Genome-Wide Association Studies of Fiber Content in Sugarcane
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Phenotypic Data Collection and Statistical Analysis
2.3. SNP-Based GWAS
2.4. Identification of Potential Candidate Genes
3. Results
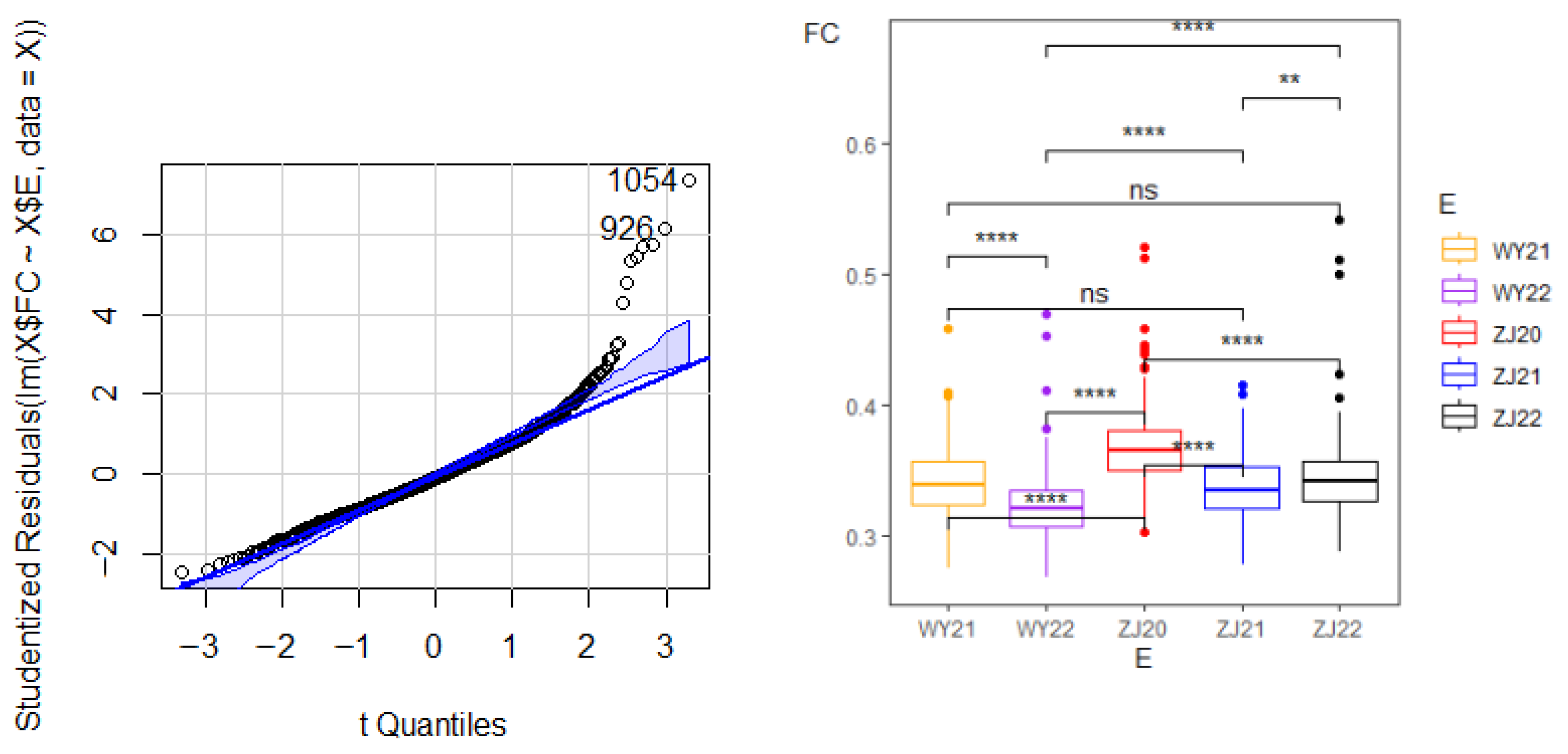
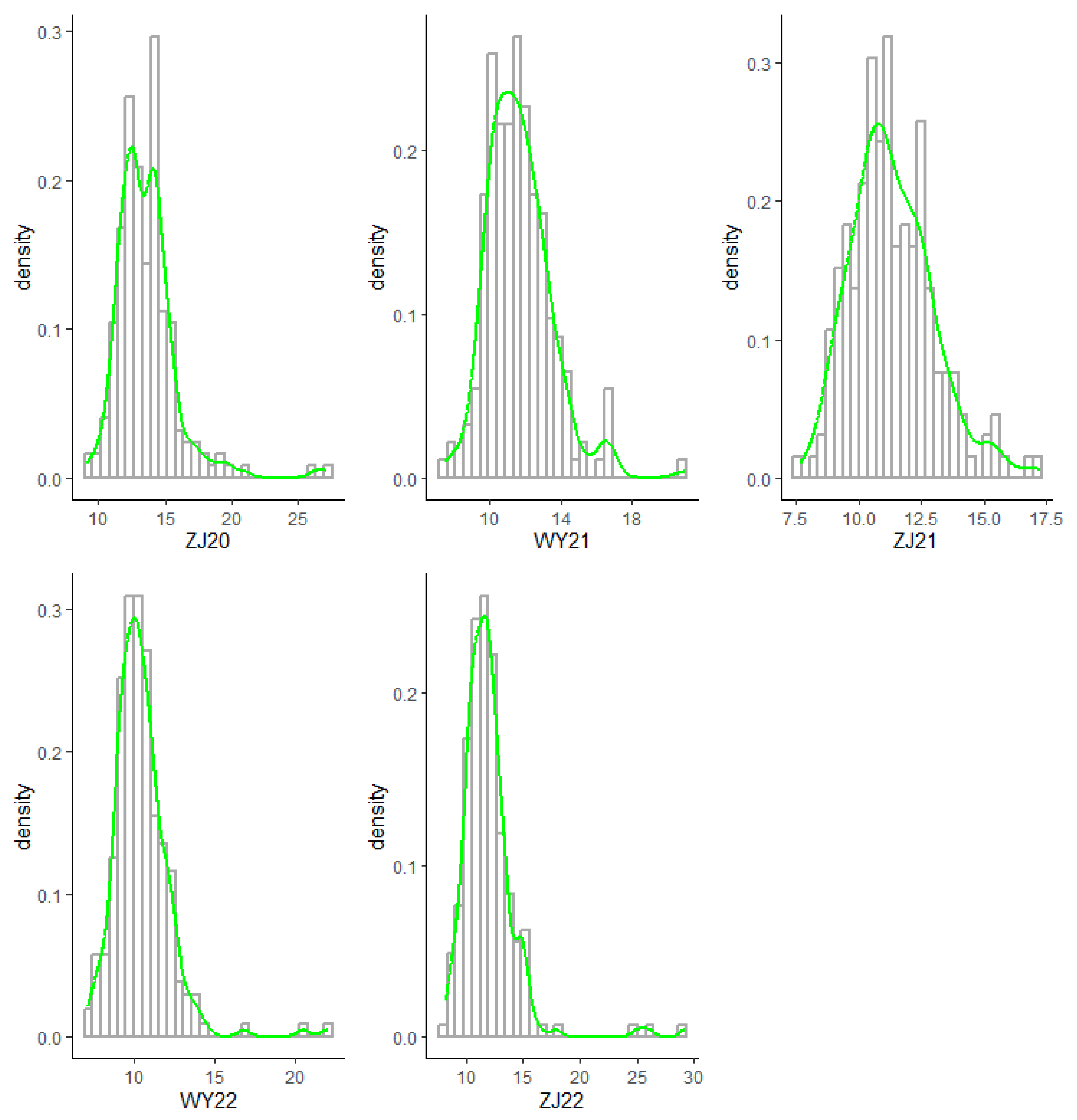
3.1. Phenotypic Variation Analysis
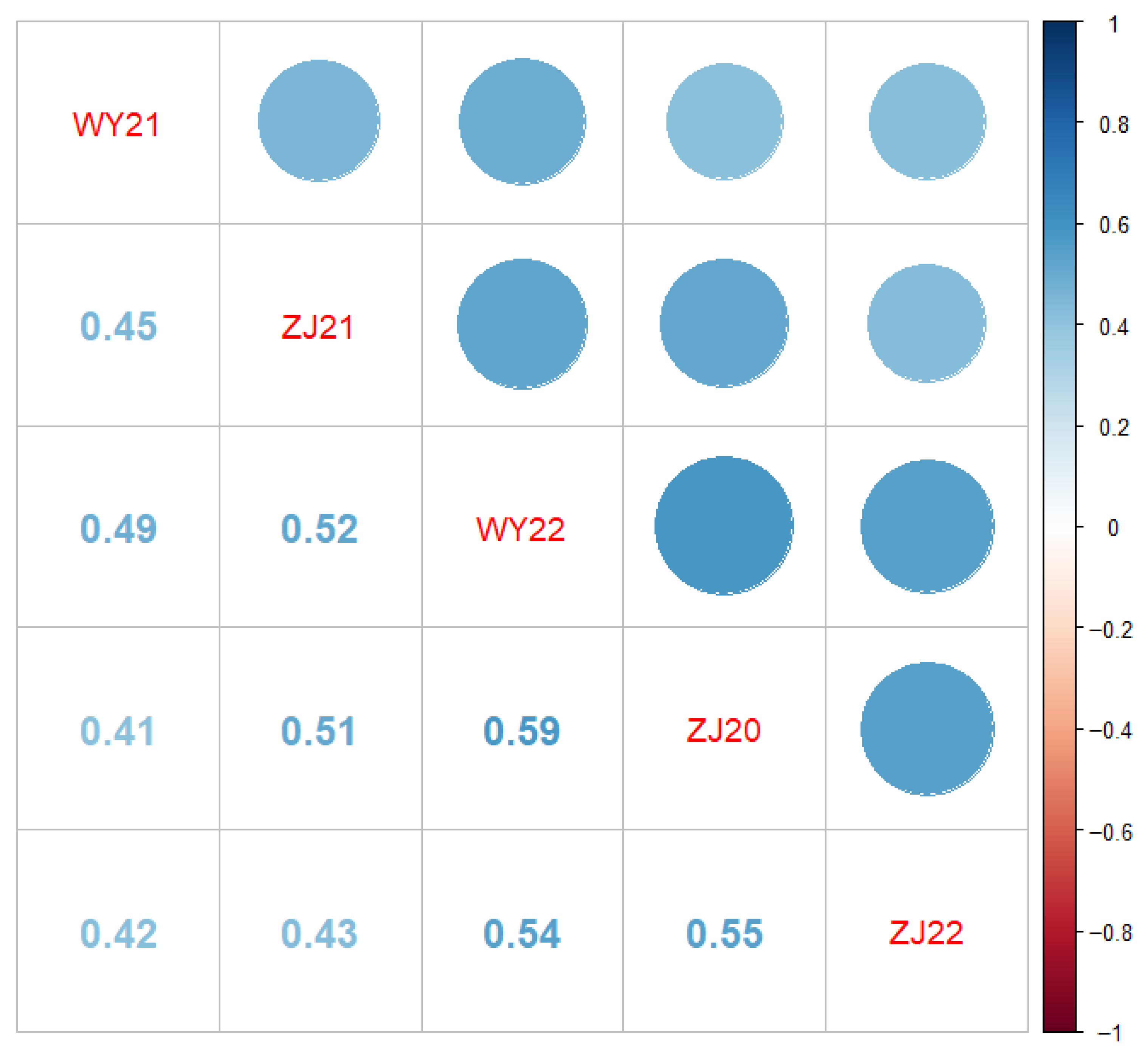
3.2. Correlation Analysis of Phenotypic Data
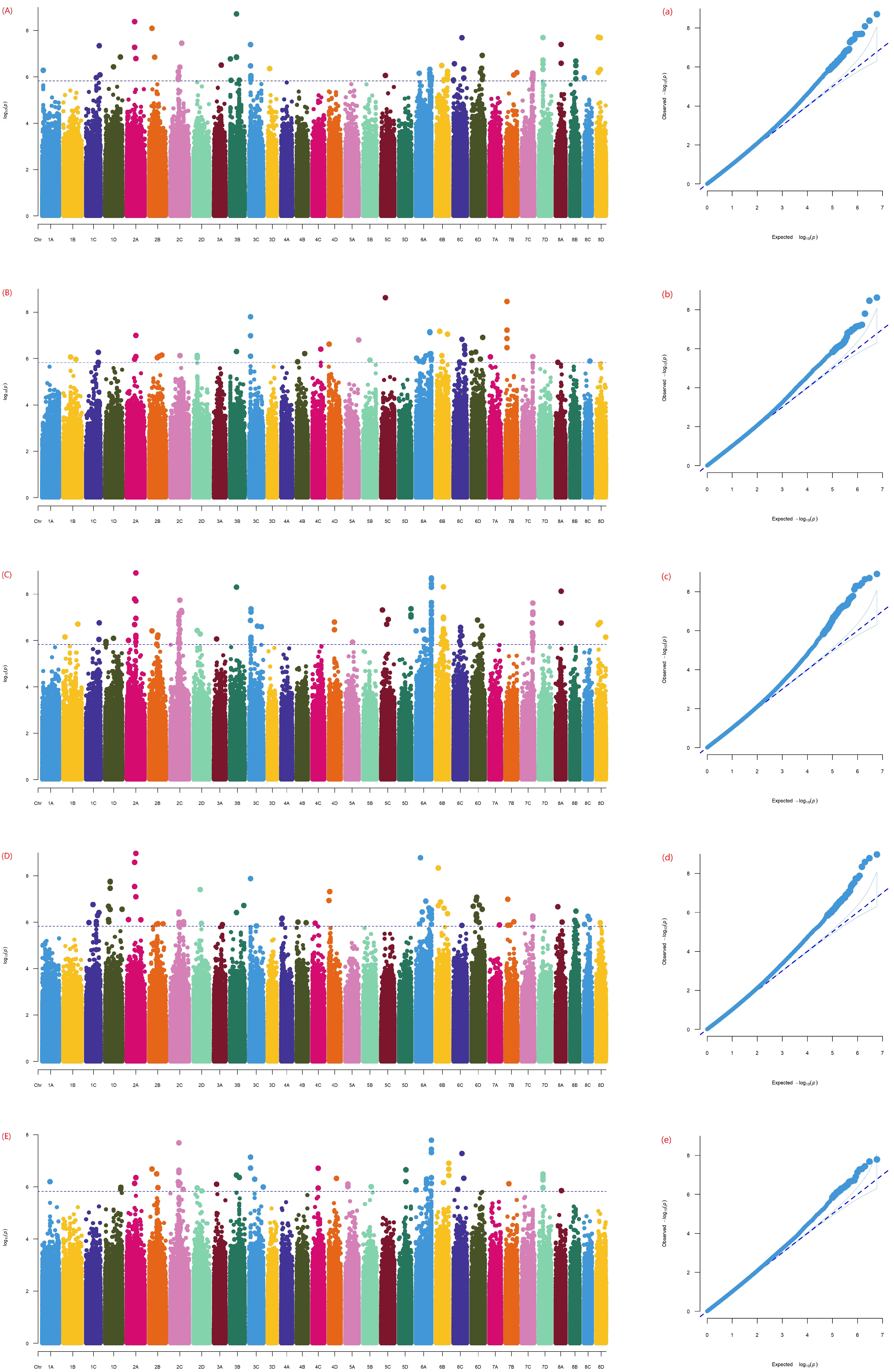
3.3. Genome-Wide Association Analysis
3.4. Identification of Candidate Genes
4. Discussion
4.1. Phenotypic Variation of Sugarcane Fiber Content Across Different Environments
4.2. QTLs Identified for Sugarcane Fiber Content
4.3. Key Candidate Genes Associated with Fiber Content in Sugarcane
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Milano, E.R.; Payne, C.E.; Wolfrum, E.; Lovell, J.; Jenkins, J.; Schmutz, J.; Juenger, T.E. Quantitative trait loci for cell wall composition traits measured using near-infrared spectroscopy in the model c4 perennial grass Panicum hallii. Biotechnol. Biofuels 2018, 11, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Sulis, D.B.; Lavoine, N.; Sederoff, H.; Jiang, X.; Marques, B.M.; Lan, K.; Cofre-Vega, C.; Barrangou, R.; Wang, J.P. Advances in lignocellulosic feedstocks for bioenergy and bioproducts. Nat. Commun. 2025, 16, 1244–1256. [Google Scholar] [CrossRef]
- Yang, X.P.; Todd, J.; Arundale, R.; Binder, J.B.; Luo, Z.L.; Islam, M.S.; Sood, S.; Wang, J.P. Identifying loci controlling fiber composition in polyploid sugarcane (saccharum spp.) Through genome-wide association study. Ind. Crops Prod. 2019, 130, 598–605. [Google Scholar] [CrossRef]
- Lee, Y.; Yang, B.; Kim, W.J.; Kim, J.; Kwon, S.; Kim, J.H.; Ahn, J.; Kim, S.H.; Rha, E.; Ha, B.; et al. Genome-wide association study (GWAS) of the agronomic traits and phenolic content in sorghum (sorghum bicolor l.) Genotypes. Agronomy 2023, 13, 1449. [Google Scholar] [CrossRef]
- Tang, R.H.; Li, X.H.; Lu, X.G.; Zhang, M.Q.; Yao, W. Transmittance vis-NIR spectroscopy for detecting fibre content of living sugarcane. Spectrosc. Spectr. Anal. 2023, 43, 2419–2425. [Google Scholar]
- Li, J.G.; Zhao, P.M.; Zhao, L.Y.; Chen, Q.; Nong, S.K.; Li, Q.; Wang, L.Q. Integrated VIS/NIR spectrum and genome-wide association study for genetic dissection of cellulose crystallinity in wheat stems. Int. J. Mol. Sci. 2024, 25, 3028. [Google Scholar] [CrossRef]
- Shah, L.; Yahya, M.; Shah, S.M.A.; Nadeem, M.; Ali, A.; Ali, A.; Wang, J.; Riaz, M.W.; Rehman, S.; Wu, W.X.; et al. Improving lodging resistance: Using wheat and rice as classical examples. Int. J. Mol. Sci. 2019, 20, 4211. [Google Scholar] [CrossRef]
- Wu, X. QTL Mapping and GWAS for Soluble Solids and Cellulose Components in Brassica Napus. Master”s Thesis, Jiangsu University, Zhenjiang, China, 2022. [Google Scholar]
- Esposito, S.; Taranto, F.; Vitale, P.; Ficco, D.B.M.; Colecchia, S.A.; Stevanato, P.; De Vita, P. Unlocking the molecular basis of wheat straw composition and morphological traits through multi-locus GWAS. BMC Plant Biol. 2022, 22, 519. [Google Scholar] [CrossRef]
- Wang, F.Q.; Ouyang, D.H.; Zhou, Z.Y.; Page, S.J.; Liu, D.H.; Zhao, X.B. Lignocellulosic biomass as sustainable feedstock and materials for power generation and energy storage. J. Energy Chem. 2021, 57, 247–280. [Google Scholar] [CrossRef]
- Bao, J.S.; Jin, L.; Shen, Y.; Xie, J.K. Genetic mapping of quantitative trait loci associated with fiber and lignin content in rice. Cereal Res. Commun. 2007, 35, 23–30. [Google Scholar] [CrossRef]
- Xie, J.K.; Kong, X.L.; Chen, J.; Hu, B.L.; Wen, P.; Zhuang, J.Y.; Bao, J.S. Mapping of quantitative trait loci for fiber and lignin contents from an interspecific cross oryza sativa × oryza rufipogon. J. Zhejiang Univ. Sci. B (Biomed. Biotechnol.) 2011, 12, 518–526. [Google Scholar] [CrossRef][Green Version]
- Cardinal, A.; Lee, M.; Moore, K. Genetic mapping and analysis of quantitative trait loci affecting fiber and lignin content in maize. Theor. Appl. Genet. 2003, 106, 866–874. [Google Scholar] [CrossRef]
- Lorenzana, R.E.; Lewis, M.F.; Jung, H.J.G.; Bernardo, R. Quantitative trait loci and trait correlations for maize stover cell wall composition and glucose release for cellulosic ethanol. Crops Sci. 2010, 50, 541–555. [Google Scholar] [CrossRef]
- Shiringani, A.L.; Friedt, W. QTL for fibre-related traits in grain × sweet sorghum as a tool for the enhancement of sorghum as a biomass crop. Theor. Appl. Genet. 2011, 123, 999–1011. [Google Scholar] [CrossRef] [PubMed]
- Devate, N.B.; Krishna, H.; Parmeshwarappa, S.K.V.; Manjunath, K.K.; Chauhan, D.; Singh, S.; Singh, J.B.; Kumar, M.; Patil, R.; Khan, H.; et al. Genome-wide association mapping for component traits of drought and heat tolerance in wheat. Front. Plant Sci. 2022, 13, 943033–943052. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Yang, X.; Zhao, H.B.; Song, X.Z.; Tsvetkov, Y.D.; Wu, Y.E.; Gao, Q.; Zhang, R.; Zhang, J.M. Genetic analysis of protein content and oil content in soybean by genome-wide association study. Front. Plant Sci. 2023, 14, 1182771–1182782. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.K.; Qu, J.Z.; Lao, Y.H.; Li, T.; Xue, J.Q.; Xu, S.T.; Zhang, X.H. Genome wide association analysis for chlorophyll index at silking stages in maize. Mol. Plant Breed. 2020, 18, 7613–7624. [Google Scholar] [CrossRef]
- Wang, R.; Ren, Y.; Cheng, Y.K.; Wang, W.; Zhang, Z.H.; Geng, H.W. Genome-wide association analysis of morphological traits of flag leaf in wheat. Acta Agron. Sin. 2023, 49, 2886–2901. [Google Scholar] [CrossRef]
- Wang, N.S.; Wang, X.M.; Qian, Y.Z.; Bai, D.; Bao, Y.L.; Zhao, X.Y.; Xu, P.; Li, K.Y.; Li, J.F.; Li, K.; et al. Genome-wide association analysis of rice leaf traits. Agronomy 2023, 13, 2687. [Google Scholar] [CrossRef]
- Wang, Z.B.; Yu, D.J.; Morota, G.; Dhakal, K.; Singer, W.; Lord, N.; Huang, H.B.; Chen, P.Y.; Mozzoni, L.; Li, S.; et al. Genome-wide association analysis of sucrose and alanine contents in edamame beans. Front. Plant Sci. 2023, 13, 1086007–1086018. [Google Scholar] [CrossRef]
- Wang, Q.; Zhu, Y.X.; Zhou, M.M.; Zhang, W.; Zhang, H.M.; Chen, X.; Chen, H.T.; Cui, X.Y. Genome-wide association analysis and candidate genes predication of leaf characteristics traits in soybean (glycine max l.). Acta Agron. Sin. 2024, 50, 623–632. [Google Scholar]
- Zhang, D.Y.; Wang, X.R.; Wu, Y.H.; He, C.Y.; Yang, L.; Jia, L.; Zhuang, J.E. Genome wide association analysis of chlorophyll content in some main spring soybean cultivars. J. Northeast. Agric. Univ. 2022, 53, 1–9. [Google Scholar] [CrossRef]
- Zhang, B.Q.; Huang, Y.X.; Zhang, L.J.; Zhou, Z.F.; Zhou, S.; Duan, W.X.; Yang, C.F.; Gao, Y.J.; Li, S.C.; Chen, M.Y.; et al. Genome-wide association study unravels quantitative trait loci and genes associated with yield-related traits in sugarcane. J. Agric. Food. Chem. 2023, 71, 16815–16826. [Google Scholar] [CrossRef]
- Zou, K.; Kim, K.; Kang, D.; Kim, M.; Ha, J.; Moon, J.; Jun, T. Genome-wide association study of leaf chlorophyll content using high-density SNP array in peanuts (arachis hypogaea l.). Agronomy 2022, 12, 152. [Google Scholar] [CrossRef]
- Healey, A.L.; Garsmeur, O.; Lovell, J.T.; Shengquiang, S.; Sreedasyam, A.; Jenkins, J.; Plott, C.B.; Piperidis, N.; Pompidor, N.; Llaca, V.; et al. The complex polyploid genome architecture of sugarcane. Nature 2024, 628, 804–810. [Google Scholar] [CrossRef]
- Dinesh Babu, K.S.; Janakiraman, V.; Palaniswamy, H.; Kasirajan, L.; Gomathi, R.; Ramkumar, T.R. A short review on sugarcane: Its domestication, molecular manipulations and future perspectives. Genet. Resour. Crop Evol. 2022, 69, 2623–2643. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.B.; Xu, Y.Z.; Khan, A.; Jiang, C.X.; Li, H.J.; Wu, Y.L.; Zhang, C.; Wang, M.Y.; Chen, J.; Zeng, L.F.; et al. Analysis of photosynthetic characteristics and screening high light-efficiency germplasm in sugarcane. Plants 2024, 13, 587. [Google Scholar] [CrossRef]
- Li, H.G.; Tan, Y.M.; Tan, F.; Wang, L.W. Correlation analysis of fiber content and agronomic process characteristics of sugarcane. J. Anhui Agric. Sci. 2010, 38, 15528–15530. [Google Scholar]
- Li, X.J.; Zi, Q.Y.; Xu, C.H.; Li, C.J.; Liu, H.B.; Lin, X.Q.; Mao, J.; Qin, W.; Zhao, L.P.; Liu, X.L. Evaluation and cluster analysis on yield and quality traits of commonly used sugarcane parental clones in China. Sugar Crops China 2022, 44, 1–8. [Google Scholar]
- Amjed, M.; Jung, H.G.; Donker, J.D. Effect of alkaline hydrogen peroxide treatment on cell wall composition and digestion kinetics of sugarcane residues and wheat straw. J. Anim. Sci. 1992, 70, 2877–2884. [Google Scholar] [CrossRef]
- Ferreira, S.S.; Hotta, C.T.; Poelking, V.G.D.C.; Leite, D.C.C.; Buckeridge, M.S.; Loureiro, M.E.; Barbosa, M.H.P.; Carneiro, M.S.; Souza, G.M. Co-expression network analysis reveals transcription factors associated to cell wall biosynthesis in sugarcane. Plant Mol. Biol. 2016, 91, 15–35. [Google Scholar] [CrossRef]
- Kuang, B.W.; Zhao, J.H.; Li, S.C.; Wei, N.; Feng, M.F.; Yang, X.P. Bioinformatics analysis and function prediction of CesA7 gene in sugarcane. Chin. J. Trop. Crops 2023, 44, 1337–1347. [Google Scholar]
- Li, X. Evaluation on Lodging Resistance and Mechanism of Lodging Resistance in Sugarcane; Guangxi University: Nanning, China, 2019. [Google Scholar]
- Lin, Q.S.; Li, T.Y.; Li, X.L.; Zhang, Z.T. Reconsider the effecting of the fiber on milling capacity. Sugarcane Canesugar 1995, 36–39. [Google Scholar]
- Fickett, N.; Gutierrez, A.; Verma, M.; Pontif, M.; Hale, A.; Kimbeng, C.; Baisakh, N. Genome-wide association mapping identifies markers associated with cane yield components and sucrose traits in the louisiana sugarcane core collection. Genomics 2019, 111, 1794–1801. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.Z.; Chen, Y.L.; Pan, Y.B.; Shi, A.N. A genome-wide association study and genomic prediction for fiber and sucrose contents in a mapping population of LCP 85-384 sugarcane. Plants 2023, 12, 1041. [Google Scholar] [CrossRef] [PubMed]
- Andru, S.; Pan, Y.B.; Thongthawee, S.; Burner, D.M.; Kimbeng, C.A. Genetic analysis of the sugarcane (saccharum spp.) Cultivar ‘LCP 85-384’. I. Linkage mapping using AFLP, SSR, and TRAP markers. Theor. Appl. Genet. 2011, 123, 77–93. [Google Scholar] [CrossRef]
- Nayak, S.N.; Song, J.; Villa, A.; Pathak, B.; Ayala-Silva, T.; Yang, X.P.; Todd, J.; Glynn, N.C.; Kuhn, D.N.; Glaz, B.; et al. Promoting utilization of saccharum spp. Genetic resources through genetic diversity analysis and core collection construction. PLoS ONE 2014, 9, e110856. [Google Scholar] [CrossRef]
- Todd, J.R.; Sandhu, H.; Binder, J.; Arundale, R.; Gordon, V.; Song, J.; Glaz, B.; Wang, J.P. Fiber composition of a diversity panel of the world collection of sugarcane (saccharum spp.) And related grasses. Bragantia 2018, 77, 48–61. [Google Scholar] [CrossRef]
- Yang, X.P.; Song, J.; Todd, J.; Peng, Z.; Paudel, D.; Luo, Z.L.; Ma, X.K.; You, Q.; Hanson, E.; Zhao, Z.F.; et al. Target enrichment sequencing of 307 germplasm accessions identified ancestry of ancient and modern hybrids and signatures of adaptation and selection in sugarcane (saccharum spp.), A ‘sweet’ crop with ‘bitter’ genomes. Plant Biotechnol. J. 2019, 17, 488–498. [Google Scholar] [CrossRef]
- Chen, X.L.; Huang, Z.H.; Fu, D.W.; Fang, J.T.; Zhang, X.B.; Feng, X.M.; Xie, J.F.; Wu, B.; Luo, Y.J.; Zhu, M.F.; et al. Identification of genetic loci for sugarcane leaf angle at different developmental stages by genome-wide association study. Front. Plant Sci. 2022, 13, 841693–841705. [Google Scholar] [CrossRef] [PubMed]
- Li, X.H.; Chen, X.L.; Fang, J.T.; Feng, X.M.; Zhang, X.B.; Lin, H.Z.; Chen, W.W.; Zhang, N.N.; He, H.Y.; Huang, Z.H.; et al. Whole-genome sequencing of a worldwide collection of sugarcane cultivars (saccharum spp.) Reveals the genetic basis of cultivar improvement. Plant J. 2024, 119, 2151–2167. [Google Scholar] [CrossRef]
- Zhang, Q.; Qi, Y.W.; Zhang, C.M.; Chen, Y.S.; Deng, H.H. Pedigree analysis of genetic relationship among core parents of sugarcane in mainland China. Guangdong Agric. Sci. 2009, 13, 44–48. (In Chinese) [Google Scholar] [CrossRef]
- Cai, Q.; Fuan, Y.H. Descriptors and Data Standard for Sugarcane; China Agriculture Press: Beijing, China, 2006. [Google Scholar]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, X.; Tang, H.; Zhang, Q.; Hua, X.; Ma, X.; Zhu, F.; Jones, T.; Zhu, X.; Bowers, J.; et al. Allele-defined genome of the autopolyploid sugarcane saccharum spontaneum L. Nat. Genet. 2018, 50, 1565–1573. [Google Scholar] [CrossRef]
- Abbas, M. Functional Characterization of Cellulose Synthase Genes in Secondary Cell Wall Thickening of Xylem Cells during Wood Formation in Populus Trichocarpa; Beijing Forestry University: Beijing, China, 2020. [Google Scholar]
- Du, J.; Vandavasi, V.G.; Molloy, K.R.; Yang, H.; Massenburg, L.N.; Singh, A.; Kwansa, A.L.; Yingling, Y.G.; Neill, H.O.; Chait, B.T.; et al. Evidence for plant-conserved region mediated trimeric CESAs in plant cellulose synthase complexes. Biomacromolecules 2022, 23, 3663–3677. [Google Scholar] [CrossRef]
- Kumar, M.; Mishra, L.; Carr, P.; Pilling, M.; Gardner, P.; Mansfield, S.D.; Turner, S. Exploiting cellulose synthase (CESA) class specificity to probe cellulose microfibril biosynthesis. Plant Physiol. 2018, 177, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Que, F.; Zha, R.F.; Qiang, W. Advances in research of cellulose synthase genes in plants. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2022, 46, 207–214. [Google Scholar]
- Su, J.; He, J.X.; Fu, C.M.; Xian, K.H.; Huang, N.Z. Cloning and preliminary expression analysis of cellulose synthase PfCesA2 and PfCesA8 from paulownia fortunei. Mol. Plant Breed. 2020, 18, 2512–2519. [Google Scholar]
- Baute, J.; Polyn, S.; De Block, J.; Blomme, J.; Van Lijsebettens, M.; Inzï, D. F-box protein FBX92 affects leaf size in Arabidopsis thaliana. Plant Cell Physiol. 2017, 58, 962–975. [Google Scholar] [CrossRef]
- Huo, D.Y.; Zheng, W.J.; Li, P.S.; Xu, Z.S.; Zhou, Y.B.; Chen, M.; Ma, Y.Z.; Min, D.H.; Zhang, X.H. Identification, classification, and drought response of f-box gene family in foxtail millet. Acta Agron. Sin. 2014, 40, 1585–1594. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.R.; Xiong, H.H.; Yu, H.Y.; Wang, C.; Zhang, S.F.; Hao, J.F.; Wang, J.H.; Zhang, H.G.; Zhang, L. Function of MYB8 in larch under PEG simulated drought stress. Sci. Rep. 2024, 14, 11290–11306. [Google Scholar] [CrossRef]
- Anusonpornpurm, S.; Lersrutaiyotin, R.; Rattanakreetakul, C.; Thamchaipenet, A.; Weerathaworn, P. Identifying QTLs for fiber content and agronomic characters in sugarcane using AFLP markers. Kasetsart J. Nat. Sci. 2008, 42, 668–675. [Google Scholar]
- Margarido, G.R.A.; Pastina, M.M.; Souza, A.P.; Garcia, A.A.F. Multi-trait multi-environment quantitative trait loci mapping for a sugarcane commercial cross provides insights on the inheritance of important traits. Mol. Breed. 2015, 35, 175–190. [Google Scholar] [CrossRef] [PubMed]
- Ming, R.; Wang, Y.; Draye, X.; Moore, P.; Irvine, J.; Paterson, A. Molecular dissection of complex traits in autopolyploids: Mapping QTLs affecting sugar yield and related traits in sugarcane. Theor. Appl. Genet. 2002, 105, 332–345. [Google Scholar] [CrossRef]
- Pastina, M.M.; Malosetti, M.; Gazaffi, R.; Mollinari, M.; Margarido, G.R.A.; Oliveira, K.M.; Pinto, L.R.; Souza, A.P.; van Eeuwijk, F.A.; Garcia, A.A.F. A mixed model QTL analysis for sugarcane multiple-harvest-location trial data. Theor. Appl. Genet. 2012, 124, 835–849. [Google Scholar] [CrossRef]
- Pinto, L.R.; Garcia, A.A.F.; Pastina, M.M.; Teixeira, L.H.M.; Bressiani, J.A.; Ulian, E.C.; Bidoia, M.A.P.; Souza, A.P. Analysis of genomic and functional RFLP derived markers associated with sucrose content, fiber and yield QTLs in a sugarcane (saccharum spp.) Commercial cross. Euphytica 2010, 172, 313–327. [Google Scholar] [CrossRef]
- Wenworn, W.; Lersrutaiyotin, R.; Rattanakreetakul, C.; Sreewongchai, T. Identifying quantitative trait loci for fiber content and fiber components in sugarcane using amplified fragment length polymorphism markers. Kasetsart J. Nat. Sci. 2013, 47, 416–423. [Google Scholar]
- Raboin, L.; Pauquet, J.; Butterfield, M.; Hont, A.D.; Glaszmann, J. Analysis of genome-wide linkage disequilibrium in the highly polyploid sugarcane. Theor. Appl. Genet. 2008, 116, 701–714. [Google Scholar] [CrossRef]
- Zhu, Y.J.; Lu, B.S.; Zhang, S.T.; Liu, P.D.; Li, X.F.; Lai, Z.X.; Lin, Y.L. Identification and expression patterns of the cellulose synthase gene family in dimocarpus longan lour. Chin. J. Appl. Environ. Biol. 2020, 26, 1235–1243. [Google Scholar]
- Li, Y.; He, Y.C.; Lin, Y.H.; Wan, T.; Li, M.; Chen, Z.Y. Bioinformatics analysis of cellulose synthase CesA gene from miscanthus lutarioriparius. Fenzi Zhiwu Yuzhong (Mol. Plant Breed.) 2021, 19, 4378–4385. [Google Scholar] [CrossRef]
- Zhang, X.Z.; Yang, K.T.; Bao, R.; Liu, X.L.; Qin, R.L.; Zhao, C.P. Identification and expression analysis of cellulose synthase (CesA csl) gene superfamily members in peach (prunus persica). J. Agric. Biotechnol. 2022, 30, 1096–1111. [Google Scholar]
- Kang, C.; Zhao, X.F.; Wang, P.; Li, Y.D.; Tian, Z.J.; Wu, Z.M. Identification and abiotic stress response analysis of CESA family genes in cucumber (cucumis sativus l.). China Veg. 2022, 29–41. [Google Scholar] [CrossRef]
- Chi, F.L. The Evolution of CesA/csl Superfamily and Gene Function of CesA in Saccharum; Fujian Normal University: Fuzhou, China, 2015. [Google Scholar]
- Hou, J.X. Function Analysis of Three Key CesAs in Sugarcane; Fujian Agriculture and Forestry University: Fuzhou, China, 2018. [Google Scholar]
- Huang, C.M.; Wu, K.C.; Wei, Y.W.; Deng, Z.N.; Lu, Z.; Cao, H.Q.; Luo, H.B.; Jiang, S.L.; Xu, L. Full-length cloning and expression analysis of ScCes3 in sugarcane (saccharum officinarum). J. Agric. Biotechnol. 2018, 26, 2047–2056. [Google Scholar]
- Wang, D.F.; Qin, Y.L.; Fang, J.J.; Yuan, S.J.; Peng, L.X.; Zhao, J.F.; Li, X.Y. A missense mutation in the zinc finger domain of OsCESA7 deleteriously affects cellulose biosynthesis and plant growth in rice. PLoS ONE 2016, 11, e0153993. [Google Scholar] [CrossRef]
- Zhu, F.J.; Cheng, H.H.; Guo, J.N.; Bai, S.M.; Liu, Z.A.; Huang, C.X.; Shen, J.Y.; Wang, K.; Yang, C.J.; Guan, Q.J. Vegetative cell wall protein OsGP1 regulates cell wall mediated soda saline-alkali stress in rice. PeerJ 2024, 12, e16790. [Google Scholar] [CrossRef]
- Zhong, R.Q.; Ye, Z.H. MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol. 2012, 53, 368–380. [Google Scholar] [CrossRef]
- Zhong, R.Q.; Ye, Z.H. Transcriptional regulation of lignin biosynthesis. Plant Signal. Behav. 2009, 4, 1028–1034. [Google Scholar] [CrossRef]
- Huang, D.B.; Wang, S.G.; Zhang, B.C.; Shang-Guan, K.K.; Shi, Y.Y.; Zhang, D.M.; Liu, X.L.; Wu, K.; Xu, Z.P.; Fu, X.D.; et al. A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 2015, 27, 1681–1696. [Google Scholar] [CrossRef]
- Wang, X.Y.; Zhu, C.L.; Yang, K.B.; Li, G.Z.; Li, Z.; Yuan, T.T.; Gao, Z.M. Identification and expression regulation of CesA genes in moso bamboo. J. Nucl. Agric. 2022, 36, 706–715. [Google Scholar]
| Environment 1 | Min 2 (%) | Max 3 (%) | Mean (%) | SD 4 (%) | CV 5 (%) | Skew 6 | Kurt 7 |
|---|---|---|---|---|---|---|---|
| ZJ20 | 9.16 | 27.09 | 13.60 | 2.29 | 16.81 | 2.22 | 9.61 |
| WY21 | 7.52 | 20.94 | 11.69 | 1.81 | 15.48 | 1.13 | 3.16 |
| ZJ21 | 7.69 | 17.24 | 11.34 | 1.64 | 14.42 | 0.67 | 0.71 |
| WY22 | 7.15 | 22.0 | 10.50 | 1.80 | 17.11 | 2.42 | 12.04 |
| ZJ22 | 8.24 | 29.26 | 12.00 | 2.46 | 20.55 | 3.44 | 18.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Feng, X.; Zhang, N.; Lei, Y.; Wu, Z.; Wu, J. Genome-Wide Association Studies of Fiber Content in Sugarcane. Agronomy 2025, 15, 2249. https://doi.org/10.3390/agronomy15102249
Chen Y, Feng X, Zhang N, Lei Y, Wu Z, Wu J. Genome-Wide Association Studies of Fiber Content in Sugarcane. Agronomy. 2025; 15(10):2249. https://doi.org/10.3390/agronomy15102249
Chicago/Turabian StyleChen, Yongsheng, Xiaomin Feng, Nannan Zhang, Yawen Lei, Zilin Wu, and Jiayun Wu. 2025. "Genome-Wide Association Studies of Fiber Content in Sugarcane" Agronomy 15, no. 10: 2249. https://doi.org/10.3390/agronomy15102249
APA StyleChen, Y., Feng, X., Zhang, N., Lei, Y., Wu, Z., & Wu, J. (2025). Genome-Wide Association Studies of Fiber Content in Sugarcane. Agronomy, 15(10), 2249. https://doi.org/10.3390/agronomy15102249






