Mining and Genetic Mapping of a Novel Powdery Mildew Resistance Gene, PmKu-2013, Identified in Aegilops tauschii
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Phenotypic Assessment of Reactions to Powdery Mildew
2.3. BSA and Molecular Markers Analysis
2.4. Genetic Analysis and Genetic Mapping
2.5. Identification of Potential Candidate Genes
2.6. Comparison Pm2 Sequences from KU-2013 and JingY303
2.7. Evaluation of the Closely Linked Markers for MAS
3. Results
3.1. Identification and Inheritance of Powdery Mildew Resistance in KU-2013
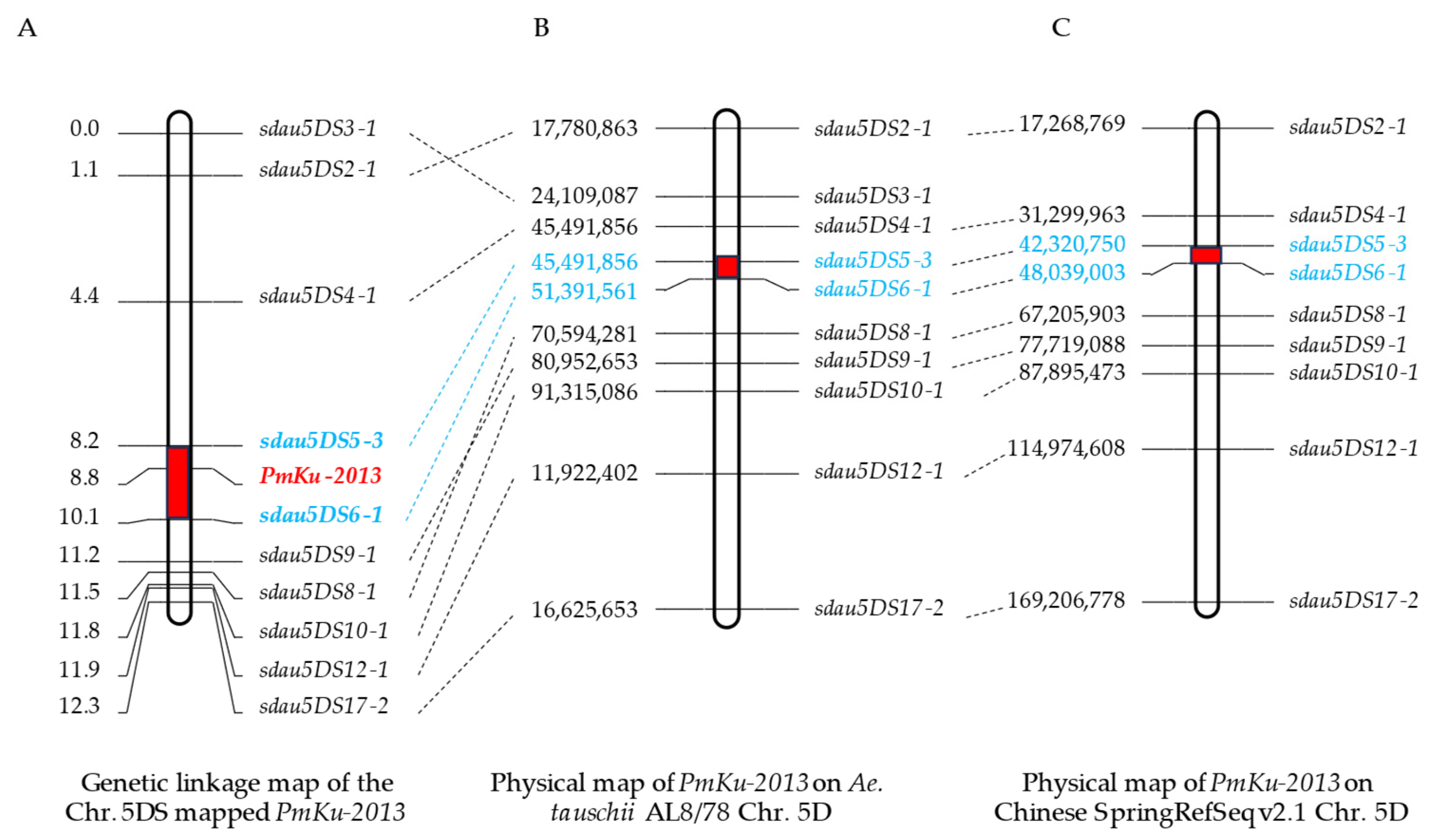
3.2. Chromosomal Localization of PmKu-2013
3.3. Comparison Pm2 between “KU-2013” and “JingY303”
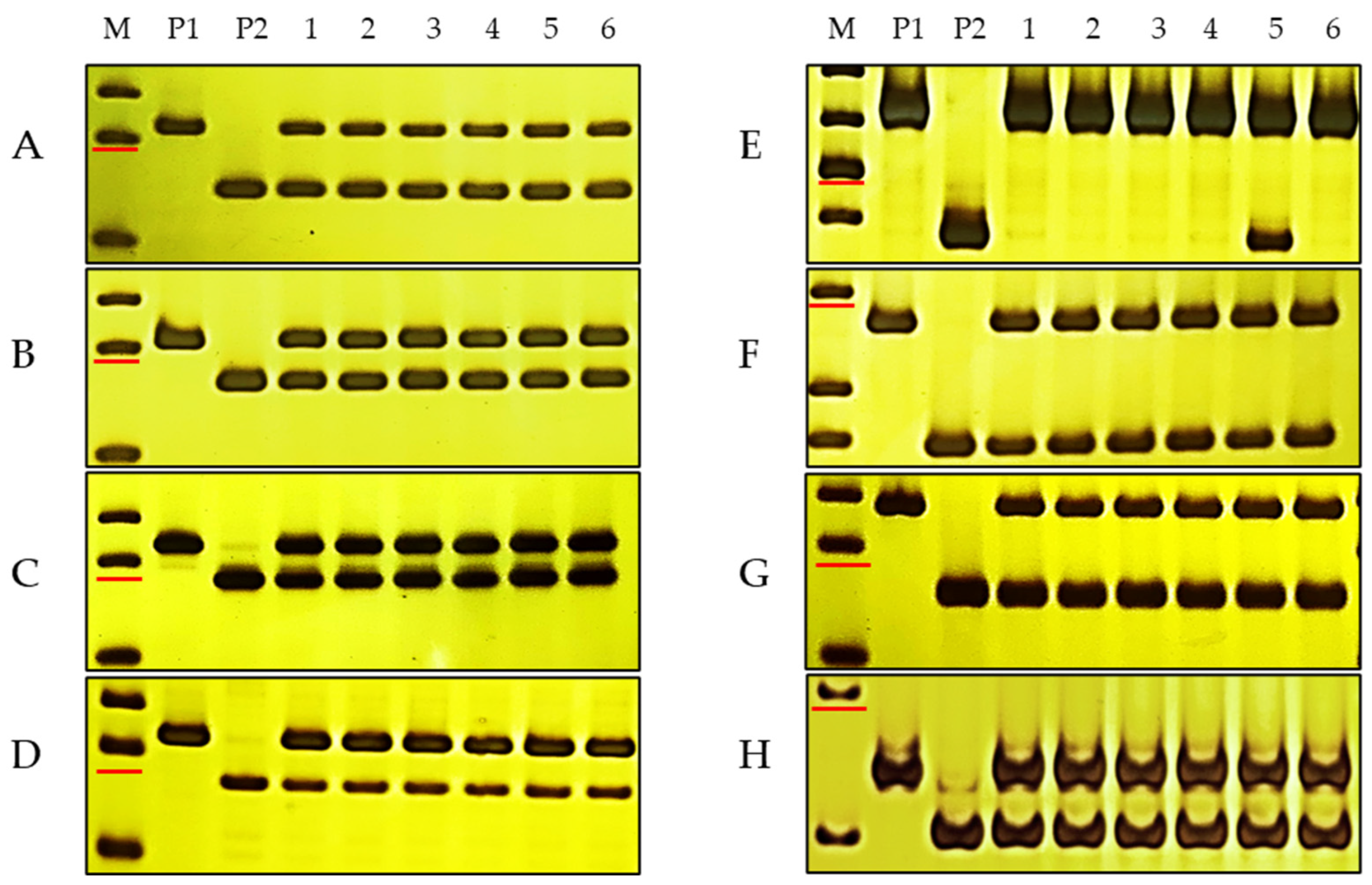
3.4. Molecular Mapping of PmKu-2013
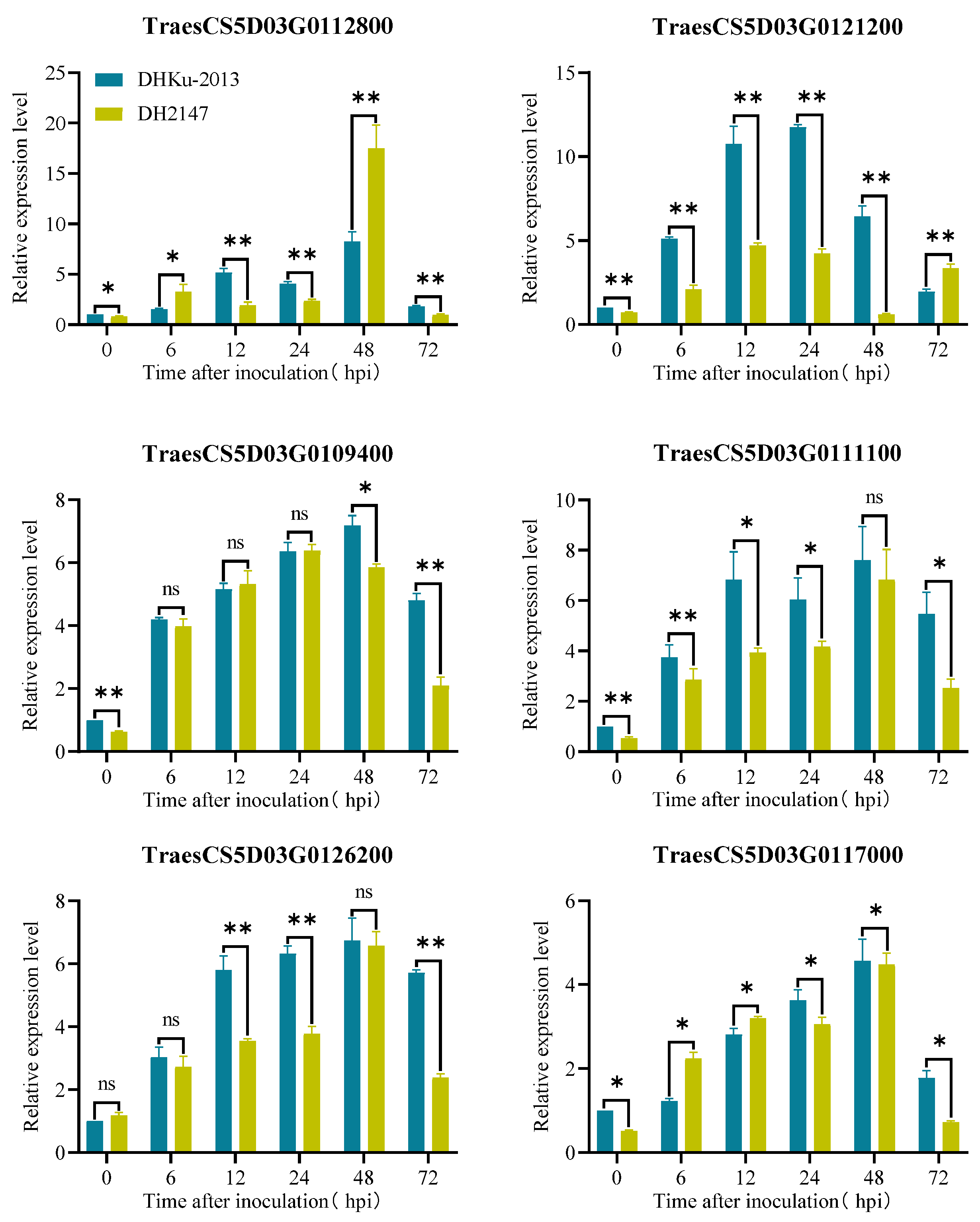
3.5. Identification of Potential Candidate Genes
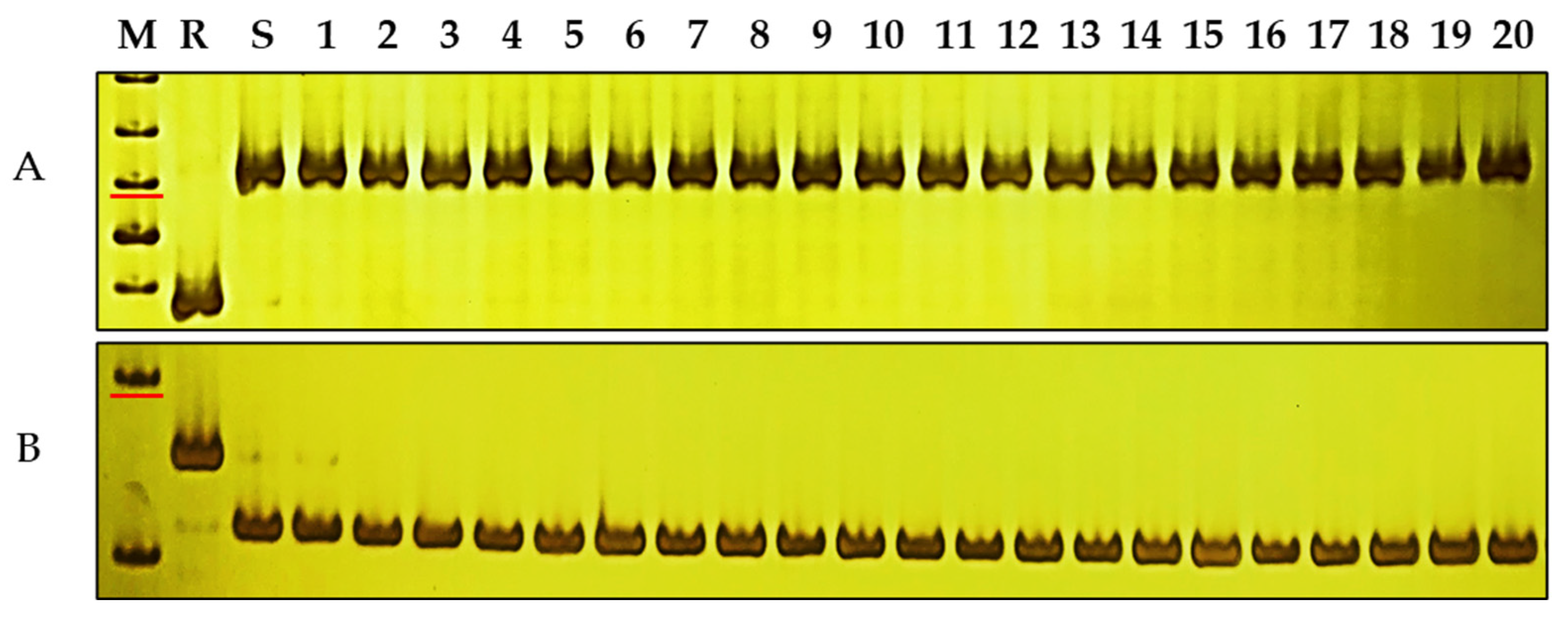
3.6. Molecular Markers Closely Linked to PmKu-2013 for MAS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Haile, J.K.; N’Diaye, A.; Walkowiak, S.; Nilsen, K.T.; Clarke, J.M.; Kutcher, H.R.; Steiner, B.; Buerstmayr, H.; Pozniak, C.J. Fusarium Head Blight in Durum Wheat: Recent Status, Breeding Directions, and Future Research Prospects. Phytopathology 2019, 109, 1664–1675. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lin, D.; Zhang, Y.; Deng, M.; Chen, Y.; Lv, B.; Li, B.; Lei, Y.; Wang, Y.; Zhao, L.; et al. Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 2022, 602, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Singh, P.K.; Rutkoski, J.; Hodson, D.P.; He, X.; Jørgensen, L.N.; Hovmøller, M.S.; Huerta-Espino, J. Disease Impact on Wheat Yield Potential and Prospects of Genetic Control. Annu. Rev. Phytopathol. 2016, 54, 303–322. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.X.; Xu, X.F.; Ma, X.; Chen, R.Z.; Li, T.Y.; Cao, Y.Y. Virulence structure and its genetic diversity analyses of Blumeria graminis f. sp. tritici isolates in China. BMC Evol. Biol. 2019, 19, 183. [Google Scholar] [CrossRef] [PubMed]
- Cu, L.; Ren, Y.; Murray, T.D.; Yan, W.; Guo, Q.; Niu, Y.; Sun, Y.; Li, H. Development of Perennial Wheat Through Hybridization Between Wheat and Wheatgrasses: A Review. Engineering 2018, 4, 507–513. [Google Scholar] [CrossRef]
- Jia, Z.; Qiu, Y.; Lin, Z.; Wang, K.; Ye, X. Research Progress on Wheat Improvement by Using Desirable Genes from Its Relative Species. Crops 2021, 37, 1–14. [Google Scholar]
- Liu, C.; Han, R.; Wang, X.L.; Gong, W.P.; Cheng, D.G.; Cao, X.Y.; Liu, A.F.; Li, H.S.; Liu, J.J. Research Progress of Wheat Wild Hybridization, Disease Resistance Genes Transfer and Utilization. Sci. Agric. Sin. 2020, 53, 1287–1308. [Google Scholar]
- Jin, Y.; Xiao, L.; Zheng, J.; Su, F.; Yu, Z.; Mu, Y.; Zhang, W.; Li, L.; Han, G.; Ma, P. Genetic Analysis and Molecular Identification of the Powdery Mildew Resistance in 116 Elite Wheat Cultivars/Lines. Plant Dis. 2023, 107, 3801–3809. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Liu, H.; Gu, T.; Xing, L.; Han, G.; Ma, P.; Li, X.; Zhou, Y.; Fan, J.; Li, L.; et al. PM2b, a CC-NBS-LRR protein, interacts with TaWRKY76-D to regulate powdery mildew resistance in common wheat. Front. Plant Sci. 2022, 26, 973065. [Google Scholar] [CrossRef]
- Yahiaoui, N.; Srichumpa, P.; Dudler, R.; Keller, B. Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J. 2004, 37, 528–538. [Google Scholar] [CrossRef]
- Hurni, S.; Brunner, S.; Buchmann, G.; Herren, G.; Jordan, T.; Krukowski, P.; Wicker, T.; Yahiaoui, N.; Mago, R.; Keller, B. Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J. 2013, 76, 957–969. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P.; Hurni, S.; Ruinelli, M.; Brunner, S.; Sanchez-Martin, J.; Krukowski, P.; Peditto, D.; Buchmann, G.; Zbinden, H.; Keller, B. Evolutionary divergence of the rye Pm17 and Pm8 resistance genes reveals ancient diversity. Plant Mol. Biol. 2018, 98, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, T.; Müller, M.C.; Molnár, I.; Mascher, M.; Holušová, K.; Šimková, H.; Kunz, L.; Zhang, J.; Li, J.; Bhatt, D.; et al. A highly differentiated region of wheat chromosome 7AL encodes a Pm1a immune receptor that recognizes its corresponding AvrPm1a effector from Blumeria graminis. New Phytol. 2021, 229, 2812–2826. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Martín, J.; Steuernagel, B.; Ghosh, S.; Herren, G.; Hurni, S.; Adamski, N.; Vrána, J.; Kubaláková, M.; Krattinger, S.G.; Wicker, T.; et al. Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 2016, 17, 221. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Guo, G.; Wang, Y.; Hu, T.; Wang, L.; Li, J.; Qiu, D.; Li, Y.; Wu, Q.; Lu, P.; et al. A rare single nucleotide variant in Pm5e confers powdery mildew resistance in common wheat. New Phytol. 2020, 228, 1011–1026. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Liu, C.; Gong, S.; Chen, Z.; Chen, R.; Liu, T.; Liu, R.; Du, H.; Guo, R.; Li, G.; et al. Orthologous genes Pm12 and Pm21 from two wild relatives of wheat show evolutionary conservation but divergent powdery mildew resistance. Plant Commun. 2023, 4, 100472. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Hu, P.; Liu, J.; Witek, K.; Zhou, S.; Xu, J.; Zhou, W.; Gao, L.; Huang, Z.; Zhang, R.; et al. Pm21 from Haynaldia villosa Encodes a CC-NBS-LRR Protein Conferring Powdery Mildew Resistance in Wheat. Mol. Plant 2018, 11, 874–878. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhong, S.; Chen, C.; Yang, H.; Chen, W.; Tan, F.; Zhang, M.; Chen, W.; Ren, T.; Li, Z.; et al. Identification and Cloning of a CC-NBS-NBS-LRR Gene as a Candidate of Pm40 by Integrated Analysis of Both the Available Transcriptional Data and Published Linkage Mapping. Int. J. Mol. Sci. 2021, 22, 10239. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Fang, T.; Zhang, H.; Xie, C.; Li, H.; Yang, T.; Nevo, E.; Fahima, T.; Sun, Q.; Liu, Z. Molecular identification of a new powdery mildew resistance gene Pm41 on chromosome 3BL derived from wild emmer (Triticum turgidum var. dicoccoides). Theor. Appl. Genet. 2009, 119, 531–539. [Google Scholar] [CrossRef]
- Zou, S.; Wang, H.; Li, Y.; Kong, Z.; Tang, D. The NB-LRR gene Pm60 confers powdery mildew resistance in wheat. New Phytol. 2018, 218, 298–309. [Google Scholar] [CrossRef]
- Wu, Q.; Chen, Y.; Li, B.; Li, J.; Zhang, P.; Xie, J.; Zhang, H.; Guo, G.; Lu, P.; Li, M.; et al. Functional characterization of powdery mildew resistance gene MlIW172, a new Pm60 allele and its allelic variation in wild emmer wheat. J. Genet. Genom. 2022, 49, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Zhao, F.; Chen, Y.; Zhang, P.; Zhang, H.; Guo, G.; Xie, J.; Dong, L.; Lu, P.; Li, M.; et al. Bulked segregant CGT-Seq-facilitated map-based cloning of a powdery mildew resistance gene originating from wild emmer wheat (Triticum dicoccoides). Plant Biotechnol. J. 2021, 19, 1288–1290. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wei, Z.Z.; Sela, H.; Govta, L.; Klymiuk, V.; Roychowdhury, R.; Chawla, H.S.; Ens, J.; Wiebe, K.; Bocharova, V.; et al. Dissection of a rapidly evolving wheat resistance gene cluster by long-read genome sequencing accelerated the cloning of Pm69. Plant Commun. 2023, 6, 100646. [Google Scholar] [CrossRef]
- Lu, P.; Guo, L.; Wang, Z.; Li, B.; Li, J.; Li, Y.; Qiu, D.; Shi, W.; Yang, L.; Wang, N.; et al. A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat. Commun. 2020, 11, 680. [Google Scholar] [CrossRef] [PubMed]
- Gaurav, K.; Arora, S.; Silva, P.; Sánchez-Martín, J.; Horsnell, R.; Gao, L.; Brar, G.S.; Widrig, V.; John Raupp, W.; Singh, N.; et al. Population genomic analysis of Aegilops tauschii identifies targets for bread wheat improvement. Nat. Biotechnol. 2022, 40, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Martín, J.; Widrig, V.; Herren, G.; Wicker, T.; Zbinden, H.; Gronnier, J.; Spörri, L.; Praz, C.R.; Heuberger, M.; Kolodziej, M.C.; et al. Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nat. Plants 2021, 7, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Krattinger, S.G.; Lagudah, E.S.; Spielmeyer, W.; Singh, R.P.; Huerta-Espino, J.; McFadden, H.; Bossolini, E.; Selter, L.L.; Keller, B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 2009, 323, 1360–1363. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.W.; Herrera-Foessel, S.; Lan, C.; Schnippenkoetter, W.; Ayliffe, M.; Huerta-Espino, J.; Lillemo, M.; Viccars, L.; Milne, R.; Periyannan, S.; et al. A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 2015, 47, 1494–1498. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; He, Z.; Appels, R.; Xia, X. Functional markers in wheat: Current status and future prospects. Theor. Appl. Genet. 2012, 125, 1–10. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, B.; Zhang, J.; Zhang, J.; Li, J. Enhancement of plant variety protection and regulation using molecular marker technology. Acta Agron. Sin. 2022, 48, 1853–1870. [Google Scholar]
- Xiang, M.; Liu, S.; Wang, X.; Zhang, M.; Yan, W.; Wu, J.; Wang, Q.; Li, C.; Zheng, W.; He, Y.; et al. Development of breeder chip for gene detection and molecular-assisted selection by target sequencing in wheat. Mol. Breed. 2023, 43, 13. [Google Scholar] [CrossRef] [PubMed]
- Si, Q.M.; Zhang, X.X.; Duan, X.Y.; Sheng, B.Q.; Zhou, Y.L. On gene analysis and classification of powdery mildew (Erysiphe graminis f. sp. tritici) resistant wheat varieties. Acta Phytopathol. Sin. 1992, 22, 349–355. [Google Scholar]
- Zhou, R.; Zhu, Z.; Kong, X.; Huo, N.; Tian, Q.; Li, P.; Jin, C.; Dong, Y.; Jia, J. Development of wheat near-isogenic lines for powdery mildew resistance. Theor. Appl. Genet. 2005, 110, 640–648. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.R.; Pena, S.D.; Epplen, J.T. Genetic and population study of a Y-linked tetranucleotide repeat DNA polymorphism with a simple non-isotopic technique. Hum. Genet. 1993, 90, 655–656. [Google Scholar] [CrossRef] [PubMed]
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugen. 1944, 12, 172–175. [Google Scholar] [CrossRef]
- Liu, R.H.; Meng, J.L. MapDraw: A microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Yi Chuan 2003, 25, 317–321. [Google Scholar] [PubMed]
- Ma, S.; Wang, M.; Wu, J.; Guo, W.; Chen, Y.; Li, G.; Wang, Y.; Shi, W.; Xia, G.; Fu, D.; et al. WheatOmics: A platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant 2021, 14, 1965–1968. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhu, T.; Rodriguez, J.C.; Deal, K.R.; Dubcovsky, J.; McGuire, P.E.; Lux, T.; Spannagl, M.; Mayer, K.F.X.; Baldrich, P.; et al. Aegilops tauschii genome assembly Aet v5.0 features greater sequence contiguity and improved annotation. G3 2021, 11, jkab325. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Wang, L.; Rimbert, H.; Rodriguez, J.C.; Deal, K.R.; De Oliveira, R.; Choulet, F.; Keeble-Gagnère, G.; Tibbits, J.; Rogers, J.; et al. Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J. 2021, 107, 303–314. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Xu, H.; Yi, Y.; Ma, P.; Qie, Y.; Fu, X.; Xu, Y.; Zhang, X.; An, D. Molecular tagging of a new broad-spectrum powdery mildew resistance allele Pm2c in Chinese wheat landrace Niaomai. Theor. Appl. Genet. 2015, 128, 2077–2084. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Jia, H.; Qiao, L.; Fu, B.; Brown-Guedira, G.; Nagarajan, R.; Yan, L. Genetic basis of resistance against powdery mildew in the wheat cultivar “Tabasco”. Mol. Breed. 2023, 43, 56. [Google Scholar] [CrossRef]
- Chen, F.; Jia, H.; Zhang, X.; Qiao, L.; Li, X.; Zheng, J.; Guo, H.; Powers, C.; Yan, L.; Chang, Z. Positional cloning of PmCH1357 reveals the origin and allelic variation of the Pm2 gene for powdery mildew resistance in wheat. Crop J. 2019, 7, 771–783. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhao, X.; Li, Y.; Xu, J.; Bi, A.; Kang, L.; Xu, D.; Chen, H.; Wang, Y.; Wang, Y.G.; et al. Triticum population sequencing provides insights into wheat adaptation. Nat. Genet. 2020, 52, 1412–1422. [Google Scholar] [CrossRef] [PubMed]
- Delorean, E.; Gao, L.; Lopez, J.F.C.; Open Wild Wheat Consortium; Wulff, B.B.H.; Ibba, M.I.; Poland, J. High molecular weight glutenin gene diversity in Aegilops tauschii demonstrates unique origin of superior wheat quality. Commun. Biol. 2021, 4, 1242. [Google Scholar] [CrossRef]
- Xu, S.; Lyu, Z.; Zhang, N.; Li, M.; Wei, X.; Gao, Y.; Cheng, X.; Ge, W.; Li, X.; Bao, Y.; et al. Genetic mapping of the wheat leaf rust resistance gene Lr19 and development of translocation lines to break its linkage with yellow pigment. Theor. Appl. Genet. 2023, 136, 200. [Google Scholar] [CrossRef] [PubMed]
- Pont, C.; Leroy, T.; Seidel, M.; Tondelli, A.; Duchemin, W.; Armisen, D.; Lang, D.; Bustos-Korts, D.; Goué, N.; Balfourier, F.; et al. Tracing the ancestry of modern bread wheats. Nat. Genet. 2019, 51, 905–911. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Luo, M.C.; Chen, Z.; You, F.M.; Wei, Y.; Zheng, Y.; Dvorak, J. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 2013, 198, 925–937. [Google Scholar] [CrossRef]
- Kerber, E.R. Suppression of rust resistance in amphiploids of Triticum. In Proceedings of the 6th International Wheat Genetics Symposium, Kyoto, Japan, 28 November–3 December 1983; Sakamoto, S., Ed.; Plant Germ-Plasm Institute, Faculty of Agriculture, Kyoto University: Kyoto, Japan, 1983; pp. 813–817. [Google Scholar]
- McIntosh, R.A.; Zhang, P.; Cowger, C.; Parks, R.; Lagudah, E.S.; Hoxha, S. Rye-derived powdery mildew resistance gene Pm8 in wheat is suppressed by the Pm3 locus. Theor. Appl. Genet. 2011, 123, 359–367. [Google Scholar] [CrossRef]
- Hiebert, C.W.; Moscou, M.J.; Hewitt, T.; Steuernagel, B.; Hernández-Pinzón, I.; Green, P.; Pujol, V.; Zhang, P.; Rouse, M.N.; Jin, Y.; et al. Stem rust resistance in wheat is suppressed by a subunit of the mediator complex. Nat. Commun. 2020, 11, 1123. [Google Scholar] [CrossRef]
- Awan, M.J.A.; Rasheed, A.; Saeed, N.A.; Mansoor, S. Aegilops tauschii presents a genetic roadmap for hexaploid wheat improvement. Trends Genet. 2022, 38, 307–309. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Hu, J.; Sun, Y.; Li, B.; Zhang, D.; Li, W.; Liu, J.; Li, D.; Gao, C.; Zhang, Y.; et al. Highly efficient heritable genome editing in wheat using an RNA virus and bypassing tissue culture. Mol. Plant 2021, 14, 1787–1798. [Google Scholar] [CrossRef] [PubMed]

| Parents and Cross | Generation | Total | Observed Ratio | Expected Ratio | χ2 a | p | |
|---|---|---|---|---|---|---|---|
| Resistant | Susceptible | ||||||
| DH2147 | PS | 18 | 0 | 18 | |||
| DHKu-2013 | PR | 18 | 18 | 0 | |||
| DH2147/DHKu-2013 | F1 | 15 | 15 | 0 | |||
| F2 | 167 | 129 | 38 | 3:1 | 0.45 | 0.5 | |
| Number | Primer Name | Forward Sequence | Reverse Sequence | Fragment Size | Chromosome Location |
|---|---|---|---|---|---|
| 1 | sdau5DS2-1 | CTTGAACCCTGCCAGCAA | ATTTGTATGGTACAGTGCTGTTCTT | 315 bp | 17,780,863–17,781,863 |
| 2 | sdau5DS3-1 | CAGCGTGTAGGACGTGCCAT | TGGAGACAGCCAAGCATCGT | 118 bp | 24,109,087–24,110,087 |
| 3 | sdau5DS4-1 | GATGAGTTTCCTGCACGAGACTG | AGCACCCTTGATCGAAAAAGTATT | 139 bp | 33,901,995–33,902,995 |
| 4 | sdau5DS5-3 | GCAACTCACCAAGGAGCACAG | TCAGCTTCAGACCATCGAACG | 230 bp | 45,491,856–45,492,856 |
| 5 | sdau5DS6-1 | GCAATAGAATAACTCGCATAGACAC | CCTCCCTAAGTACCGCTGCTAC | 184 bp | 51,391,561–51,392,561 |
| 6 | sdau5DS8-1 | TGAAGACCTTTTGATGGCAGCT | GCCGTGGTTGATGTGGGTAC | 241 bp | 70,594,281–70,595,281 |
| 7 | sdau5DS9-1 | AGTGCTCAAAAGTCAAATCTATGAC | ATATTGATGTGCGACGTAATCCT | 310 bp | 80,952,653–80,953,653 |
| 8 | sdau5DS10-1 | TCTAGGGTTCGTCTCGTTCGG | TACGGGAATCTATTTTGAAAACAGT | 193 bp | 91,315,086–91,316,086 |
| 9 | sdau5DS12-1 | AGCTCCTCCACCGCCGCT | CCTGGACACTCGGTGTGCTGT | 356 bp | 11,922,402–119,225,023 |
| 10 | sdau5DS17-2 | GTTCATAATGTAGTGTTAGTTTGCG | CAAGAGCAAGATAACAACAGCG | 225 bp | 16,625,653–166,257,539 |
| Number | Database | Type | Note Description | Confidence | Location Start 1 | Location End 1 | Gene ID 2 |
|---|---|---|---|---|---|---|---|
| 1 | pgsb | mRNA | Wall-associated receptor kinase galacturonan-binding | High | 46,300,602 | 46,303,096 | TraesCS5D03G0109400 |
| 2 | pgsb | mRNA | NB-ARC domain | High | 46,770,391 | 46,774,346 | TraesCS5D03G0111100 |
| 3 | pgsb | mRNA | NB-ARC domain (homologous sequence of Pm2 allele) | High | 46,864,873 | 46,869,229 | TraesCS5D03G0111700 |
| 4 | pgsb | mRNA | Protein kinase domain | High | 47,624,255 | 47,625,253 | TraesCS5D03G0112800 |
| 5 | pgsb | mRNA | Oxidation of toxic reductants and pathogen attack | High | 48,672,840 | 48,674,639 | TraesCS5D03G0117000 |
| 6 | pgsb | mRNA | LRR-repeat protein | High | 49,494,627 | 49,496,016 | TraesCS5D03G0121200 |
| 7 | pgsb | mRNA | LRR-repeat protein | High | 50,790,007 | 50,791,794 | TraesCS5D03G0126200 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.; Li, J.; Fan, L.; Qi, D.; Zhang, H.; Hao, Y.; Liang, M.; Bo, C.; Sun, S.; Wang, X.; et al. Mining and Genetic Mapping of a Novel Powdery Mildew Resistance Gene, PmKu-2013, Identified in Aegilops tauschii. Agronomy 2024, 14, 744. https://doi.org/10.3390/agronomy14040744
Chen W, Li J, Fan L, Qi D, Zhang H, Hao Y, Liang M, Bo C, Sun S, Wang X, et al. Mining and Genetic Mapping of a Novel Powdery Mildew Resistance Gene, PmKu-2013, Identified in Aegilops tauschii. Agronomy. 2024; 14(4):744. https://doi.org/10.3390/agronomy14040744
Chicago/Turabian StyleChen, Wuying, Jing Li, Lijun Fan, Dandan Qi, Honglu Zhang, Yongchao Hao, Mingmin Liang, Cunyao Bo, Silong Sun, Xiaoqian Wang, and et al. 2024. "Mining and Genetic Mapping of a Novel Powdery Mildew Resistance Gene, PmKu-2013, Identified in Aegilops tauschii" Agronomy 14, no. 4: 744. https://doi.org/10.3390/agronomy14040744
APA StyleChen, W., Li, J., Fan, L., Qi, D., Zhang, H., Hao, Y., Liang, M., Bo, C., Sun, S., Wang, X., Li, A., Wang, H., Kong, L., & Ma, X. (2024). Mining and Genetic Mapping of a Novel Powdery Mildew Resistance Gene, PmKu-2013, Identified in Aegilops tauschii. Agronomy, 14(4), 744. https://doi.org/10.3390/agronomy14040744





