In-Depth Characterization of Crown Gall Disease of Tobacco in Serbia
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Bacterial Isolation
2.2. Molecular Characterization
2.2.1. DNA Isolation
2.2.2. Multiplex PCR Detection of Agrobacterium Species/Biovars
2.2.3. Detection of Virulence Genes
2.2.4. Multi-Locus Sequence Analysis (MLSA)
2.3. Phenotypic Characterization
2.3.1. Pathogenicity
2.3.2. Biochemical Tests
3. Results
3.1. Symptoms and Bacterial Isolation
3.2. Molecular Characterization
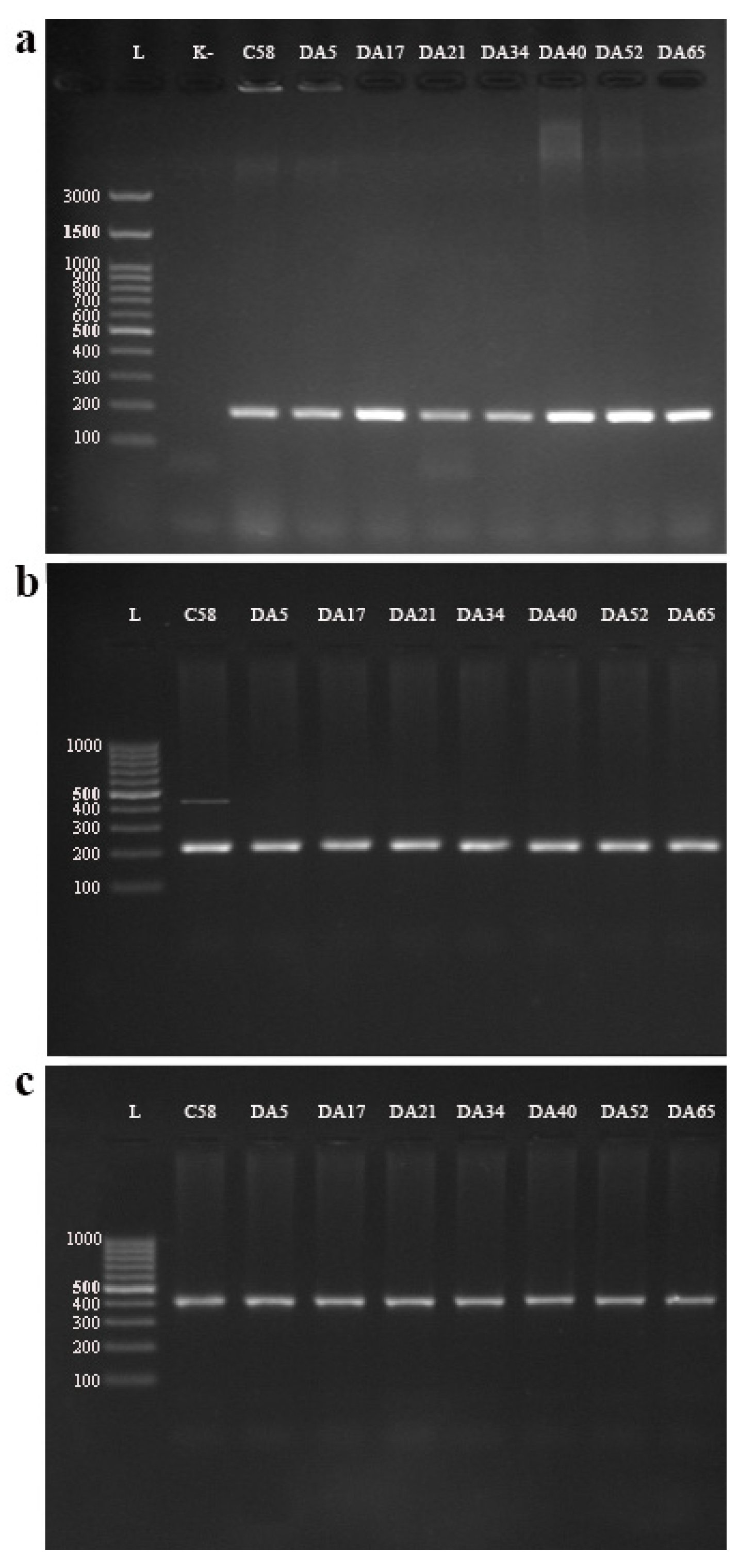
3.2.1. Multiplex PCR Detection of Agrobacterium Species/Biovars
3.2.2. Detection of Virulence Genes
3.2.3. Multi-locus Sequence Analysis (MLSA)
3.3. Phenotypic Characterization of Tobacco Isolates
3.3.1. Pathogenicity
3.3.2. Biochemical Tests
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- CABI. Nicotiana tabacum (tobacco). 2014. Available online: https://www.cabidigitallibrary.org/doi/10.1079/cabicompendium.36326 (accessed on 10 December 2023).
- SYRS. Statistical Yearbook of the Republic of Serbia; Statistical Office of the Republic of Serbia: Belgrade, Serbia, 2023.
- Hooykaas, M.J.G.; Hooykaas, P.J.J. Complete genomic sequence and phylogenomics analysis of Agrobacterium strain AB2/73: A new Rhizobium species with a unique mega-Ti plasmid. BMC Microbiol. 2021, 21, 295. [Google Scholar] [CrossRef] [PubMed]
- Tekiner, N.; Kotan, R. Pathogenicity of Different Rhizobium radiobacter (Agrobacterium tumefaciens) Isolates and Their Diagnosis with Classical Methods. Kahramanmaraş Sütçü İmam Üniversitesi Tarım ve Doğa Derg. 2022, 25, 149–157. [Google Scholar] [CrossRef]
- Portier, P.; Fischer-Le Saux, M.; Mougel, C.; Lerondelle, C.; Chapulliot, D.; Thioulouse, J.; Nesme, X. Identification of genomic species in Agrobacterium biovar 1 by AFLP genomic markers. Appl. Environ. Microbiol. 2006, 72, 7123–7131. [Google Scholar] [CrossRef][Green Version]
- Tiwari, M.; Mishra, A.K.; Chakrabarty, D. Agrobacterium-mediated gene transfer: Recent advancements and layered immunity in plants. Planta 2022, 256, 37. [Google Scholar] [CrossRef]
- Nabi, N.; Hafsa, A.B.; Zellama, M.S.; Chaouachi, M. Pathogenicity, Phylogenetic relationship and NGS based identifcation and assembly of tumorigenic Agrobacterium radiabacter plasmidic and chromosomic reads isolated from Prunus duclcis. Genomics 2019, 111, 1423–1430. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Vijayanand, K.G.; Reddy, M.P.; Singh, A.S.; Naraynan, S. Agrobacterium tumefaciens-Mediated Genetic Transformation: Mechanism and Factors. J. For. Environ. Sci. 2009, 25, 195–204. [Google Scholar]
- Hooykaas, P.J. The Ti plasmid, driver of Agrobacterium pathogenesis. Phytopathology 2023, 113, 594–604. [Google Scholar] [CrossRef]
- Mougel, C.; Thioulouse, J.; Perrie‘re, G.; Nesme, X. A mathematical method for determining genome divergence and species delineation using AFLP. Int. J. Syst. Evol. Microbiol. 2002, 52, 573–586. [Google Scholar] [CrossRef]
- Costechareyre, D.; Bertolla, F.; Nesme, X. Homologous recombination in Agrobacterium: Potential implications for the genomic species concept in bacteria. Mol. Biol. Evol. 2009, 26, 167–176. [Google Scholar] [CrossRef]
- Lassalle, F.; Campillo, T.; Vial, L.; Baude, J.; Costechareyre, D.; Chapulliot, D.; Shams, M.; Abrouk, D.; Lavirre, C.; Oger-Desfeux, C.; et al. Genomic species are ecological species as revealed by comparative genomics in Agrobacterium tumefaciens. Genome Biol. Evol. 2011, 3, 762–781. [Google Scholar] [CrossRef]
- Mafakheri, H.; Taghavi, S.M.; Puławska, J.; De Lajudie, P.; Lassalle, F.; Osdaghi, E. Two novel genomospecies in the Agrobacterium tumefaciens species complex associated with rose crown gall. Phytopathology 2019, 109, 1859–1868. [Google Scholar] [CrossRef] [PubMed]
- Flores-Félix, J.D.; Menéndez, E.; Peix, A.; García-Fraile, P.; Velázquez, E. History and current taxonomic status of genus Agrobacterium. Syst. Appl. Microbiol. 2020, 43, 126046. [Google Scholar] [CrossRef]
- Singh, N.K.; Lavire, C.; Nesme, J.; Vial, L.; Nesme, X.; Mason, C.E.; Lassalle, F.; Venkateswaran, K. Comparative Genomics of Novel Agrobacterium G3 Strains Isolated From the International Space Station and Description of Agrobacterium tomkonis sp. nov. Front. Microbiol. 2021, 12, 765943. [Google Scholar] [CrossRef] [PubMed]
- Weisberg, A.J.; Wu, Y.; Chang, J.H.; Lai, E.M.; Kuo, C.H. Virulence and Ecology of Agrobacteria in the Context of Evolutionary Genomics. Annu. Rev. Phytopathol. 2023, 61, 1–23. [Google Scholar] [CrossRef]
- Furuya, N.; Shimokusuzono, F.; Nakamura, Y.; Nishimura, K.; Takeshita, M.; Matsuyama, N.; Manabe, K.; Takanami, Y. Crown gall of tobacco caused by Agrobacterium tumefaciens biovar 1 in tobacco fields. J. Gen. Pant Pathol. 2004, 70, 39–44. [Google Scholar] [CrossRef]
- Kado, C.I.; Heskett, M.G. Selective media for isolation of Agrobacterium, Corynebacterium, Erwinia, Pseudomonas and Xanthomonas. Phytopathology 1970, 60, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Puławska, J.; Willems, A.; Sobiczewski, P. Rapid and specific identification of four Agrobacterium species and biovars using multiplex PCR. Syst. Appl. Microbiol. 2006, 29, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Haas, J.H.; Moore, L.W.; Ream, W. Universal PCR primers for detection of phytopathogenic Agrobacterium strains. Appl. Environ. Microbiol. 1995, 61, 2879–2884. [Google Scholar] [CrossRef] [PubMed]
- Szegedi, E.; Bottka, S. Detection of Agrobacterium vitis by polymerase chain reaction in grapevine bleeding sap after isolation on a semiselective medium. Vitis 2002, 41, 37–42. [Google Scholar]
- Suzaki, K.; Yoshida, K.; Sawada, H. Detection of tumorigenic Agrobacterium strains from infected apple saplings by colony PCR with improved PCR primers. J. Gen. Plant Pathol. 2004, 70, 342–347. [Google Scholar] [CrossRef]
- Kuzmanović, N.; Ivanović, M.; Ćalić, A.; Gašić, K.; Obradović, A. Differentiation of Phytopathogenic agrobacterium spp. Pestic. Phytomed. 2011, 26, 245–253. [Google Scholar] [CrossRef]
- Aujoulat, F.; Jumas-Bilak, E.; Masnou, A.; Sallé, F.; Faure, D.; Segonds, C.; Marchandin, H.; Teyssier, C. Multilocus sequence-based analysis delineates a clonal population of Agrobacterium (Rhizobium) radiobacter (Agrobacterium tumefaciens) of human origin. J. Bacteriol. 2011, 193, 2608–2618. [Google Scholar] [CrossRef] [PubMed]
- Caccamo, D.; Di Cello, F.; Fani, R.; Gugliandolo, C.; Maugeri, T.L. Polyphasic approach to the characterisation of marine luminous bacteria. Res. Microbiol. 1999, 150, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Ryder, M.H.; Tate, M.E.; Kerr, A. Virulence Properties of Strains of Agrobacterium on the Apical and Basal Surfaces of Carrot Root Discs. Plant Physiol. 1985, 77, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Tolba, I.H.; Soliman, M.A. Efficacy of Native Antagonistic Bacterial Isolates in Biological Control of Crown Gall Disease in Egypt. Ann. Agric. Sci. 2013, 58, 43–49. [Google Scholar] [CrossRef]
- Schaad, N.W.; Jones, J.B.; Chun, W. Laboratory Guide for Identification of Plant Pathogenic Bacteria; The American Phytopathological Society: St. Paul, MN, USA, 2001. [Google Scholar]
- Puławska, J. Crown gall of stone fruits and nuts, economic significance and diversity of its causal agents: Tumorigenic Agrobacterium spp. J. Plant Pathol. 2010, 92, S87–S98. [Google Scholar]
- Holmes, J.E.; Sanghera, H.; Punja, Z.K. Crown gall development on cannabis (Cannabis sativa L., marijuana) plants caused by Agrobacterium tumefaciens species-complex. Can. J. Plant Pathol. 2023, 45, 433–445. [Google Scholar] [CrossRef]
- Alexandre, A.; Laranjo, M.; Young, J.P.; Oliveira, S. dnaJ is a useful phylogenetic marker for Alphaproteobacteria. Int. J. Syst. Evol. Microbiol. 2008, 58, 2839–2849. [Google Scholar] [CrossRef] [PubMed]
- Puławska, J.; Kałuzna, M. Phylogenetic relationship and genetic diversity of Agrobacterium spp. isolated in Poland based on gyrB gene sequence analysis and RAPD. Eur. J. Plant Pathol. 2012, 133, 379–390. [Google Scholar] [CrossRef]
- Pérez-Yépez, J.; Armas-Capote, N.; Velázquez, E.; Pérez-Galdona, R.; Rivas, R.; León-Barrios, M. Evaluation of seven housekeeping genes for multilocus sequence analysis of the genus Mesorhizobium: Resolving the taxonomic affiliation of the Cicer canariense rhizobia. Syst. Appl. Microbiol. 2014, 37, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Kuzmanović, N.; Puławska, J.; Smalla, K.; Nesme, X. Agrobacterium rosae sp. nov., isolated from galls on different agricultural crops. Syst. Appl. Microbiol. 2018, 41, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Kuzmanović, N.; Behrens, P.; Idczak, E.; Wagner, S.; Götz, M.; Spröer, C.; Smalla, K. A novel group of Rhizobium tumorigenes-like agrobacteria associated with crown gall disease of rhododendron and blueberry. Phytopathology 2019, 109, 1840–1848. [Google Scholar] [CrossRef] [PubMed]
- Mysore, K.S.; Bassuner, B.; Deng, X.B.; Darbinian, N.S.; Motchoulski, A.; Ream, W.; Gelvin, S.B. Role of the Agrobacterium tumefaciens VirD2 protein in T-DNA transfer and integration. Mol. Plant Microbe Interact. 1998, 11, 668–683. [Google Scholar] [CrossRef] [PubMed]
- Zoina, A.; Raio, A.; Peluso, R.; Spasiano, A. Characterisation of agrobacteria from weeping fig (Ficus benjamina). Plant Pathol. 2001, 50, 620–627. [Google Scholar] [CrossRef]
- Peluso, R.; Raio, A.; Morra, F.; Zoina, A. Physiological, biochemical and molecular analyses of an Italian collection of Agrobacterium tumefaciens strains. Eur. J. Plant Pathol. 2003, 109, 291–300. [Google Scholar] [CrossRef]




| Primer | Primer Sequence | Region | Fragment Length (bp) |
|---|---|---|---|
| UF/ B1R | 5′-GTAAGAAGCGAACGCAGGGAACT-3′ 5′-GACAATGACTGTTCTACGCGTAA-3′ | Chromosomal (23S rRNA) gene, A. tumefaciens/biovar 1 | 184 |
| UF/ B2R | 5′-TCCGATACCTCCAGGGCCCCTCACA-3′ | Chromosomal (23S rRNA) gene, A. rhizogenes/biovar 2 | 1066 |
| UF/ AvR | 5′-AACTAACTCAATCGCGCTATTAAC-3′ | Chromosomal (23S rRNA) gene, A. vitis | 478 |
| UF/ ArR | 5′-AAAACAGCCACTACGACTGTCTT-3′ | Chromosomal (23S rRNA) gene, A. rubi | 1006 |
| A/ C | 5′-ATGCCCGATCGAGCTCAAGT-3′ 5′-TCGTCTGGCTGACTTTCGTCATAA-3′ | Ti and Ri plasmid virD2 gene | 224 |
| CYT/ CYT | 5′-GATCG(G/C)GTCCAATG(C/T)TGT-3′ 5′-GATATCCATCGATC(T/C)CTT-3′ | Ti plasmid ipt gene | 427 |
| VCF3/ VCR3 | 5′-GGCGGGCGYGCYGAAAGRAARACYT-3′ 5′-AAGAACGYGGNATGTTGCATCTYAC-3′ | Ti and Ri plasmid virC gene | 414 |
| PGF/ PGR | 5′-GGGGCAGGATGCGTTTTTGAG-3′ 5′-GACGGCACTGGGGCTAAGGAT-3′ | Chromosomal pehA gene, A. vitis | 466 |
| atpD (800F/ 1350R) | 5′-GGCCAGGACGTTCTGTTCTT-3′ 5′-CTTGAAGCCCTTGATCGTGT-3′ | F0-F1 ATP synthase subunit beta | 465 |
| dnaK (720F/ 1400R) | 5′-GAAGACTTCGACATGCGTCT-3′ 5′-GCCGAGCAGCTTGTTGTC-3′ | Heat shock protein, 70 kDa | 480 |
| glnA (144F/ 900R) | 5′-GTCATGTTCGACGGCTCCT-3′ 5′-CCTTGGCATGCTTGATGAT-3′ | Glutamine synthetase | 474 |
| rpoB (2040F/ 2718R) | 5′-GAAAACGACGACGCCAAC-3′ 5′-GCGCAGAAGCTTTTCTTCC-3′ | Beta subunit RNA polymerase | 534 |
| 27F 1495R | 5′-GAGAGTTTGATCCTGGCTCAG-3′ 5′-CTACGGCTACCTTGTTACGA-3′ | 16S rRNA | 1550 |
| Species | Strain | Isolation Source | Country | Acc. No. |
|---|---|---|---|---|
| A. tumefaciens | HAMBI 105 | soil | USA | CP139997 |
| Gle002 | walnut | USA: California | CP048564 | |
| Yol001 | walnut | USA: California | CP048477 | |
| Yol002 | walnut | USA: California | CP048473 | |
| 12D1 | - | - | CP033031 | |
| 183 | almond | Tunisia | CP029044 | |
| O54/95 | cherry | - | CP124967 | |
| BIM B-1315G | root endosphere of soybean | Belarus: Minsk | CP061003 | |
| A. larrymoorei | AF3.44 | Ficus benjamina | USA: Florida | CP072167 |
| CFBP5477 | - | Italy | CP124733 | |
| A. leguminum | CFBP4996 | - | - | CP120211 |
| A. fabrum | C58 | USA | AE007869 | |
| 1D132 | - | CP033022 | ||
| A. vaccinii | B7.6 | blueberry | Poland | CP054150 |
| A. pusense | 76 | hyphae Fusarium oxysporum f. sp. cucumerinum | China: Beijing | CP053856 |
| M. huakuii a | NZP2235 | Lotus japonicus | New Zealand | CP139858 |
| Isolate | ||||||||
|---|---|---|---|---|---|---|---|---|
| Test | DA5 | DA17 | DA21 | DA34 | DA40 | DA52 | DA65 | C58 |
| Gram reaction | − | − | − | − | − | − | − | − |
| Catalase | + | + | + | + | + | + | + | + |
| Oxidase | − | − | − | − | − | − | − | + |
| Production of fluorescent pigment | − | − | − | − | − | − | − | − |
| Fermentation of: | ||||||||
| glycerol | + | + | + | + | + | + | + | − |
| d-arabinose | + | + | + | + | + | + | + | + |
| l-arabinose | + | + | + | + | + | + | + | + |
| d-ribose | + | + | + | + | + | + | + | + |
| d-xylose | + | + | + | + | + | + | + | + |
| d-adonitol | + | + | + | + | + | + | + | + |
| methyl-β-d-xylopyranoside | + | + | + | + | + | + | + | − |
| d-galactose | + | + | + | + | + | + | + | − |
| d-glucose | + | + | + | + | + | + | + | + |
| d-fructose | + | + | + | + | + | + | + | + |
| d-mannose | + | + | + | + | + | + | + | + |
| l-rhamnose | + | + | + | + | + | + | + | + |
| dulcitol | + | + | + | + | + | + | + | + |
| inositol | + | + | + | + | + | + | + | + |
| d-mannitol | + | + | + | + | + | + | + | + |
| d-sorbitol | + | + | + | + | + | + | + | + |
| aesculin ferric citrate | + | + | + | + | + | + | + | + |
| salicin | + | + | + | + | + | + | + | + |
| d-cellobiose | + | + | + | + | + | + | + | + |
| d-maltose | + | + | + | + | + | + | + | + |
| d-lactose | + | + | + | + | + | + | + | + |
| d-melibiose | + | + | + | + | + | + | + | + |
| d-sucrose | + | + | + | + | + | + | + | + |
| d-trehalose | + | + | + | + | + | + | + | + |
| d-raffinose | + | + | + | + | + | + | + | + |
| d-turanose | + | + | + | + | + | + | + | + |
| d-lyxose | + | + | + | + | + | + | + | + |
| d-tagatose | + | + | + | + | + | + | + | + |
| d-fucose | + | + | + | + | + | + | + | − |
| l-fucose | + | + | + | + | + | + | + | − |
| d-arabitol | + | + | + | + | + | + | + | − |
| l-arabitol | + | + | + | + | + | + | + | + |
| erythritol | − | − | − | − | − | − | − | − |
| l-xylose | − | − | − | − | − | − | − | − |
| l-sorbose | − | − | − | − | − | − | − | + |
| methyl α-d-mannopyranoside | − | − | − | − | − | − | − | − |
| methyl α-d-glucopyranoside | − | − | − | − | − | − | − | − |
| N-acetylglucosamine | − | − | − | − | − | − | − | − |
| amygdalin | − | − | − | − | − | − | − | − |
| arbutin | − | − | − | − | − | − | − | − |
| inulin | − | − | − | − | − | − | − | − |
| d-melezitose | − | − | − | − | − | − | − | − |
| amidon (starch) | − | − | − | − | − | − | − | − |
| glycogen | − | − | − | − | − | − | − | − |
| xylitol | − | − | − | − | − | − | − | − |
| gentiobiose | − | − | − | − | − | − | − | − |
| potassium gluconate | − | − | − | − | − | − | − | − |
| potassium 2-ketogluconate | − | − | − | − | − | − | − | − |
| potassium 5-ketogluconate | − | − | − | − | − | − | − | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iličić, R.; Jelušić, A.; Barać, G.; Nikolić, D.; Stošić, N.; Scortichini, M.; Milovanović, T.P. In-Depth Characterization of Crown Gall Disease of Tobacco in Serbia. Agronomy 2024, 14, 851. https://doi.org/10.3390/agronomy14040851
Iličić R, Jelušić A, Barać G, Nikolić D, Stošić N, Scortichini M, Milovanović TP. In-Depth Characterization of Crown Gall Disease of Tobacco in Serbia. Agronomy. 2024; 14(4):851. https://doi.org/10.3390/agronomy14040851
Chicago/Turabian StyleIličić, Renata, Aleksandra Jelušić, Goran Barać, Dušan Nikolić, Nemanja Stošić, Marco Scortichini, and Tatjana Popović Milovanović. 2024. "In-Depth Characterization of Crown Gall Disease of Tobacco in Serbia" Agronomy 14, no. 4: 851. https://doi.org/10.3390/agronomy14040851
APA StyleIličić, R., Jelušić, A., Barać, G., Nikolić, D., Stošić, N., Scortichini, M., & Milovanović, T. P. (2024). In-Depth Characterization of Crown Gall Disease of Tobacco in Serbia. Agronomy, 14(4), 851. https://doi.org/10.3390/agronomy14040851








