to-use, and highly customizable R package to generate Venn Diagram. Front. Genet. 2021, 12, e706907. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Wu, D.; Guo, R.; Li, H.; Wei, R.; Zhang, J.; Wei, Z.; Meng, X.; Yu, H.; Xie, L.; et al. Chromosome-Scale Genomics, Metabolomics, and Transcriptomics Provide Insight into the Synthesis and Regulation of Phenols in Vitis Adenoclada Grapes. Front. Plant Sci. 2023, 14, e1124046. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ren, H.; Yao, X.; Wang, K.; Chang, J. Comparative Transcriptome Analysis Reveals Differential Regulation of Flavonoids Biosynthesis Between Kernels of Two Pecan Cultivars. Front. Plant Sci. 2022, 13, e804968. [Google Scholar] [CrossRef]
- Zhang, W.; Han, Y.; Liao, L. Phenomics and Transcriptomic Profiling of Fruit Development in Distinct Apple Varieties. Sci. Data 2024, 11, e390. [Google Scholar] [CrossRef]
- Liu, L.; Long, C.; Hao, X.R.; Zhang, R.; Li, C.Q.; Song, Y.P. Identification of key genes involved in lignin and flavonoid accumulation during Tilia tuan seed maturation. Plant Cell Rep. 2024, 43, e205. [Google Scholar] [CrossRef]
- Bak, S.; Paquette, S.M.; Morant, M.; Morant, A.V.; Saito, S.; Bjarnholt, N.; Zagrobelny, M.; Jørgensen, K.; Osmani, S.; Simonsen, H.T.; et al. Cyanogenic glycosides: A case study for evolution and application of cytochromes P450. Phytochem. Rev. 2006, 5, 309–329. [Google Scholar] [CrossRef]
- Clausen, M.; Kannangara, R.M.; Olsen, C.E.; Blomstedt, C.K.; Gleadow, R.M.; Jorgensen, K.; Bak, S.; Motawie, M.S.; Moller, B.L. The bifurcation of the cyanogenic glucoside and glucosinolate biosynthetic pathways. Plant J. 2015, 84, 558–573. [Google Scholar] [CrossRef]
- Del Giudice, R.; Putkaradze, N.; dos Santos, B.M.; Hansen, C.C.; Crocoll, C.; Motawia, M.S.; Fredslund, F.; Laursen, T.; Welner, D.H. Structure-guided engineering of key amino acids in UGT85B1 controlling substrate and stereo-specificity in aromatic cyanogenic glucoside biosynthesis. Plant J. 2022, 111, 1539–1549. [Google Scholar] [CrossRef]
- Hansen, C.C.; Sorensen, M.; Veiga, T.A.M.; Zibrandtsen, J.F.S.; Heskes, A.M.; Olsen, C.E.; Boughton, B.A.; Moller, B.L.; Neilsono, E.H.J. Reconfigured cyanogenic glucoside biosynthesis in Eucalyptus cladocalyx involves a cytochrome P450 CYP706C55. Plant Physiol. 2018, 178, 1081–1095. [Google Scholar] [CrossRef]
- Ruegenberg, S.; Horn, M.; Pichlo, C.; Allmeroth, K.; Baumann, U.; Denzel, M.S. Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis. Nat. Commun. 2020, 11, e687. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.M.; Yu, H.X.; Ye, W.W.; Shan, J.X.; Dong, N.Q.; Guo, T.; Xiang, Y.H.; Zhang, H.; Yang, Y.B.; Li, Y.C.; et al. A rice QTL GS3.1 regulates grain size through metabolic-flux distribution between flavonoid and lignin metabolons without affecting stress tolerance. Commun. Biol. 2021, 4, e1171. [Google Scholar] [CrossRef]
- He, B.S.; Zhu, R.R.; Yang, H.D.; Lu, Q.Q.; Wang, W.W.; Song, L.; Sun, X.; Zhang, G.D.; Li, S.J.; Yang, J.L.; et al. Assessing the impact of data preprocessing on analyzing next generation sequencing data. Front. Bioeng. Biotechnol. 2020, 8, e817. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, M.; Sato, F. Unusual P450 Reactions in Plant Secondary Metabolism. Arch. Biochem. Biophys. 2011, 507, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T.; Bishop, G.J. Cytochrome P450s in Plant Steroid Hormone Synthesis and Metabolism. Phytochem. Rev. 2006, 5, 421–432. [Google Scholar] [CrossRef]
- Gao, J.; Ma, B.; Lu, Y.; Zhang, Y.; Tong, Y.; Guo, S.; Gao, W.; Huang, L. Investigating the Catalytic Activity of Glycosyltransferase on Quercetin from Tripterygium Wilfordii. ACS Omega 2020, 5, 1414–1421. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, X.; Zhang, J.; Tang, L.; Yuan, H.; Yao, D.; Jiang, W.; Wu, G.; Cheng, L.; Liu, D.; Yang, L.; et al. Transcriptome Analysis Revealed the Molecular Mechanism of Cyanogenic Glycoside Synthesis in Flax. Agronomy 2025, 15, 2327. https://doi.org/10.3390/agronomy15102327
Song X, Zhang J, Tang L, Yuan H, Yao D, Jiang W, Wu G, Cheng L, Liu D, Yang L, et al. Transcriptome Analysis Revealed the Molecular Mechanism of Cyanogenic Glycoside Synthesis in Flax. Agronomy. 2025; 15(10):2327. https://doi.org/10.3390/agronomy15102327
Chicago/Turabian StyleSong, Xixia, Jinhao Zhang, Lili Tang, Hongmei Yuan, Dandan Yao, Weidong Jiang, Guangwen Wu, Lili Cheng, Dandan Liu, Lie Yang, and et al. 2025. "Transcriptome Analysis Revealed the Molecular Mechanism of Cyanogenic Glycoside Synthesis in Flax" Agronomy 15, no. 10: 2327. https://doi.org/10.3390/agronomy15102327
APA StyleSong, X., Zhang, J., Tang, L., Yuan, H., Yao, D., Jiang, W., Wu, G., Cheng, L., Liu, D., Yang, L., Sun, Z., Qiu, C., Zhang, J., Yi, L., & Kang, Q. (2025). Transcriptome Analysis Revealed the Molecular Mechanism of Cyanogenic Glycoside Synthesis in Flax. Agronomy, 15(10), 2327. https://doi.org/10.3390/agronomy15102327





