Phytochemical and Gene Network Analysis Elucidating the Key Genes Involved in the Biosynthesis of Gomisin J in Schisandra sphenanthera
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Condition
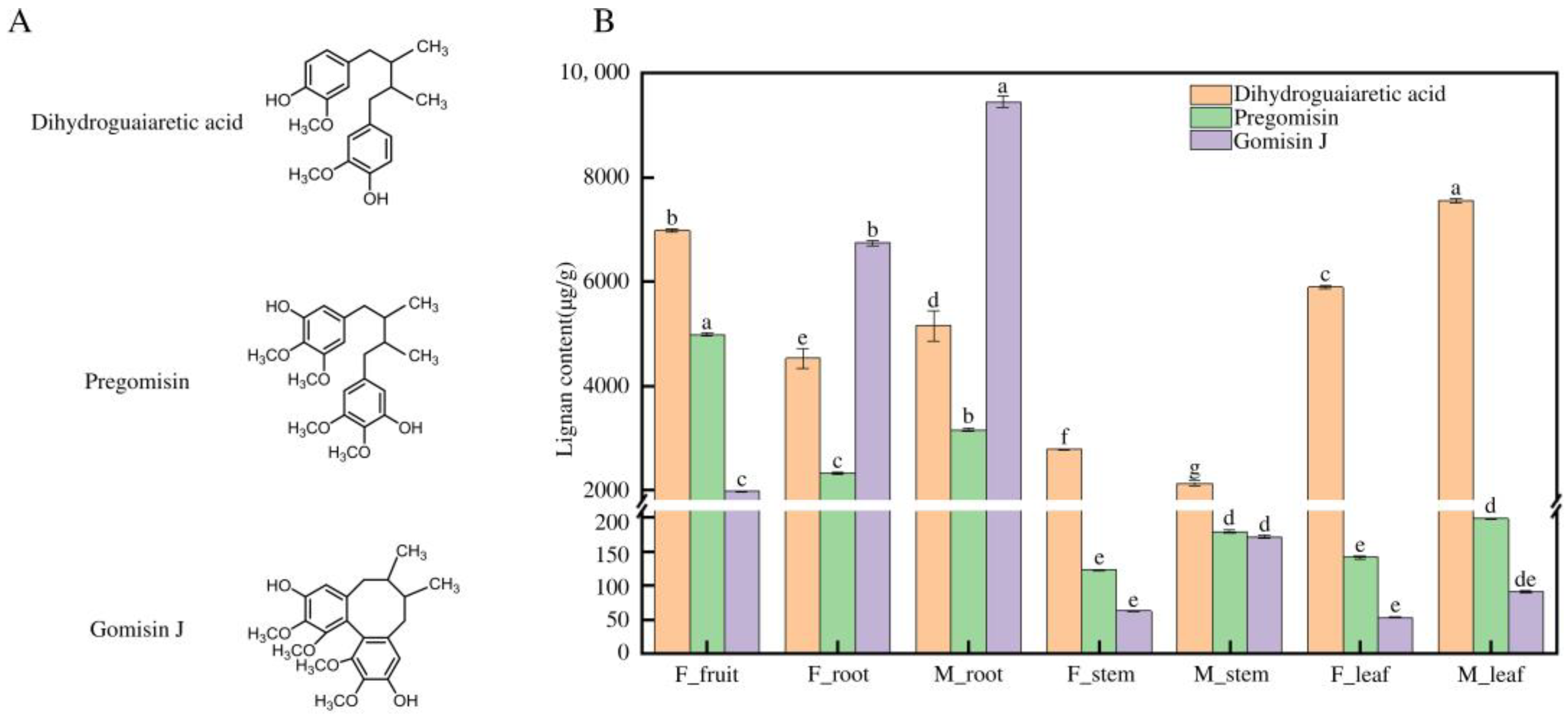
2.2. HPLC Analysis of Lignan Content in Different Tissues of Schisandra sphenanthera
2.3. RNA Extraction and RNA-Seq Workflow in Schisandra sphenanthera
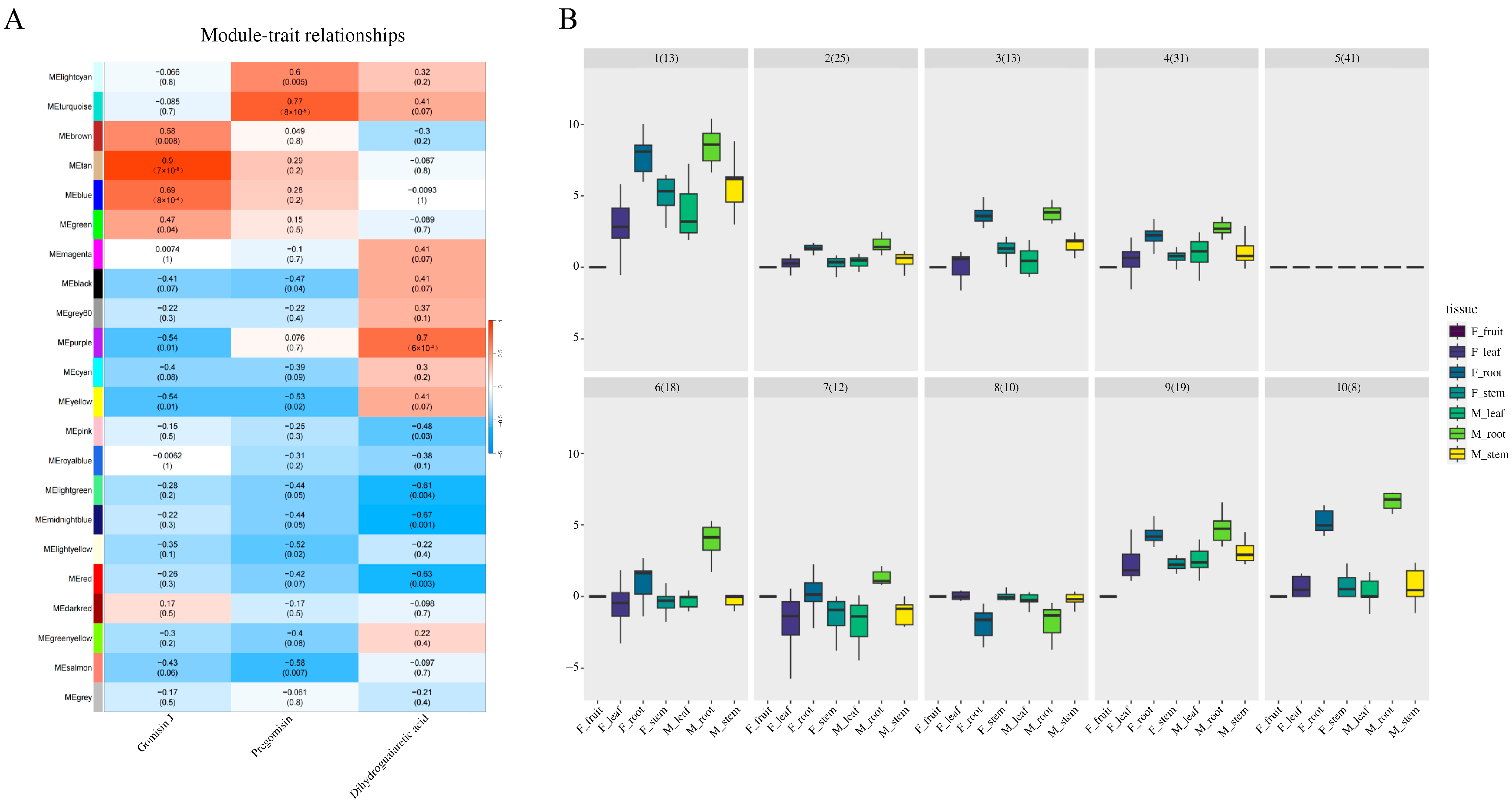
2.4. WGCNA Methodology and Clustering Analysis in Schisandra sphenanthera
2.5. Basic Bioinformatics Analysis of Cytochrome P450
3. Results
3.1. Tissue-Specific Distribution of Lignans in Schisandra sphenanthera
3.2. Elucidating Tissue-Specific Gene Expression Patterns in Schisandra sphenanthera with DEG Analysis
3.3. WGCNA Analysis Deciphering the Gene Expression Patterns in Lignan Biosynthesis
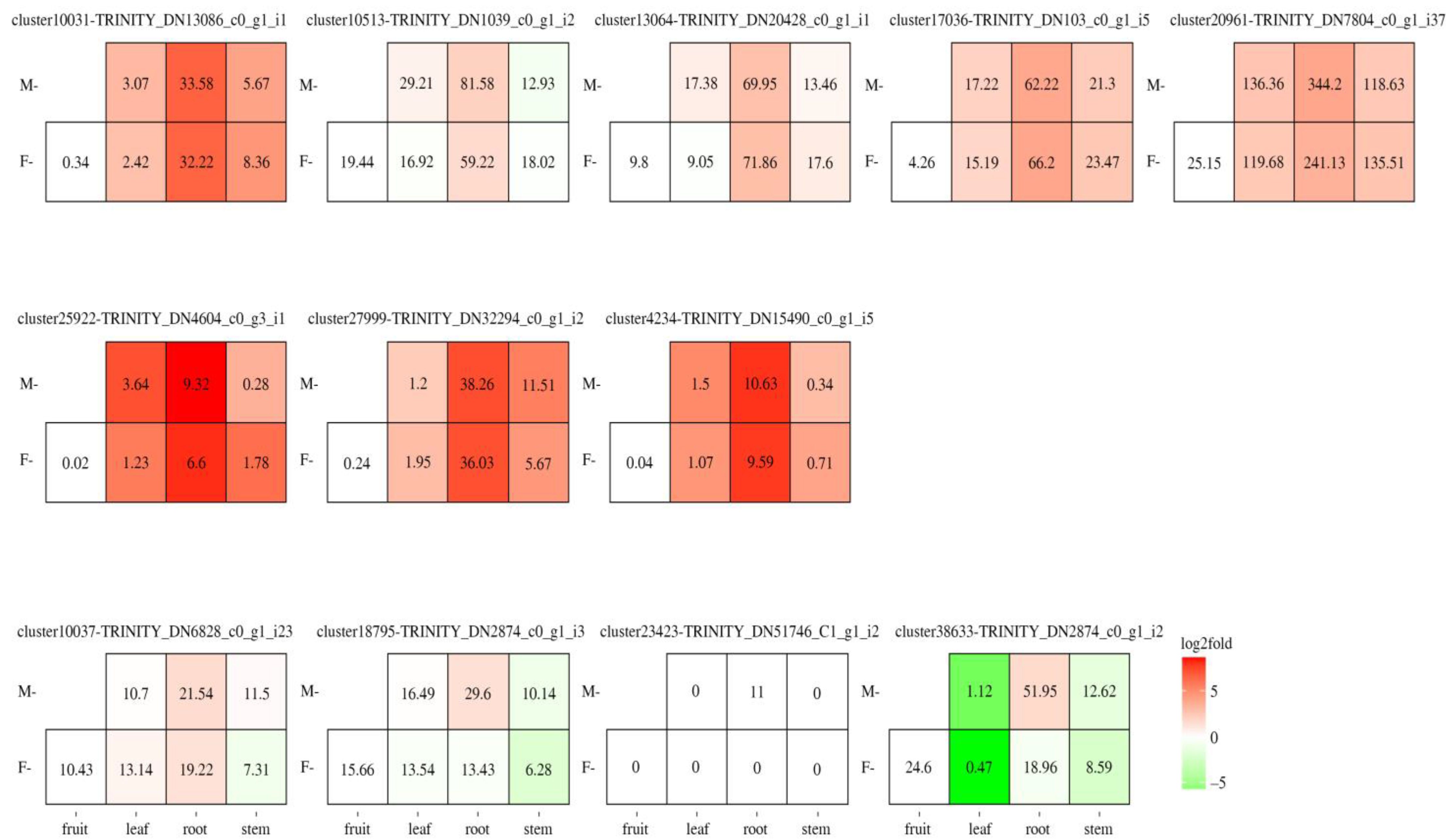
3.4. Deciphering the Tan Module Genes to Uncover Key Genes in Gomisin J Biosynthesis
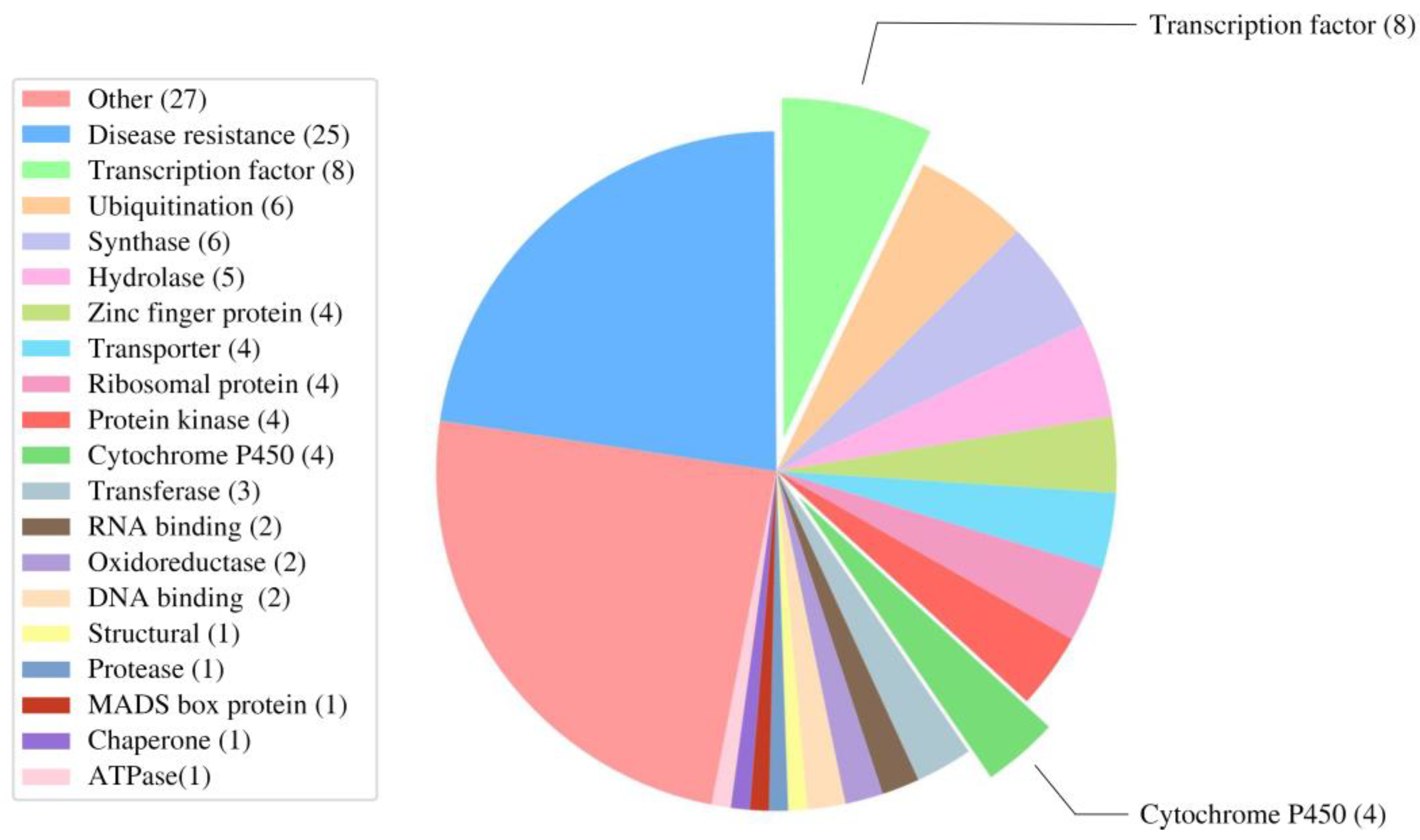
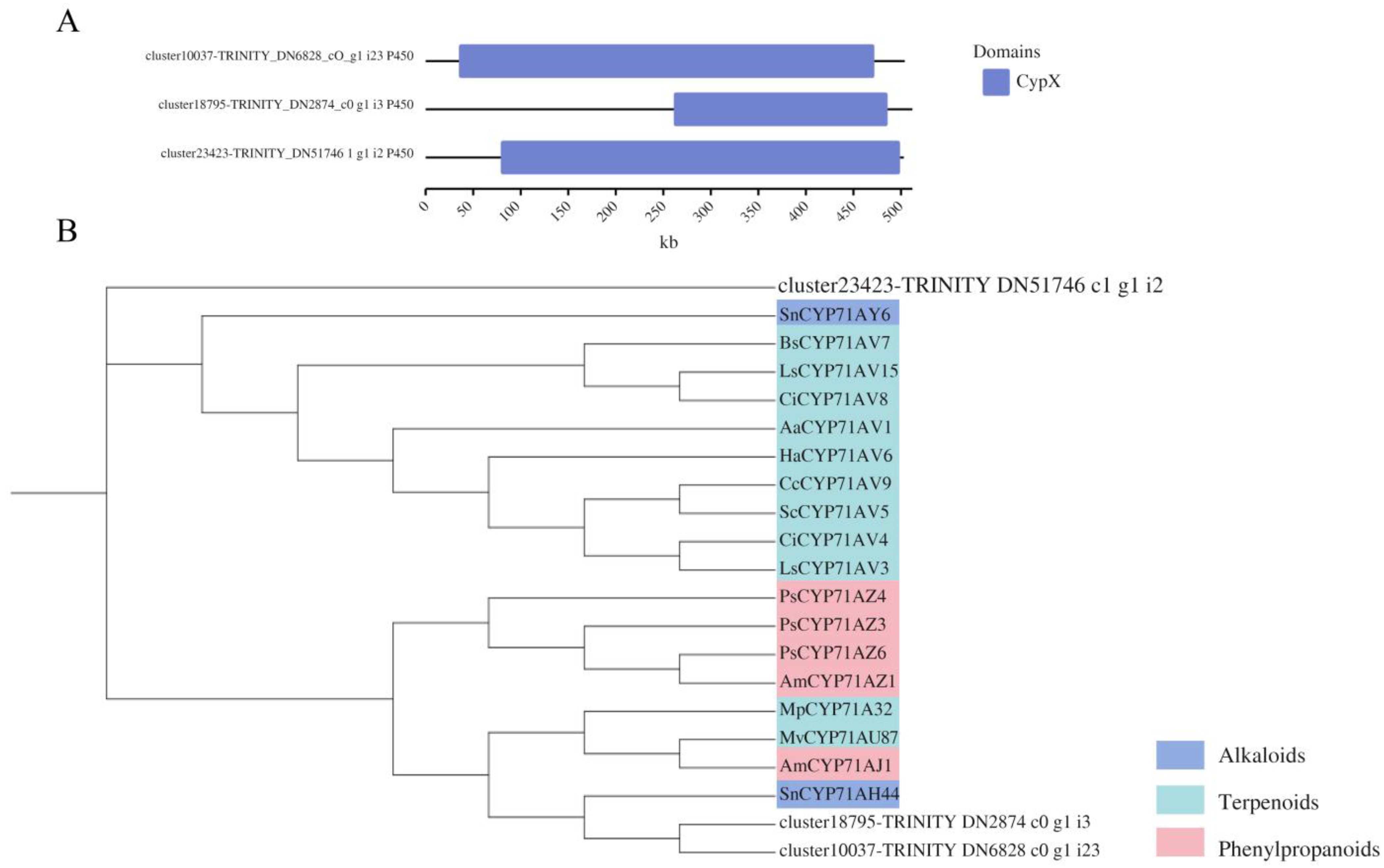
3.5. Investigating Transcription Factors and Cytochrome P450 Roles in Gomisin J Biosynthesis in Schisandra sphenanthera
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen, M.; Kilgore, N.; Lee, K.; Chen, D.F. Rubrisandrins A and B, lignans and related anti-HIV compounds from Schisandra rubriflora. J. Nat. Prod. 2006, 69, 1697–1701. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.; Ci, X.; Li, H.; Luo, G.; Li, R.T.; Deng, X.M. New Dibenzocyclooctadiene Lignans from Schisandra sphenanthera and Their Proinflammatory Cytokine Inhibitory Activities. Z. Naturforsch. B 2010, 65, 1–8. [Google Scholar] [CrossRef]
- Li, M.; Zhao, L.; Shi, Y.; Wang, J.; Lv, H.; Song, X.; Zhao, Q.; Zhao, Q.; Jing, R.; Hu, J. Gomisin J attenuates cerebral ischemia/reperfusion injury by inducing anti-apoptotic, anti-inflammatory, and antioxidant effects in rats. Bioengineered 2022, 13, 6908–6918. [Google Scholar]
- Li, R.; Yang, W. Gomisin J inhibits the glioma progression by inducing apoptosis and reducing HKII-regulated glycolysis. Biochem. Biophys. Res. Commun. 2020, 529, 15–22. [Google Scholar] [CrossRef]
- Kim, M.; Lim, S.J.; Lee, H.J.; Kim, S.Y.; Nho, C.W. Gomisin J inhibits oleic acid-induced hepatic lipogenesis by activation of the AMPK-dependent pathway and inhibition of the hepatokine fetuin-A in HepG2 cells. J. Agric. Food Chem. 2015, 63, 9729–9739. [Google Scholar] [CrossRef]
- Samil, J.; Moon, H.; Kim, S.; Quỳnh, N.T.; Yu, J.; Sandag, Z.; Le, D.T.; Lee, H.; Lee, H.; Lee, M. Anticancer of gomisin J from Schisandra chinensis fruit. Oncol. Rep. 2018, 41, 711–717. [Google Scholar]
- Song, J.H.; Cui, L.; An, L.B.; Li, W.T.; Fang, Z.; Zhang, Y.Y.; Dong, P.; Wu, X.; Wang, L.X.; Gonzalez, F.; et al. Inhibition of UDP-Glucuronosyltransferases (UGTs) Activity by constituents of Schisandra chinensis. Phytother. Res. 2015, 29, 1658–1664. [Google Scholar] [CrossRef]
- Satake, H.; Shiraishi, A.; Koyama, T.; Matsumoto, E.; Morimoto, K.; Bahabadi, S.E.; Ono, E.; Murata, J. Lignan Biosynthesis for Food Bioengineering. In Food Biosynthesis; Academic Press: Cambridge, MA, USA, 2017; pp. 351–379. [Google Scholar]
- Costa, M.A.; Xia, Z.; Davin, L.; Lewis, N. Toward Engineering the Metabolic Pathways of Cancer-Preventing Lignans in Cereal Grains and Other Crops. In Phytochemicals in Human Health Protection, Nutrition, and Plant Defense; Springer: Boston, MA, USA, 1999; pp. 67–87. [Google Scholar]
- Ehlting, J.; Hamberger, B.; Million-Rousseau, R.; Werck-Reichhart, D. Cytochromes P450 in phenolic metabolism. Phytoch. Rev. 2006, 5, 239–270. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, Y.; Gou, M.; Liu, C. Tissue-preferential recruitment of electron transfer chains for cytochrome P450-catalyzed phenolic biosynthesis. Sci. Adv. 2023, 9, 438. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Cui, H.; Huang, Y.; Yan, H.; Zhou, Y.; Wan, Y. Molecular docking and in vitro evaluations reveal the role of human cytochrome P450 3A4 in the cross-coupling metabolism of phenolic xenobiotics. Environ. Res. 2023, 10, 115256. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Ono, E.; Morimoto, K.; Yamagaki, T.; Okazawa, A.; Kobayashi, A.; Satake, H. Metabolic engineering of lignan biosynthesis in Forsythia cell culture. Plant Cell Phys. 2009, 50, 2200–2209. [Google Scholar] [CrossRef]
- Kuhlmann, S.; Kranz, K.; Lücking, B.; Alfermann, A.; Petersen, M. Aspects of cytotoxic lignan biosynthesis in suspension cultures of Linum nodiflorum. Phytoch. Rev. 2002, 1, 37–43. [Google Scholar] [CrossRef]
- Ono, E.; Nakai, M.; Fukui, Y.; Tomimori, N.; Fukuchi-Mizutani, M.; Saito, M.; Satake, H.; Tanaka, T.; Katsuta, M.; Umezawa, T.; et al. Formation of two methylenedioxy bridges by a Sesamum CYP81Q protein yielding a furofuran lignan, (+)-sesamin. Proc. Natl. Acad. Sci. USA 2006, 103, 10116–10121. [Google Scholar] [CrossRef] [PubMed]
- Schuler, M.; Werck-Reichhart, D. Functional genomics of P450s. Annu. Rev. Plant Biol. 2003, 54, 629–667. [Google Scholar] [CrossRef]
- Schuler, M. Plant Cytochrome P450 Monooxygenases. Crit. Rev. Plant Sci. 1996, 15, 235–284. [Google Scholar] [CrossRef]
- Seo, H.J.; Ji, S.B.; Kim, S.E.; Lee, G.M.; Park, S.Y.; Wu, Z.; Jang, D.; Liu, K.H. Inhibitory Effects of Schisandra Lignans on Cytochrome P450s and Uridine 5′-Diphospho-Glucuronosyl Transferases in Human Liver Microsomes. Pharmaceutics 2021, 13, 371. [Google Scholar] [CrossRef] [PubMed]
- Giddings, L.; Liscombe, D.; Hamilton, J.; Childs, K.; DellaPenna, D.; Buell, C.; O’Connor, S. A Stereoselective Hydroxylation Step of Alkaloid Biosynthesis by a Unique Cytochrome P450 in Catharanthus roseus. J. Biol. Chem. 2011, 286, 16751–16757. [Google Scholar] [CrossRef]
- Kang, K.; Lee, K.M.; Yoo, J.H.; Lee, H.J.; Kim, C.; Nho, C. Dibenzocyclooctadiene lignans, gomisins J and N inhibit the Wnt/β-catenin signaling pathway in HCT116 cells. Biochem. Biophys. Res. Commun. 2012, 428, 285–291. [Google Scholar] [CrossRef]
- Decembrino, D.; Ricklefs, E.; Wohlgemuth, S.; Girhard, M.; Schullehner, K.; Jach, G.; Urlacher, V. Assembly of Plant Enzymes in E. coli for the Production of the Valuable (-)-Podophyllotoxin Precursor (-)-Pluviatolide. ACS Synth. Biol. 2020, 9, 3091–3103. [Google Scholar] [CrossRef]
- Gorinova, N.; Nedkovska, M.; Bakalova, E.; Atanassov, A.; Ohkawa, H. Biotransformation of Xenobiotics and Endogenous Substrates by Plant Cytochrome P450s. Biotechnol. Biotechnol. Equip. 1999, 13, 13–19. [Google Scholar] [CrossRef]
- Xin, A.; Herburger, K. Precursor biosynthesis regulation of lignin, suberin and cutin. Protoplasma 2021, 258, 1171–1178. [Google Scholar] [CrossRef] [PubMed]
- Attoumbré, J.; Bienaimé, C.; Dubois, F.; Fliniaux, M.; Chabbert, B.; Baltora-Rosset, S. Development of antibodies against secoisolariciresinol—Application to the immunolocalization of lignans in Linum usitatissimum seeds. Phytochemistry 2010, 71, 1979–1987. [Google Scholar] [CrossRef] [PubMed]
- Kulik, T.; Buśko, M.; Pszczółkowska, A.; Perkowski, J.; Okorski, A. Plant lignans inhibit growth and trichothecene biosynthesis in Fusarium graminearum. Lett. Appl. Microbiol. 2014, 59, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Satake, H.; Ono, E.; Murata, J. Recent advances in the metabolic engineering of lignan biosynthesis pathways for the production of transgenic plant-based foods and supplements. J. Agric. Food Chem. 2013, 61, 11721–11729. [Google Scholar] [CrossRef]
- Köse, L.P.; Gulcin, I. Inhibition effects of some lignans on carbonic anhydrase, acetylcholinesterase and butyrylcholinesterase enzymes. Rec. Nat. Prod. 2017, 11, 558–561. [Google Scholar] [CrossRef]
- Fu, H.; Guo, C.; Peng, J.; Shao, F.; Sheng, S.; Wang, S. Transcriptomic insights and cytochrome P450 gene analysis in Kadsura coccinea for lignan biosynthesis. Genes 2024, 15, 270. [Google Scholar] [CrossRef]
- Baldi, A.; Farkya, S.; Jain, A.; Gupta, N.; Mehra, R.; Datta, V.; Srivastava, A.; Bisaria, V. Enhanced production of podophyllotoxins by co-culture of transformed Linum album cells with plant growth-promoting fungi. Pure Appl. Chem. 2010, 82, 227–241. [Google Scholar] [CrossRef]
- Saarinen, N.; Wärri, A.; Airio, M.; Smeds, A.; Mäkelä, S. Role of dietary lignans in the reduction of breast cancer risk. Mol. Nutr. Food Res. 2007, 51, 857–866. [Google Scholar] [CrossRef]
- Ji, F.; Sadreyev, R.I. RNA-seq: Basic bioinformatics analysis. Curr. Protoc. Mol. Biol. 2018, 124, e68. [Google Scholar] [CrossRef]
- Morant, M.; Bak, S.; Møller, B.L.; Werck-Reichhart, D. Plant cytochromes P450: Tools for pharmacology, plant protection and phytoremediation. Curr. Opin. Biotechnol. 2003, 14, 151–162. [Google Scholar] [CrossRef]
- Liu, X.; Zhu, X.; Wang, H.; Liu, T.; Cheng, J.; Jiang, H. Discovery and modification of cytochrome P450 for plant natural products biosynthesis. Synth. Syst. Biotechnol. 2020, 5, 187–199. [Google Scholar] [CrossRef]
- Mizutani, M.; Ohta, D. Diversification of P450 genes during land plant evolution. Annu. Rev. Plant Biol. 2010, 61, 291–315. [Google Scholar] [CrossRef]
- Nomura, T.; Bishop, G. Cytochrome P450s in plant steroid hormone synthesis and metabolism. Phytochem. Rev. 2006, 5, 421–432. [Google Scholar] [CrossRef]
- Wu, J.J.; Cao, Y.F.; Feng, L.; He, Y.Q.; Hong, J.Y.; Dou, T.; Wang, P.; Hao, D.; Ge, G.; Yang, L. A Naturally Occurring Isoform-Specific Probe for Highly Selective and Sensitive Detection of Human Cytochrome P450 3A5. J. Med. Chem. 2017, 60, 3804–3813. [Google Scholar] [CrossRef]
- Yang, C.Q.; Fang, X.; Wu, X.M.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Transcriptional regulation of plant secondary metabolism F. J. Integr. Plant Biol. 2012, 54, 703–712. [Google Scholar] [CrossRef]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple functions of MYB transcription factors in abiotic stress responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef] [PubMed]
- Schluttenhofer, C.M.; Yuan, L. Regulation of Specialized Metabolism by WRKY Transcription Factors. Plant Physiol. 2015, 167, 295–306. [Google Scholar] [CrossRef]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Xiao, Y.; Lv, Z.; Tan, H.; Chen, R.; Li, Q.; Chen, J.; Wang, Y.; Yin, J.; Zhang, L.; et al. AP2/ERF transcription factor, Ii049, positively regulates lignan biosynthesis in Isatis indigotica through activating salicylic acid signaling and lignan/lignin pathway genes. Front. Dev. Plant Sci. 2017, 8, 1361. [Google Scholar] [CrossRef] [PubMed]
- Ražná, K.; Harenčár, Ľ.; Kučka, M. The Involvement of microRNAs in Plant Lignan Biosynthesis—Current View. Cells 2022, 11, 2151. [Google Scholar] [CrossRef] [PubMed]
- Tamagnone, L.; Merida, A.; Parr, A.; Mackay, S.; Culianez-Macia, F.A.; Roberts, K.; Martin, C. The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 1998, 10, 135–154. [Google Scholar] [CrossRef]
- Yu, X.; Feng, Y.; Zhai, W.; Chen, M.; Wu, G. The complete mitochondrial genome of Schisandra sphenanthera (Schisandraceae). Mitochondrial DNA B Resour. 2018, 3, 1246–1247. [Google Scholar] [CrossRef]
- Ikeya, Y.; Sugama, K.; Okada, M.; Mitsuhashi, H. Two lignans from Schisandra sphenanthera. Phytochemistry 1991, 30, 975–980. [Google Scholar] [CrossRef]
- Jiang, K.; Song, Q.; Peng, S.J.; Zhao, Q.Q.; Li, G.; Li, Y.; Gao, K. New lignans from the roots of Schisandra sphenanthera. Fitoterapia 2015, 103, 63–70. [Google Scholar] [CrossRef]
- Gao, J.P.; Wang, Y.H.; Yu, Y.Q.; Chen, D.F. Determination by HPLC and Variation Regularity of Lignan Constituents in Chinese Crude Drug Fructus Schisandrae sphenantherae. Chin. J. Nat. Med. 2003, 2, 28–32. [Google Scholar]
- Rasool, S.; Mohamed, R. Plant cytochrome P450s: Nomenclature and involvement in natural product biosynthesis. Protoplasma 2016, 253, 1197–1209. [Google Scholar] [CrossRef] [PubMed]
- Dimaano, N.G.; Iwakami, S. Cytochrome P450 enzymes involved in herbicide metabolism in plants. Pest Manag. Sci. 2020, 76, 3346–3355. [Google Scholar]
| FMFruit | FMLeaf | FMRoot | FMStem | MLeaf | MRoot | MStem | |
|---|---|---|---|---|---|---|---|
| FMFruit | 0 | 2987/3961 6948 | 3728/5124 8852 | 3196/4375 7571 | 2337/3291 5628 | 3162/6037 9199 | 2424/3085 5509 |
| FMLeaf | 0 | 3065/3601 6666 | 1521/1181 2702 | 12/15 27 | 2131/3261 5392 | 1320/685 2005 | |
| FMRoot | 0 | 1852/1159 3011 | 2085/1944 4029 | 111/135 246 | 1524/522 2046 | ||
| FMStem | 0 | 832/1125 1957 | 1486/3902 5388 | 44/63 107 | |||
| MLeaf | 0 | 1696/2704 4400 | 1116/433 1549 | ||||
| MRoot | 0 | 2937/524 3461 | |||||
| MStem | 0 |
| Gene ID | CDS (bp) | Number of Amino Acids | Mw (kDa) | pI | CYP450 Subfamily Prediction (NCBI) | Instability Index | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| cluster10037-TRINITY_DN6828_c0_g1_i23 | 1512 | 503 | 56.72 | 6.38 | CYP71A1-like | 44.96 | −0.063 | Endoplasmic reticulum |
| cluster18795-TRINITY_DN2874_c0_g1_i3 | 1536 | 511 | 58.02 | 6.59 | CYP71A1 | 49.75 | −0.122 | Endoplasmic reticulum |
| cluster23423-TRINITY_DN51746_c1_g1_i2 | 1509 | 502 | 57.02 | 8.4 | CYP | 48.26 | −0.169 | Endoplasmic reticulum |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, B.; Peng, J.; Fu, H.; Shao, F.; Sheng, S.; Wang, S. Phytochemical and Gene Network Analysis Elucidating the Key Genes Involved in the Biosynthesis of Gomisin J in Schisandra sphenanthera. Agronomy 2024, 14, 576. https://doi.org/10.3390/agronomy14030576
Wu B, Peng J, Fu H, Shao F, Sheng S, Wang S. Phytochemical and Gene Network Analysis Elucidating the Key Genes Involved in the Biosynthesis of Gomisin J in Schisandra sphenanthera. Agronomy. 2024; 14(3):576. https://doi.org/10.3390/agronomy14030576
Chicago/Turabian StyleWu, Bolin, Jiqing Peng, Hanyu Fu, Fengxia Shao, Song Sheng, and Sen Wang. 2024. "Phytochemical and Gene Network Analysis Elucidating the Key Genes Involved in the Biosynthesis of Gomisin J in Schisandra sphenanthera" Agronomy 14, no. 3: 576. https://doi.org/10.3390/agronomy14030576
APA StyleWu, B., Peng, J., Fu, H., Shao, F., Sheng, S., & Wang, S. (2024). Phytochemical and Gene Network Analysis Elucidating the Key Genes Involved in the Biosynthesis of Gomisin J in Schisandra sphenanthera. Agronomy, 14(3), 576. https://doi.org/10.3390/agronomy14030576







