Diversity and Evolution of the Avirulence Gene AvrPi54 in Yunnan Rice Fields
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Isolates, Rice Cultivars, Culture, and Pathogenicity Assay
2.2. DNA Preparation, PCR Amplification, and DNA Sequencing
2.3. Data Analysis
3. Results
3.1. The Distribution and Pathogenicity Assay of Avirulence Gene AvrPi54 in Yunnan Province
3.2. Variance, Distribution of AvrPi54 Haplotypes
3.3. AvrPi54 Haplotypes Diversity and Network
3.4. Phylogenetic Analysis of AvrPi54
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zeng, D.; Tian, Z.; Rao, Y.; Dong, G.; Yang, Y.; Huang, L.; Leng, Y.; Xu, J.; Sun, C.; Zhang, G.; et al. Rational design of high-yield and superior-quality rice. Nat. Plants 2017, 3, 17031. [Google Scholar] [CrossRef]
- Dean, R.A.; Talbot, N.J.; Ebbole, D.J.; Farman, M.L.; Mitchell, T.K.; Orbach, M.J.; Thon, M.; Kulkarni, R.; Xu, J.R.; Pan, H.; et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 2005, 434, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.H. Rice Disease, 2nd ed.; Commonwealth Mycological Institute: Kew, UK, 1985. [Google Scholar]
- Couch, B.C.; Kohn, L.M. A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia 2002, 94, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Takabayashi, N.; Tosa, Y.; Oh, H.S.; Mayama, S. A gene-for-gene relationship underlying the species-specific parasitism of Avena/Triticum isolates of Magnaporthe grisea on wheat cultivars. Phytopathology 2002, 92, 1182–1188. [Google Scholar] [CrossRef] [PubMed]
- Murakami, J.; Tomita, R.; Kataoka, T.; Nakayashiki, H.; Tosa, Y.; Mayama, S. Analysis of host species specificity of Magnaporthe grisea toward foxtail millet using a genetic cross between isolates from wheat and foxtail millet. Phytopathology 2003, 93, 42–45. [Google Scholar] [CrossRef] [PubMed]
- Couch, B.C.; Fudal, I.; Lebrun, M.H.; Tharreau, D.; Valent, B.; van Kim, P.; Nottéghem, J.L.; Kohn, L.M. Origins of host-specific populations of the blast pathogen Magnaporthe oryzae in crop domestication with subsequent expansion of pandemic clones on rice and weeds of rice. Genetics 2005, 170, 613–630. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.C.; Fraser, R.W.; Paini, D.R.; Warden, A.C.; Lonsdale, W.M.; De Barro, P.J. Biosecurity and yield improvement technologies are strategic complements in the fight against food insecurity. PLoS ONE 2011, 6, e26084. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Park, S.Y.; Kim, B.R.; Roh, J.H.; Oh, I.S.; Han, S.S.; Lee, Y.H. Comparative analysis of pathogenicity and phylogenetic relationship in Magnaporthe grisea species complex. PLoS ONE 2013, 8, e57196. [Google Scholar] [CrossRef] [PubMed]
- Orbach, M.J.; Farrall, L.; Sweigard, J.A.; Chumley, F.G.; Valent, B. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 2000, 12, 2019–2032. [Google Scholar] [CrossRef]
- Dai, Y.; Jia, Y.; Correll, J.; Wang, X.; Wang, Y. Diversification and evolution of the avirulence gene AVR-Pita1 in field isolates of Magnaporthe oryzae. Fungal Genet. Biol. 2010, 47, 973–980. [Google Scholar] [CrossRef]
- Jia, Y.; Zhou, E.; Lee, S.; Bianco, T. Coevolutionary dynamics of rice blast resistance gene Pi-ta and Magnaporthe oryzae avirulence gene AVR-Pita1. Phytopathology 2016, 106, 676–683. [Google Scholar] [CrossRef]
- Singh, P.K.; Ray, S.; Thakur, S.; Rathour, R.; Sharma, V.; Sharma, T.R. Co-evolutionary interactions between host resistance and pathogen avirulence genes in rice-Magnaporthe oryzae pathosystem. Fungal Genet. Biol. 2018, 115, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.G.; Lawrence, G.J.; Luck, J.E.; Dodds, P.N. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 1999, 11, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Lawrence, G.J.; Catanzariti, A.M.; Ayliffe, M.A.; Ellis, J.G. The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 2004, 16, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Lawrence, G.J.; Catanzariti, A.M.; Teh, T.; Wang, C.I.; Ayliffe, M.A.; Kobe, B.; Ellis, J.G. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 2006, 103, 8888–8893. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.L.; McDowell, J.M. Two modes of pathogen recognition by plants. Proc. Natl. Acad. Sci. USA 2006, 103, 8575–8576. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, H.; Yoshida, K.; Saitoh, H.; Fujisaki, K.; Hirabuchi, A.; Alaux, L.; Fournier, E.; Tharreau, D.; Terauchi, R. Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J. 2012, 72, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Stahl, E.A.; Dwyer, G.; Mauricio, R.; Kreitman, M.; Bergelson, J. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 1999, 400, 667–671. [Google Scholar] [CrossRef] [PubMed]
- Stahl, E.A.; Bishop, J.G. Plant-pathogen arms races at the molecular level. Curr. Opin. Plant Biol. 2000, 3, 299–304. [Google Scholar] [CrossRef]
- Flor, H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Zeigler, R.S.; Leong, S.A.; Teng, P.S. Rice Blast Disease; Commonwealth Agricultural Bureau International: Wallingford, UK, 1994. [Google Scholar]
- Silué, D.; Notteghem, J.L.; Tharreau, D. Evidence of a gene for gene relationship in the Oryza sativa-Magnaporthe grisea pathosystem. Phytopathology 1992, 82, 577–580. [Google Scholar] [CrossRef]
- Devanna, B.N.; Jain, P.; Solanke, A.U.; Das, A.; Thakur, S.; Singh, P.K.; Kumari, M.; Dubey, H.; Jaswal, R.; Pawar, D.; et al. Understanding the dynamics of blast resistance in rice-Magnaporthe oryzae interactions. J. Fungi 2022, 8, 584. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.H.; Ebbole, D.J.; Wang, Z.H. The arms race between Magnaporthe oryzae and rice: Diversity and interaction of Avr and R genes. J. Integr. Agric. 2017, 16, 2746–2760. [Google Scholar] [CrossRef]
- Wu, W.; Wang, L.; Zhang, S.; Li, Z.; Zhang, Y.; Lin, F.; Pan, Q. Stepwise arms race between AvrPik and Pik alleles in the rice blast pathosystem. Mol. Plant. Microbe Interact. 2014, 27, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, C.Y.; Park, S.Y.; Kim, K.T.; Jeon, J.; Chung, H.; Choi, G.; Kwon, S.; Choi, J.; Jeon, J.; et al. Two nuclear effectors of the rice blast fungus modulate host immunity via transcriptional reprogramming. Nat. Commun. 2020, 11, 5845. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Singh, P.K.; Gupta, D.K.; Mahato, A.K.; Sarkar, C.; Rathour, R.; Singh, N.K.; Sharma, T.R. Analysis of Magnaporthe oryzae genome reveals a fungal effector, which is able to induce resistance response in transgenic rice line containing resistance gene, Pi54. Front. Plant Sci. 2016, 7, 1140. [Google Scholar] [CrossRef] [PubMed]
- Sharma, T.R.; Madhav, M.S.; Singh, B.K.; Shanker, P.; Jana, T.K.; Dalal, V.; Pandit, A.; Singh, A.; Gaikwad, K.; Upreti, H.C. High-resolution mapping, cloning and molecular characterization of the Pi-kh gene of rice, which confers resistance to Magnaporthe grisea. Mol. Genet. Genom. 2005, 274, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.K.; Kumar, S.P.; Gupta, S.K.; Gautam, N.; Singh, N.K.; Sharma, T.R. Functional complementation of rice blast resistance gene Pi-kh (Pi54) conferring resistance to diverse strains of Magnaporthe oryzae. J. Plant Biochem. Biotechnol. 2011, 20, 55–65. [Google Scholar] [CrossRef]
- Kumari, M.; Devanna, B.N.; Singh, P.K.; Rajashekara, H.; Sharma, V.; Sharma, T.R. Stacking of blast resistance orthologue genes in susceptible indica rice line improves resistance against Magnaporthe oryzae. 3 Biotech 2017, 8, 37. [Google Scholar] [CrossRef]
- Kumari, M.; Rai, A.K.; Devanna, B.N.; Singh, P.K.; Kapoor, R.; Rajashekara, H.; Prakash, G.; Sharma, V.; Sharma, T.R. Co-transformation mediated stacking of blast resistance genes Pi54 and Pi54rh in rice provides broad spectrum resistance against Magnaporthe oryzae. Plant Cell Rep. 2017, 36, 1747–1755. [Google Scholar] [CrossRef]
- Gupta, S.K.; Rai, A.K.; Kanwar, S.S.; Chand, D.; Singh, N.K.; Sharma, T.R. The single functional blast resistance gene Pi54 activates a complex defence mechanism in rice. J. Exp. Bot. 2012, 63, 757–772. [Google Scholar] [CrossRef]
- Singh, J.; Gupta, S.K.; Devanna, B.N.; Singh, S.; Upadhyay, A.; Sharma, T.R. Blast resistance gene Pi54 over-expressed in rice to understand its cellular and sub-cellular localization and response to different pathogens. Sci. Rep. 2020, 10, 5243. [Google Scholar] [CrossRef]
- Das, A.; Soubam, D.; Singh, P.K.; Thakur, S.; Singh, N.K.; Sharma, T.R. A novel blast resistance gene, Pi54rh cloned from wild species of rice, Oryza rhizomatis confers broad spectrum resistance to Magnaporthe oryzae. Funct. Integr. Genom. 2012, 12, 215–228. [Google Scholar] [CrossRef]
- Devanna, N.B.; Vijayan, J.; Sharma, T.R. The blast resistance gene Pi54of cloned from Oryza officinalis interacts with Avr-Pi54 through its novel non-LRR domains. PLoS ONE 2014, 9, e104840. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Das, A.; Devanna, B.N.; Thakur, S.; Singh, P.K.; Singh, N.K.; Sharma, T.R. Mining of rice blast resistance gene Pi54 shows effect of single nucleotide polymorphisms on phenotypic expression of the alleles. Eur. J. Plant Pathol. 2013, 137, 55–65. [Google Scholar] [CrossRef]
- Thakur, S.; Singh, P.K.; Das, A.; Rathour, R.; Sharma, T.R. Extensive sequence variation in rice blast resistance gene Pi54 makes it broad spectrum in nature. Front. Plant Sci. 2015, 6, 345. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vasudevan, K.; Gruissem, W.; Bhullar, N.K. Identification of novel alleles of the rice blast resistance gene Pi54. Sci. Rep. 2015, 5, 15678. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, G.; Madhav, M.S.; Rama Devi, S.J.S.; Umakanth, B.; Pandey, M.K.; Prasad, M.S.; Sundaram, R.M.; Viraktamath, B.C.; Ravindra Babu, V. Identification and validation of novel alleles of rice blast resistant gene Pi54, and analysis of their nucleotide diversity in landraces and wild Oryza species. Euphytica 2016, 209, 725–737. [Google Scholar] [CrossRef]
- Zhang, L.; Nakagomi, Y.; Endo, T.; Teranishi, M.; Hidema, J.; Sato, S.; Higashitani, A. Divergent evolution of rice blast resistance Pi54 locus in the genus Oryza. Rice 2018, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Singh, V.K.; Singh, S.P.; Pandian, R.T.; Ellur, R.K.; Singh, D.; Bhowmick, P.K.; Gopala Krishnan, S.; Nagarajan, M.; Vinod, K.K.; et al. Molecular breeding for the development of multiple disease resistance in Basmati rice. AoB Plants 2012, 2012, pls029. [Google Scholar] [CrossRef] [PubMed]
- Arunakumari, K.; Durgarani, C.V.; Satturu, V.; Sarikonda, K.R.; Chittoor, P.D.R.; Vutukuri, B.; Laha, G.S.; Nelli, A.P.K.; Gattu, S.; Jamal, M.; et al. Marker-assisted pyramiding of genes conferring resistance against bacterial blight and blast diseases into indian rice variety MTU1010. Rice Sci. 2016, 23, 306–316. [Google Scholar] [CrossRef]
- Jiang, H.; Li, Z.; Liu, J.; Shen, Z.; Gao, G.; Zhang, Q.; He, Y. Development and evaluation of improved lines with broad-spectrum resistance to rice blast using nine resistance genes. Rice 2019, 12, 29. [Google Scholar] [CrossRef]
- Jamaloddin, M.; Durga Rani, C.V.; Swathi, G.; Anuradha, C.; Vanisri, S.; Rajan, C.P.D.; Krishnam Raju, S.; Bhuvaneshwari, V.; Jagadeeswar, R.; Laha, G.S.; et al. Marker Assisted Gene Pyramiding (MAGP) for bacterial blight and blast resistance into mega rice variety “Tellahamsa”. PLoS ONE 2020, 15, e0234088. [Google Scholar] [CrossRef]
- Ramalingam, J.; Palanisamy, S.; Alagarasan, G.; Renganathan, V.G.; Ramanathan, A.; Saraswathi, R. Improvement of stable restorer lines for blast resistance through functional marker in rice (Oryza sativa L.). Genes 2020, 11, 1266. [Google Scholar] [CrossRef]
- Wu, Y.; Xiao, N.; Yu, L.; Pan, C.; Li, Y.; Zhang, X.; Liu, G.; Dai, Z.; Pan, X.; Li, A. Combination patterns of major R genes determine the level of resistance to the M. oryzae in rice (Oryza sativa L.). PLoS ONE 2015, 10, e0126130. [Google Scholar]
- Chen, F.; Xu, J.D.; Jiang, M.S.; Liang, S.M.; Zhang, Q.F.; Wang, W.L.; Li, G.X.; Yang, L.Q.; Zhu, W.Y.; Zhou, X.B. Molecular detection of rice blast resistance genes Pi-ta, Pi-b, Pi54 and Pikm in japonica rice of Huang-huai region. Curr. Biotechnol. 2018, 8, 46–54. [Google Scholar]
- Wan, B.; Liu, K.; Zhao, S.; Zhu, J.; Liu, Y.; Zhang, G.; Zhu, G.; Wang, A.; Tang, H.; Sun, M.; et al. Distribution of rice blast resistance genes Pi-ta, Pi-b, Pigm and Pi54 in backbone parent and their relationships with neck blast resistance. Southwest China J. Agric. Sci. 2020, 33, 1–6. [Google Scholar]
- Singh, R.P.; Hodson, D.P.; Huerta-Espino, J.; Jin, Y.; Bhavani, S.; Njau, P.; Herrera-Foessel, S.; Singh, P.K.; Singh, S.; Govindan, V. The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu. Rev. Phytopathol. 2011, 49, 465–481. [Google Scholar] [CrossRef] [PubMed]
- McDonald, B.A.; Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 2002, 40, 349–379. [Google Scholar] [CrossRef] [PubMed]
- Stukenbrock, E.H.; McDonald, B.A. Population genetics of fungal and oomycete effectors involved in gene-for-gene interactions. Mol. Plant. Microbe Interact. 2009, 22, 371–380. [Google Scholar] [CrossRef]
- Zhu, H. Biogeographical divergence of the flora of Yunnan, southwestern China initiated by the uplift of Himalaya and extrusion of Indochina block. PLoS ONE 2012, 7, e45601. [Google Scholar]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; da Fonseca, G.A.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Huang, X.; Kurata, N.; Wei, X.; Wang, Z.X.; Wang, A.; Zhao, Q.; Zhao, Y.; Liu, K.; Lu, H.; Li, W.; et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 2012, 490, 497–501. [Google Scholar] [CrossRef]
- Huang, Y.; Sun, X.; Wang, X. Study on the center of genetic diversity and its origin of cultivated rice in China. J. Plant Genet. Resour. 2005, 6, 125–129. [Google Scholar]
- Saleh, D.; Milazzo, J.; Adreit, H.; Fournier, E.; Tharreau, D. South-East Asia is the center of origin, diversity and dispersion of the rice blast fungus, Magnaporthe oryzae. New Phytol. 2014, 201, 1440–1456. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lu, L.; Jia, Y.; Li, C. Effectiveness and durability of the rice Pi-ta gene in Yunnan province of China. Phytopathology 2014, 104, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Valent, B.; Lee, F.N. Determination of host responses to Magnaporthe grisea on detached rice leaves using a spot inoculation method. Plant Dis. 2003, 87, 129–133. [Google Scholar] [CrossRef]
- Tai, T.H.; Tanksley, S.D. A rapid and inexpensive method for isolation of total DNA from dehydrated plant tissue. Plant Mol. Biol. Report. 1990, 8, 297–303. [Google Scholar] [CrossRef]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. In Bioinformatics Methods and Protocols. Methods in Molecular BiologyTM.; Misener, S., Krawetz, S.A., Eds.; Humana Press: Totowa, NJ, USA, 2000; pp. 71–91. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Nei, M.; Miller, J.C. A simple method for estimating average number of nucleotide substitutions within and between populations from restriction data. Genetics 1990, 125, 873–879. [Google Scholar] [CrossRef]
- Rozas, J.; Sanchez-DelBarrio, J.C.; Messeguer, X.; Rozas, R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 2003, 19, 2496–2497. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, J.; Lu, L.; He, C.; Li, C. Novel variation and evolution of AvrPiz-t of Magnaporthe oryzae in field isolates. Front. Genet. 2020, 11, 746. [Google Scholar] [CrossRef] [PubMed]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger dataset. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Kato, H.; Yamamoto, M.; Yamaguchi-Ozaki, T.; Kadouchi, H.; Iwamoto, Y.; Nakayashiki, H. Pathogenicity, mating ability and DNA restriction fragment length polymorphisms of Pyricularia populations isolated from Gramineae, Bambusideae and Zingiberaceae plants. J. Gen. Plant Pathol. 2000, 66, 30–47. [Google Scholar] [CrossRef]
- Farman, M.L. Pyricularia grisea isolates causing gray leaf spot on perennial ryegrass (Lolium perenne) in the United States: Relationship to P. grisea isolates from other host plants. Phytopathology 2002, 92, 245–254. [Google Scholar]
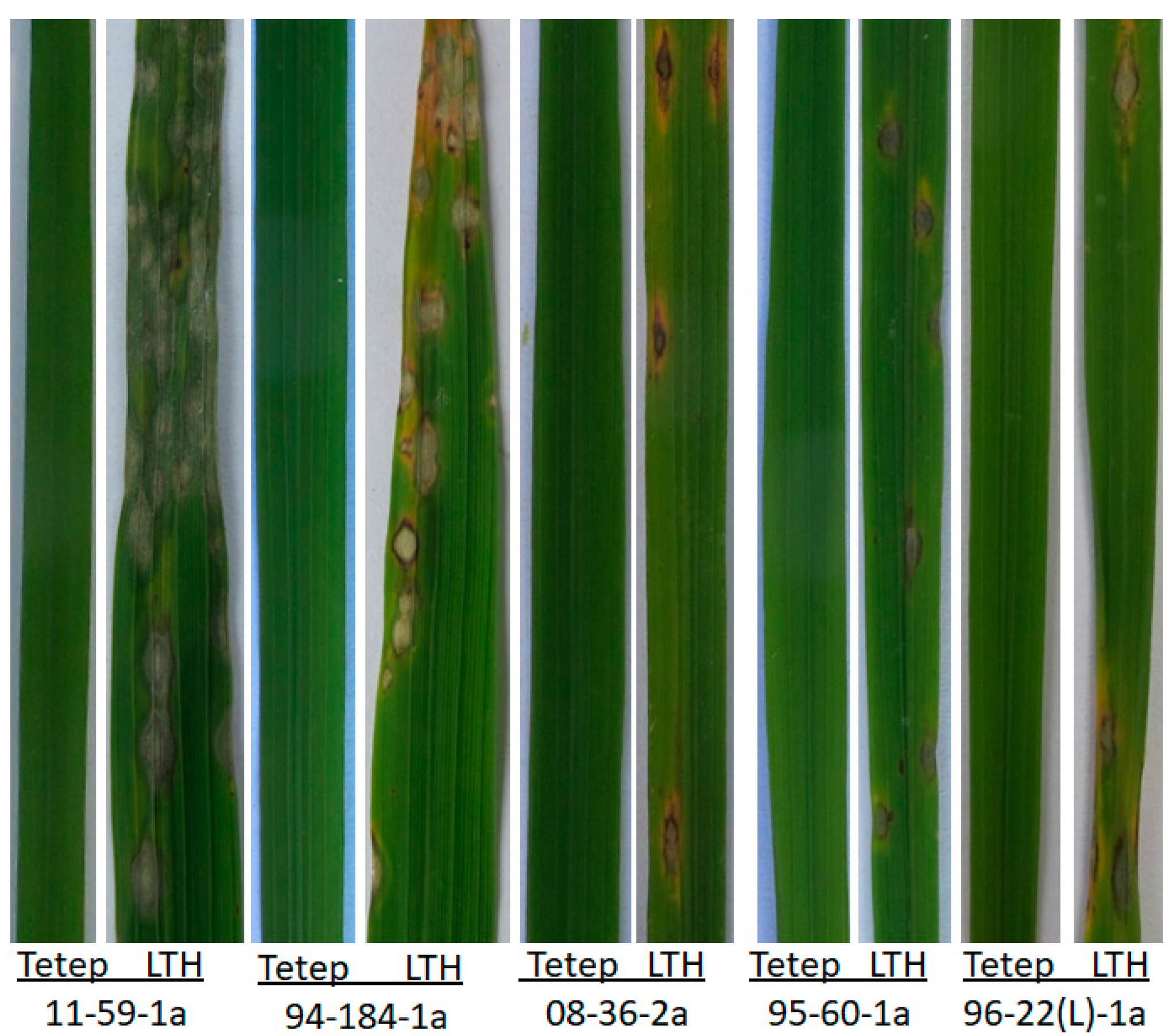
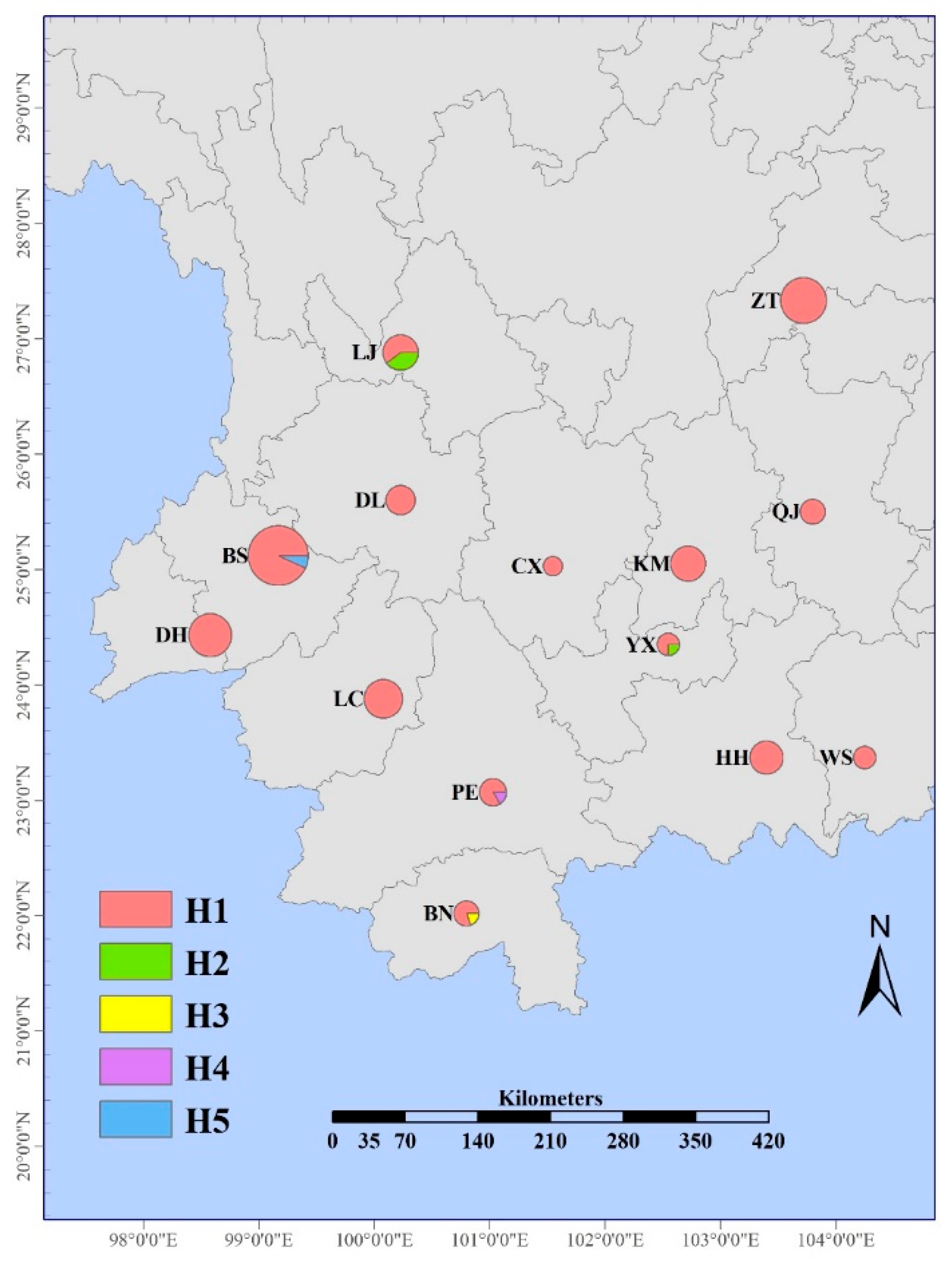

| Isolates Groups | No. | PCR Amplification of AvrPi54 | No. Seq | Pathogenicity Assay to Tetep | ||
|---|---|---|---|---|---|---|
| + (%) | - (%) | A (%) | V (%) | |||
| Central | 62 | 41 (66.1) | 21 (33.9) | 22 | 50 (80.6) | 12 (19.4) |
| KM | 28 | 20 (71.4) | 8 (28.6) | 10 | 25 (89.3) | 3 (11.7) |
| YX | 5 | 4 (80.0) | 1 (20.0) | 4 | 4 (80) | 1 (20) |
| CX | 16 | 8 (50.0) | 8 (50.0) | 3 | 11 (68.8) | 5 (31.2) |
| QJ | 13 | 9 (69.2) | 4 (30.8) | 5 | 10 (76.9) | 3 (23.1) |
| West | 191 | 93 (48.7) | 98 (51.3) | 51 | 135 (70.7) | 56 (29.3) |
| BS | 102 | 59 (57.8) | 43 (42.2) | 29 | 70 (68.6) | 32 (31.4) |
| DL | 34 | 8 (23.5) | 26 (76.5) | 7 | 19 (55.9) | 15 (44.1) |
| DH | 55 | 26 (47.3) | 29 (52.7) | 15 | 46 (83.6) | 9 (16.4) |
| Northwest | 26 | 11 (42.3) | 15 (57.7) | 10 | 15 (57.7) | 11 (42.3) |
| LJ | 26 | 11 (42.3) | 15 (57.7) | 10 | 15 (57.7) | 11 (42.3) |
| Southwest | 35 | 19 (54.3) | 16 (45.7) | 18 | 27 (77.1) | 8 (22.9) |
| LC | 22 | 13 (59.1) | 9 (40.9) | 12 | 19 (86.4) | 3 (13.6) |
| PE | 13 | 6 (46.2) | 7 (53.8) | 6 | 8 (61.5) | 5 (38.5) |
| Southeast | 16 | 6 (37.5) | 10 (62.5) | 4 | 12 (75) | 4 (25) |
| WS | 16 | 6 (37.5) | 10 (62.5) | 4 | 12 (75) | 4 (25) |
| South | 42 | 19 (45.2) | 23 (54.8) | 14 | 25 (59.5) | 17 (40.5) |
| BN | 21 | 9 (42.9) | 12 (57.1) | 5 | 12 (57.1) | 9 (42.9) |
| HH | 21 | 10 (47.6) | 11 (52.4) | 9 | 13 (61.9) | 8 (38.1) |
| Northeast | 52 | 21 (40.4) | 31 (59.6) | 17 | 45 (86.5) | 7 (13.5) |
| ZT | 52 | 21 (40.4) | 31 (59.6) | 17 | 45 (86.5) | 7 (13.5) |
| Rice host | 424 | 210 (49.5) | 214 (50.5) | 136 | 309 (72.9) | 115 (27.1) |
| GJ | 276 | 140 (50.7) | 136 (49.3) | 85 | 200 (72.5) | 76 (27.5) |
| XI | 148 | 70 (47.3) | 78 (52.7) | 51 | 109 (74.3) | 39 (25.7) |
| Non-rice host | 27 | 8 (29.6) | 19 (70.4) | 6 | 27 (100) | 0 (0) |
| Oryza rufipogon | 18 | 7 (38.9) | 11 (61.1) | 5 | 18 (100) | 0 (0) |
| Eleusine coracana | 1 | 1 (100.0) | 0 (0.0) | 1 | 1 (100) | 0 (0) |
| E. indica | 1 | 0 (0.0) | 1 (100.0) | 0 | 1 (100) | 0 (0) |
| Digitaria sanguinalis | 1 | 0 (0.0) | 1 (100.0) | 0 | 1 (100) | 0 (0) |
| Musa sp. | 6 | 0 (0.0) | 6 (100.0) | 0 | 6 (100) | 0 (0) |
| Total | 451 | 218 (48.3) | 233 (51.7) | 142 | 336 (74.5) | 115 (25.5) |
| Haplotype | Frequency | Non-CDS | CDS | Non-CDS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 140 | 141 | 223 | 619 | 683 | 732 | 795 | 841 | 918–935 | 1151 | 1159 | 1171 | 1180 | ||
| HF545677 | C | C | G | G | T | T | G | C | TGTAGATGGAGTGGTTGA | G | G | G | A | |
| H1 | 132 | C | C | G | G | T | T | G | C | TGTAGATGGAGTGGTTGA | G | G | G | A |
| H2 | 5 | C | C | G | G | T | T | G | C | TGTAGATGGAGTGGTTGA | G | G | A | A |
| H3 | 1 | C | C | G | G | T | T | G | C | TGTAGATGGAGTGGTTGA | G | C | G | A |
| H4 | 1 | C | C | G | A | T | T | G | C | TGTAGATGGAGTGGTTGA | G | G | G | A |
| H5 | 3 | T | T | A | G | C | C | A | A | ------------------ | A | G | G | C |
| Protein Type | Frequency | Amino Acid Sites | |
|---|---|---|---|
| 5 | 137 | ||
| CCN97897 | - | A | V |
| P1 | 140 | A | V |
| P2 | 1 | A | I |
| P3 | 1 | T | V |
| Groups | H1 | H2 | H3 | H4 | H5 | No. Samples | No. Variable Sites |
|---|---|---|---|---|---|---|---|
| Central | 21 | 1 | 0 | 0 | 0 | 22 | 1 |
| KM | 10 | 0 | 0 | 0 | 0 | 10 | 0 |
| YX | 3 | 1 | 0 | 0 | 0 | 4 | 1 |
| QJ | 5 | 0 | 0 | 0 | 0 | 5 | 0 |
| CX | 3 | 0 | 0 | 0 | 0 | 3 | 0 |
| West | 49 | 0 | 0 | 0 | 2 | 51 | 9 |
| DL | 7 | 0 | 0 | 0 | 0 | 7 | 0 |
| BS | 27 | 0 | 0 | 0 | 2 | 29 | 9 |
| DH | 15 | 0 | 0 | 0 | 0 | 15 | 0 |
| Northwest | 6 | 4 | 0 | 0 | 0 | 10 | 1 |
| LJ | 6 | 4 | 0 | 0 | 0 | 10 | 1 |
| Northeast | 17 | 0 | 0 | 0 | 0 | 17 | 0 |
| ZT | 17 | 0 | 0 | 0 | 0 | 17 | 0 |
| South | 13 | 0 | 1 | 0 | 0 | 14 | 1 |
| HH | 9 | 0 | 0 | 0 | 0 | 9 | 0 |
| BN | 4 | 0 | 1 | 0 | 0 | 5 | 1 |
| Southwest | 17 | 0 | 0 | 1 | 0 | 18 | 1 |
| LC | 12 | 0 | 0 | 0 | 0 | 12 | 0 |
| PE | 5 | 0 | 0 | 1 | 0 | 6 | 1 |
| Southeast | 4 | 0 | 0 | 0 | 0 | 4 | 0 |
| WS | 4 | 0 | 0 | 0 | 0 | 4 | 0 |
| Rice host | 127 | 5 | 1 | 1 | 2 | 136 | 12 |
| GJ | 78 | 5 | 0 | 0 | 2 | 85 | 10 |
| XI | 49 | 0 | 1 | 1 | 0 | 51 | 2 |
| Non-rice host | 5 | 0 | 0 | 0 | 1 | 6 | 9 |
| Oryza rufipogon | 5 | 0 | 0 | 0 | 0 | 5 | 0 |
| Eleusine coracana | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| Total | 132 | 5 | 1 | 1 | 3 | 142 | 12 |
| Groups | Total | No. Hap | Hd | π × 10−4 | Tajima’s D |
|---|---|---|---|---|---|
| Rice host | 136 | 5 | 0.127 | 2.9 | −2.1254 * |
| GJ | 85 | 3 | 0.156 | 4.3 | −1.92196 * |
| XI | 51 | 3 | 0.078 | 0.6 | −1.46227 |
| Central | 22 | 2 | 0.091 | 0.7 | −1.1624 |
| West | 51 | 2 | 0.077 | 5.6 | −1.8244 * |
| Northwest | 10 | 2 | 0.533 | 4.2 | 1.30268 |
| Northeast | 17 | 1 | 0 | 0 | - |
| South | 14 | 2 | 0.143 | 1.1 | −1.1552 |
| Southwest | 18 | 2 | 0.111 | 0.9 | −1.1647 |
| Southeast | 4 | 1 | 0 | 0 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; He, C.; Dong, C.; Lu, L.; He, C.; Bi, Y.; Shi, Z.; Fan, H.; Shi, J.; Wang, K.; et al. Diversity and Evolution of the Avirulence Gene AvrPi54 in Yunnan Rice Fields. Agronomy 2024, 14, 454. https://doi.org/10.3390/agronomy14030454
Li J, He C, Dong C, Lu L, He C, Bi Y, Shi Z, Fan H, Shi J, Wang K, et al. Diversity and Evolution of the Avirulence Gene AvrPi54 in Yunnan Rice Fields. Agronomy. 2024; 14(3):454. https://doi.org/10.3390/agronomy14030454
Chicago/Turabian StyleLi, Jinbin, Chengxing He, Chao Dong, Lin Lu, Chi He, Yunqing Bi, Zhufeng Shi, Huacai Fan, Junyi Shi, Kaibo Wang, and et al. 2024. "Diversity and Evolution of the Avirulence Gene AvrPi54 in Yunnan Rice Fields" Agronomy 14, no. 3: 454. https://doi.org/10.3390/agronomy14030454
APA StyleLi, J., He, C., Dong, C., Lu, L., He, C., Bi, Y., Shi, Z., Fan, H., Shi, J., Wang, K., Zeng, Z., Luo, H., & Wang, Q. (2024). Diversity and Evolution of the Avirulence Gene AvrPi54 in Yunnan Rice Fields. Agronomy, 14(3), 454. https://doi.org/10.3390/agronomy14030454





