Classification of Plant Leaf Disease Recognition Based on Self-Supervised Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets
2.1.1. The Dataset We Collected
2.1.2. The CCMT Dataset
2.2. Construction of the Model
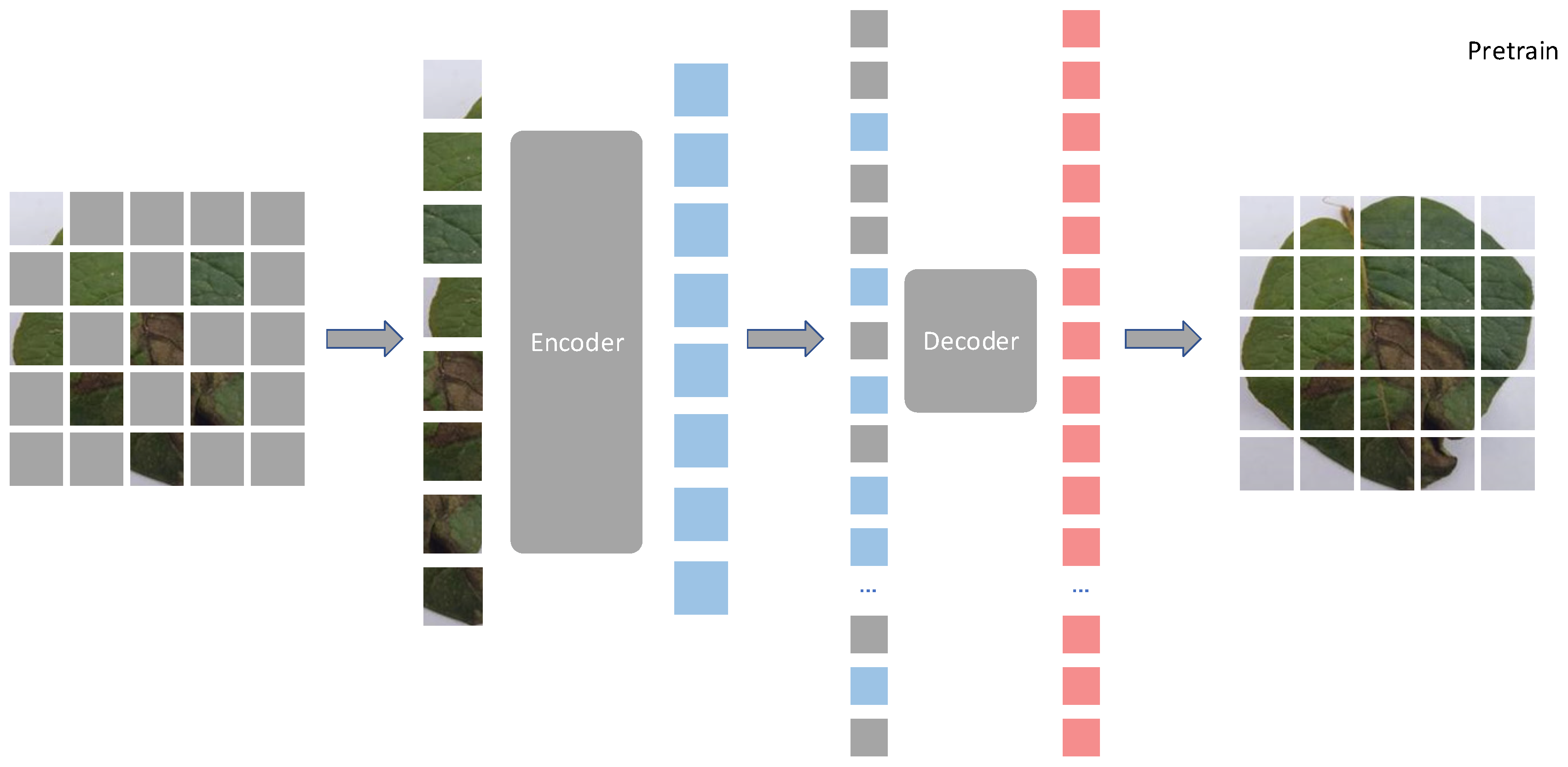
2.2.1. The Masked Autoencoder Model
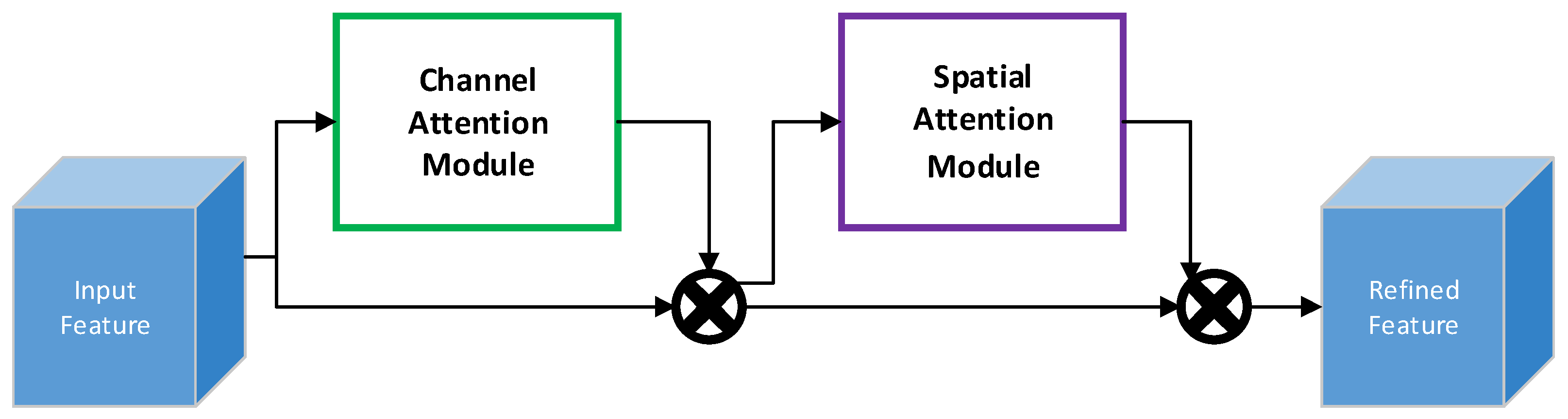
2.2.2. Convolutional Block Attention Module
2.2.3. Gate Recurrent Unit Module
2.2.4. Improved Model Architecture
2.3. Test Platform and Parameters
2.4. Evaluation Indicators of Experimental Results
3. Results
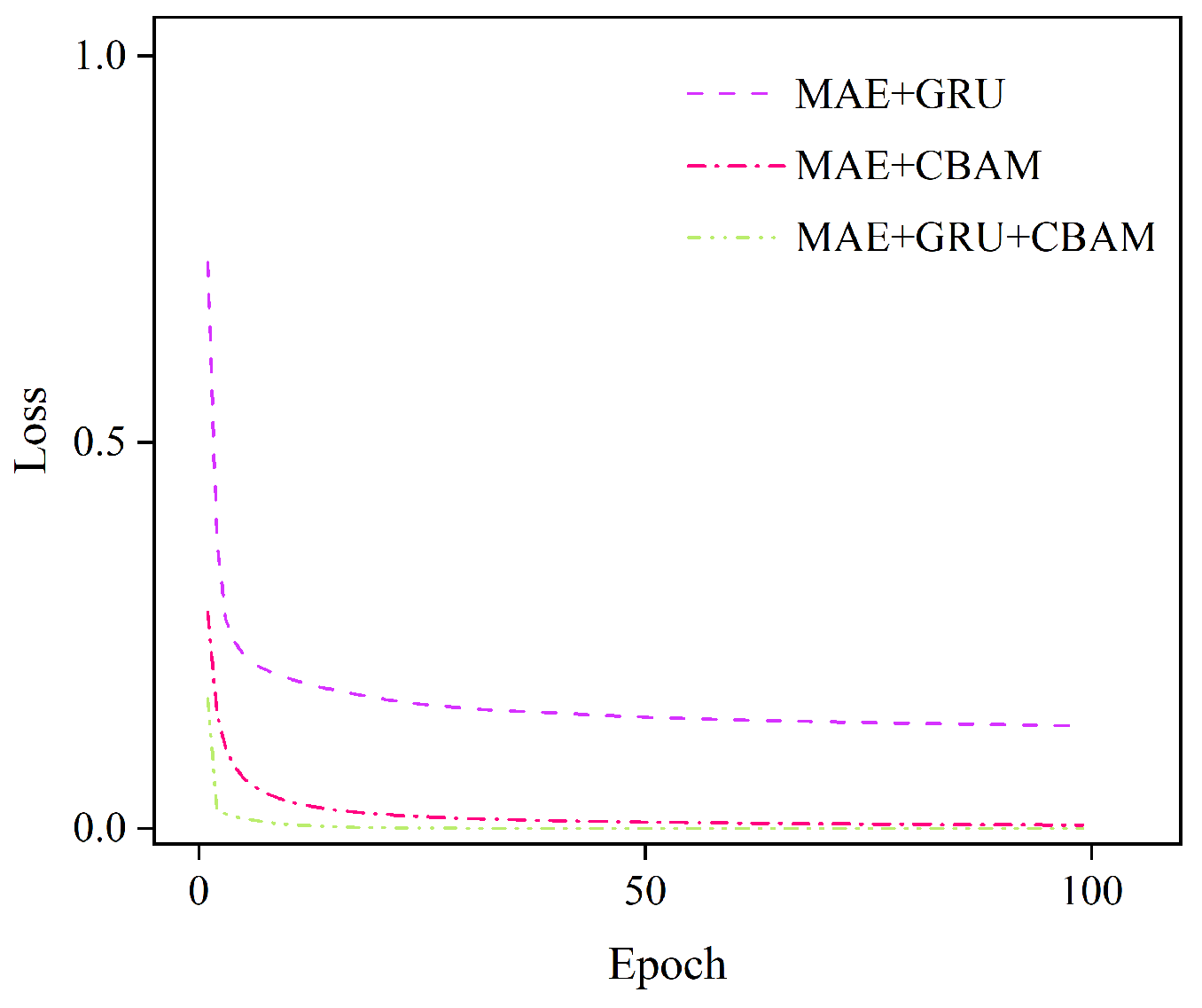
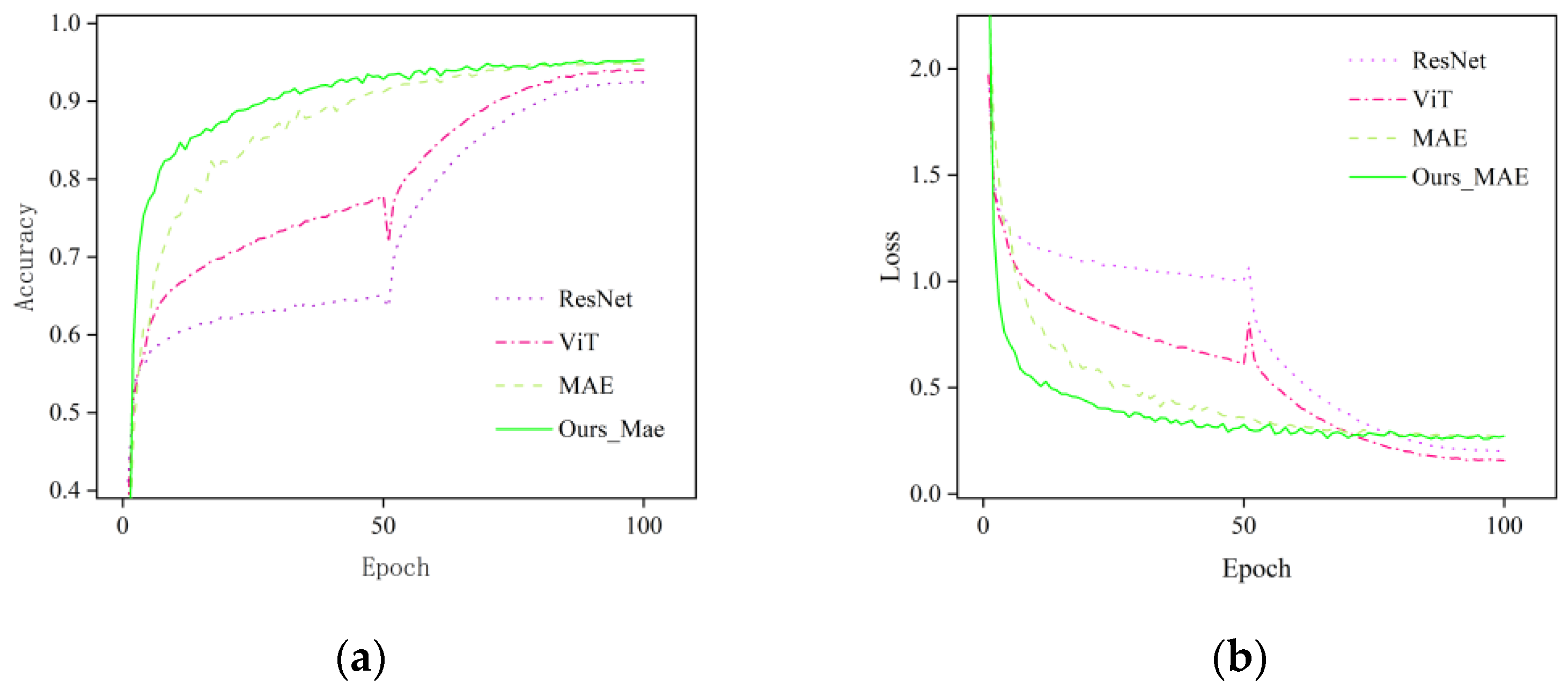
3.1. Comparison and Analysis of Different Improved Models
3.2. Comparison and Analysis of the Improved Model with Other Models
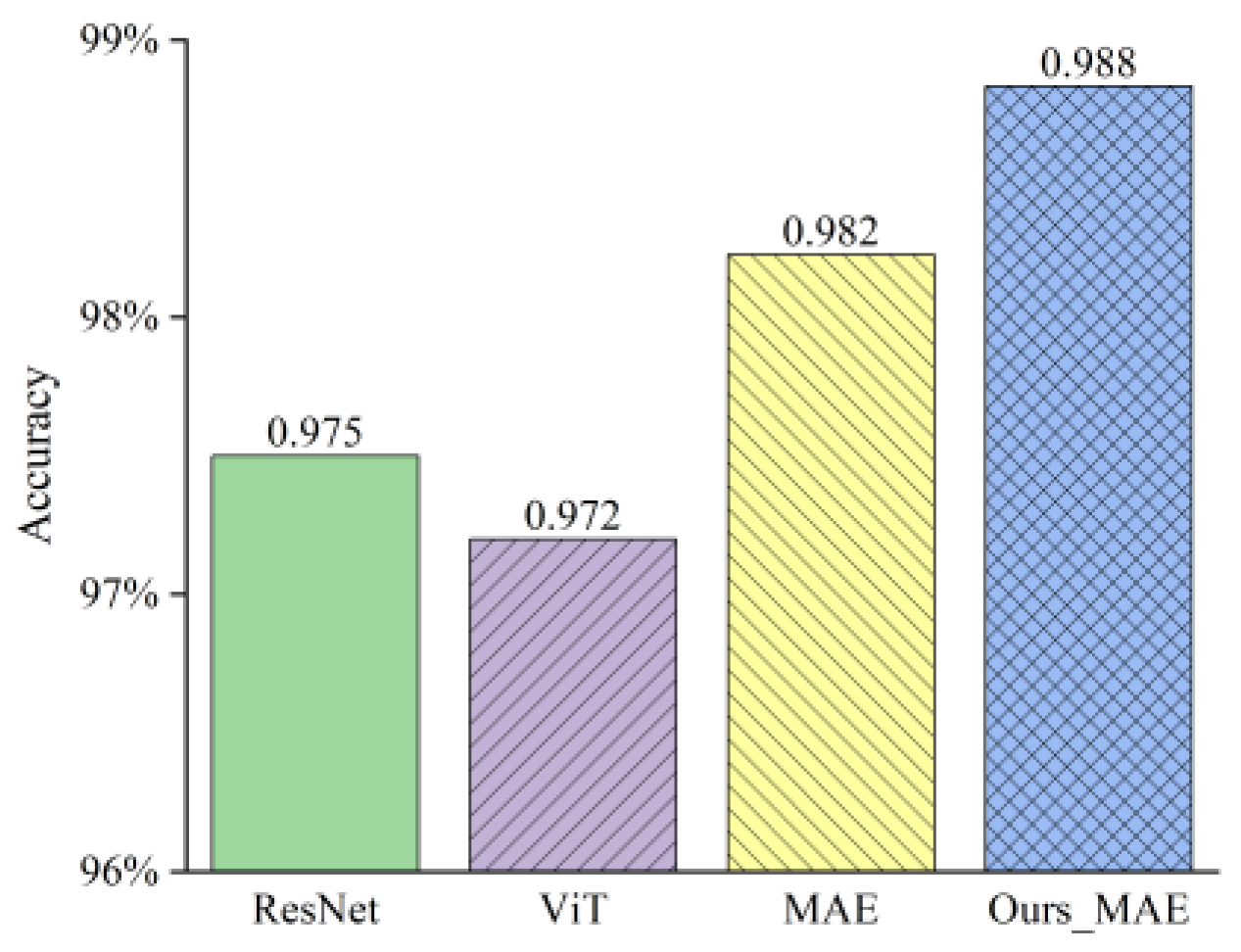
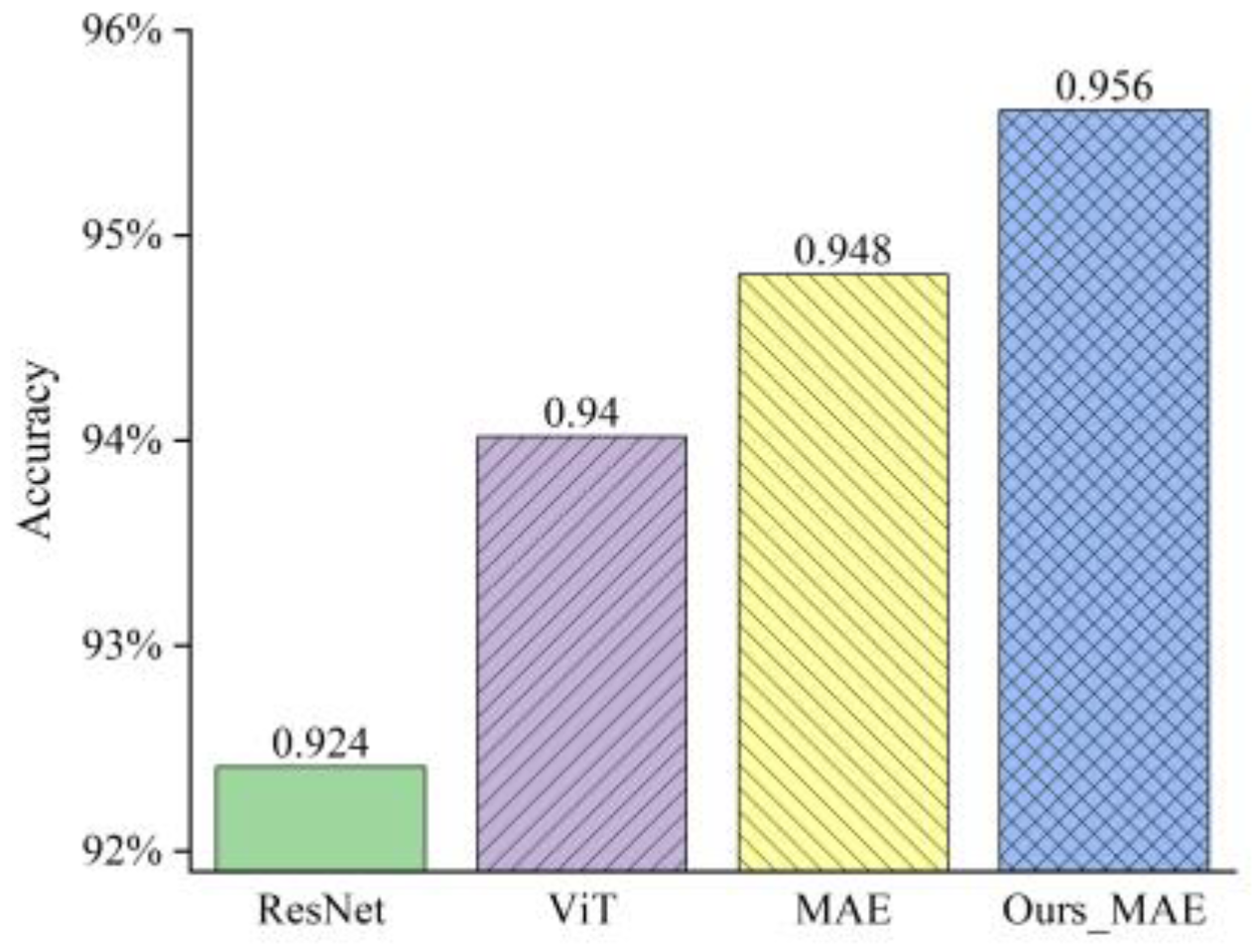
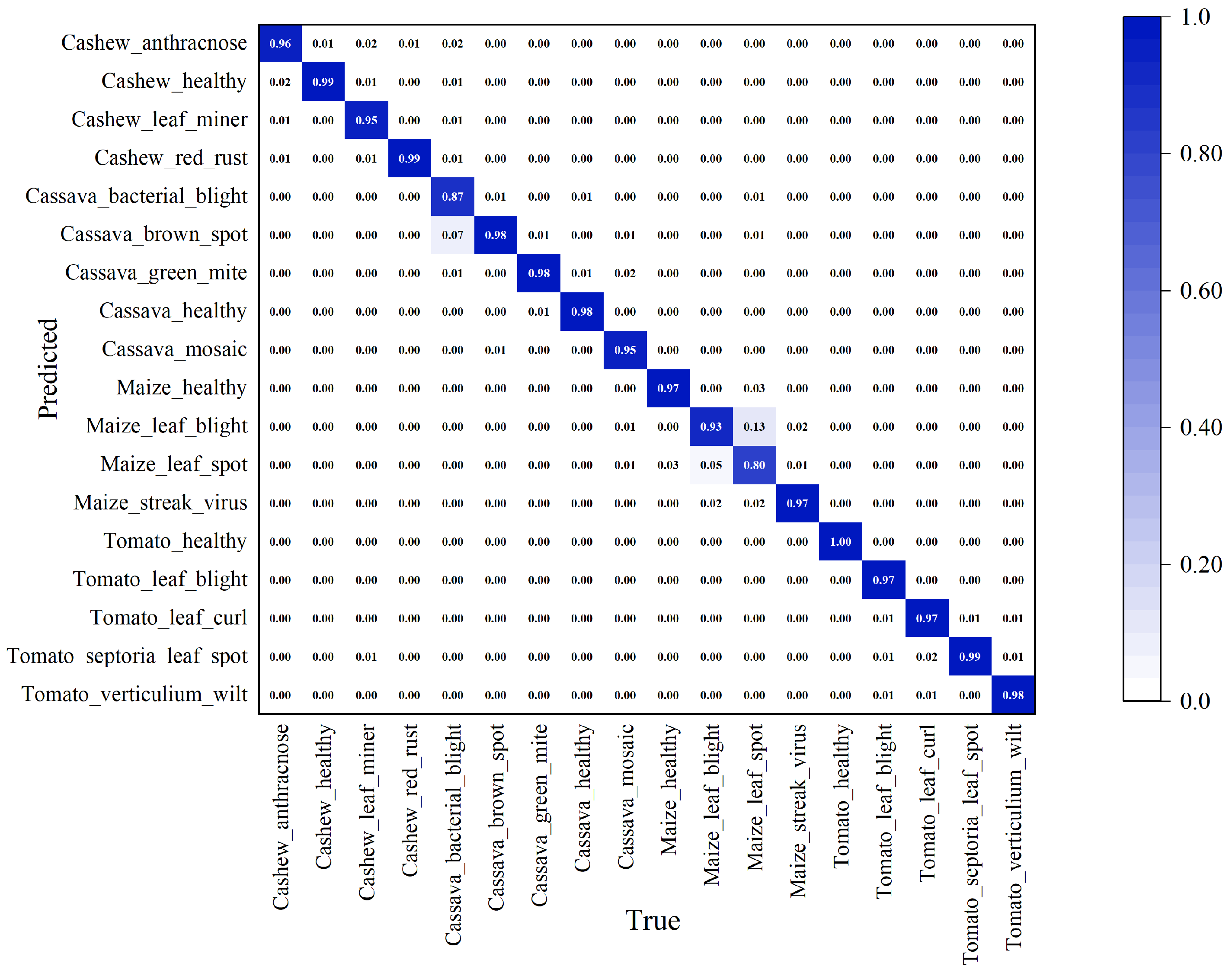
3.2.1. Comparison and Analysis on Our Collected Datasets
3.2.2. Comparison and Analysis on the CCMT Dataset
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nigam, S.; Jain, R. Plant disease identification using Deep Learning: A review. Indian J. Agric. Sci. 2020, 90, 249–257. [Google Scholar] [CrossRef]
- Jin, H.B.; Chu, X.Q.; Qi, J.F.; Zhang, X.X.; Mu, W.S. CWAN: Self-supervised learning for deep grape disease image composition. Eng. Appl. Artif. Intell. 2023, 123, 106458. [Google Scholar] [CrossRef]
- Zeng, Y.X.; Shi, J.S.; Ji, Z.J.; Wen, Z.H.; Liang, Y.; Yang, C.D. Genotype by Environment Interaction: The Greatest Obstacle in Precise Determination of Rice Sheath Blight Resistance in the Field. Plant Dis. 2017, 101, 1795–1801. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.M.; Qi, F.; Sun, M.H.; Qu, J.H.; Xue, J. Identification of Tomato Disease Types and Detection of Infected Areas Based on Deep Convolutional Neural Networks and Object Detection Techniques. Comput. Intell. Neurosci. 2019, 2019, 9142753. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Wang, L.Y.; Ding, H.X.; Xu, M. Strawberry Verticillium Wilt Detection Network Based on Multi-Task Learning and Attention. IEEE Access 2019, 7, 170003–170011. [Google Scholar] [CrossRef]
- Sunil, C.K.; Jaidhar, C.D.; Patil, N. Systematic study on deep learning-based plant disease detection or classification. Artif. Intell. Rev. 2023, 56, 14955–15052. [Google Scholar] [CrossRef]
- Khan, A.T.; Jensen, S.M.; Khan, A.R.; Li, S. Plant disease detection model for edge computing devices. Front. Plant Sci. 2023, 14, 1308528. [Google Scholar] [CrossRef] [PubMed]
- Craze, H.A.; Pillay, N.; Joubert, F.; Berger, D.K. Deep Learning Diagnostics of Gray Leaf Spot in Maize under Mixed Disease Field Conditions. Plants 2022, 11, 1942. [Google Scholar] [CrossRef]
- Li, Y.; Sun, S.Y.; Zhang, C.S.; Yang, G.S.; Ye, Q.B. One-Stage Disease Detection Method for Maize Leaf Based on Multi-Scale Feature Fusion. Appl. Sci. 2022, 12, 7960. [Google Scholar] [CrossRef]
- Li, J.W.; Qiao, Y.L.; Liu, S.; Zhang, J.H.; Yang, Z.C.; Wang, M.L. An improved YOLOv5-based vegetable disease detection method. Comput. Electron. Agric. 2022, 202, 107345. [Google Scholar] [CrossRef]
- Memon, M.S.; Kumar, P.; Iqbal, R. Meta Deep Learn Leaf Disease Identification Model for Cotton Crop. Computers 2022, 11, 102. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, Y.; Zhang, T.S.; Wang, H.G.; Jia, Y.J.; Gao, R.; Su, Z.B. Maize leaf disease identification using deep transfer convolutional neural networks. Int. J. Agric. Biol. Eng. 2022, 15, 187–195. [Google Scholar] [CrossRef]
- Jing, L.; Tian, Y. Self-supervised visual feature learning with deep neural networks: A survey. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 43, 4037–4058. [Google Scholar] [CrossRef]
- Yan, J.; Wang, X.F. Unsupervised and semi-supervised learning: The next frontier in machine learning for plant systems biology. Plant J. 2022, 111, 1527–1538. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Chen, L.; Yuan, Y. Multimodal Fine-Grained Transformer Model for Pest Recognition. Electronics 2023, 12, 2620. [Google Scholar] [CrossRef]
- Gong, X.J.; Zhang, X.H.; Zhang, R.W.; Wu, Q.F.; Wang, H.; Guo, R.C.; Chen, Z.R. U3-YOLOXs: An improved YOLOXs for Uncommon Unregular Unbalance detection of the rape subhealth regions. Comput. Electron. Agric. 2022, 203, 107461. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, F.J.; Hou, Z.Y.; Mian, L.; Wang, Z.Y.; Zhang, J.; Tang, J. Self-Supervised Learning: Generative or Contrastive. IEEE Trans. Knowl. Data Eng. 2023, 35, 857–876. [Google Scholar] [CrossRef]
- Ohri, K.; Kumar, M. Review on self-supervised image recognition using deep neural networks. Knowl.-Based Syst. 2021, 224, 107090. [Google Scholar] [CrossRef]
- Yang, G.F.; Yang, Y.; He, Z.K.; Zhang, X.Y.; He, Y. A rapid, low-cost deep learning system to classify strawberry disease based on cloud service. J. Integr. Agric. 2022, 21, 460–473. [Google Scholar] [CrossRef]
- Tomasev, N.; Bica, I.; McWilliams, B.; Buesing, L.; Pascanu, R.; Blundell, C.; Mitrovic, J. Pushing the limits of self-supervised ResNets: Can we outperform supervised learning without labels on ImageNet? arXiv 2022, arXiv:2201.05119. [Google Scholar]
- Lin, X.F.; Li, C.T.; Adams, S.; Kouzani, A.Z.; Jiang, R.C.; He, L.G.; Hu, Y.J.; Vernon, M.; Doeven, E.; Webb, L.; et al. Self-Supervised Leaf Segmentation under Complex Lighting Conditions. Pattern Recognit. 2023, 135, 109021. [Google Scholar] [CrossRef]
- Gai, R.L.; Wei, K.; Wang, P.F. SSMDA: Self-Supervised Cherry Maturity Detection Algorithm Based on Multi-Feature Contrastive Learning. Agriculture 2023, 13, 939. [Google Scholar] [CrossRef]
- Xiao, B.J.; Nguyen, M.; Yan, W.Q. Fruit ripeness identification using transformers. Appl. Intell. 2023, 53, 22488–22499. [Google Scholar] [CrossRef]
- Liu, Y.S.; Zhou, S.B.; Wu, H.M.; Han, W.; Li, C.; Chen, H. Joint optimization of autoencoder and Self-Supervised Classifier: Anomaly detection of strawberries using hyperspectral imaging. Comput. Electron. Agric. 2022, 198, 107007. [Google Scholar] [CrossRef]
- Zheng, H.; Wang, G.H.; Li, X.C. Swin-MLP: A strawberry appearance quality identification method by Swin Transformer and multi-layer perceptron. J. Food Meas. Charact. 2022, 16, 2789–2800. [Google Scholar] [CrossRef]
- Bi, C.G.; Hu, N.; Zou, Y.Q.; Zhang, S.; Xu, S.Z.; Yu, H.L. Development of Deep Learning Methodology for Maize Seed Variety Recognition Based on Improved Swin Transformer. Agronomy 2022, 12, 1843. [Google Scholar] [CrossRef]
- He, K.; Chen, X.; Xie, S.; Li, Y.; Dollár, P.; Girshick, R. Masked autoencoders are scalable vision learners. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 16000–16009. [Google Scholar]
- Wang, S.P.; Cai, J.Y.; Lin, Q.H.; Guo, W.Z. An Overview of Unsupervised Deep Feature Representation for Text Categorization. IEEE Trans. Comput. Soc. Syst. 2019, 6, 504–517. [Google Scholar] [CrossRef]
- Li, P.Z.; Pei, Y.; Li, J.Q. A comprehensive survey on design and application of autoencoder in deep learning. Appl. Soft Comput. 2023, 138, 110176. [Google Scholar] [CrossRef]
- Rashid, J.; Khan, I.; Ali, G.; Almotiri, S.H.; AlGhamdi, M.A.; Masood, K. Multi-Level Deep Learning Model for Potato Leaf Disease Recognition. Electronics 2021, 10, 2064. [Google Scholar] [CrossRef]
- Mensah, P.K.; Akoto-Adjepong, V.; Adu, K.; Ayidzoe, M.A.; Bediako, E.A.; Nyarko-Boateng, O.; Boateng, S.; Donkor, E.F.; Bawah, F.U.; Awarayi, N.S. CCMT: Dataset for crop pest and disease detection. Data Brief 2023, 49, 109306. [Google Scholar] [CrossRef]
- Arnab, A.; Dehghani, M.; Heigold, G.; Sun, C.; Lučić, M.; Schmid, C. Vivit: A video vision transformer. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Virtual, 11–17 October 2021; pp. 6836–6846. [Google Scholar]
- Voita, E.; Talbot, D.; Moiseev, F.; Sennrich, R.; Titov, I. Analyzing multi-head self-attention: Specialized heads do the heavy lifting, the rest can be pruned. arXiv 2019, arXiv:1905.09418. [Google Scholar]
- Tang, J.; Deng, C.; Huang, G.-B. Extreme learning machine for multilayer perceptron. IEEE Trans. Neural Netw. Learn. Syst. 2015, 27, 809–821. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 6000–6010. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Park, J.; Woo, S.; Lee, J.-Y.; Kweon, I.S. A simple and light-weight attention module for convolutional neural networks. Int. J. Comput. Vis. 2020, 128, 783–798. [Google Scholar] [CrossRef]
- Dey, R.; Salem, F.M. Gate-variants of gated recurrent unit (GRU) neural networks. In Proceedings of the 2017 IEEE 60th International Midwest Symposium on Circuits and Systems (MWSCAS), Boston, MA, USA, 6–9 August 2017; pp. 1597–1600. [Google Scholar]
- Medsker, L.R.; Jain, L. Recurrent neural networks. Des. Appl. 2001, 5, 2. [Google Scholar]
- Shu, W.; Cai, K.; Xiong, N.N. A short-term traffic flow prediction model based on an improved gate recurrent unit neural network. IEEE Trans. Intell. Transp. Syst. 2021, 23, 16654–16665. [Google Scholar] [CrossRef]
- Zhou, G.-B.; Wu, J.; Zhang, C.-L.; Zhou, Z.-H. Minimal gated unit for recurrent neural networks. Int. J. Autom. Comput. 2016, 13, 226–234. [Google Scholar] [CrossRef]
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A simple framework for contrastive learning of visual representations. In Proceedings of the International Conference on Machine Learning, Virtual, 13–18 July 2020; pp. 1597–1607. [Google Scholar]
- He, K.; Fan, H.; Wu, Y.; Xie, S.; Girshick, R. Momentum contrast for unsupervised visual representation learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020; pp. 9729–9738. [Google Scholar]
- Wang, X.; Qi, G.-J. Contrastive learning with stronger augmentations. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 45, 5549–5560. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Tan, X.; Zhang, P.; Wang, X. A CBAM based multiscale transformer fusion approach for remote sensing image change detection. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2022, 15, 6817–6825. [Google Scholar] [CrossRef]
- Bi, L.N.; Hu, G.P.; Raza, M.M.; Kandel, Y.; Leandro, L.; Mueller, D. A Gated Recurrent Units (GRU)-Based Model for Early Detection of Soybean Sudden Death Syndrome through Time-Series Satellite Imagery. Remote Sens. 2020, 12, 3621. [Google Scholar] [CrossRef]
- Alirezazadeh, P.; Schirrmann, M.; Stolzenburg, F. Improving Deep Learning-based Plant Disease Classification with Attention Mechanism. Gesunde Pflanz. 2023, 75, 49–59. [Google Scholar] [CrossRef]
- Dong, X.Y.; Wang, Q.; Huang, Q.D.; Ge, Q.L.; Zhao, K.J.; Wu, X.C.; Wu, X.; Lei, L.; Hao, G.F. PDDD-PreTrain: A Series of Commonly Used Pre-Trained Models Support Image-Based Plant Disease Diagnosis. Plant Phenomics 2023, 5, 0054. [Google Scholar] [CrossRef] [PubMed]
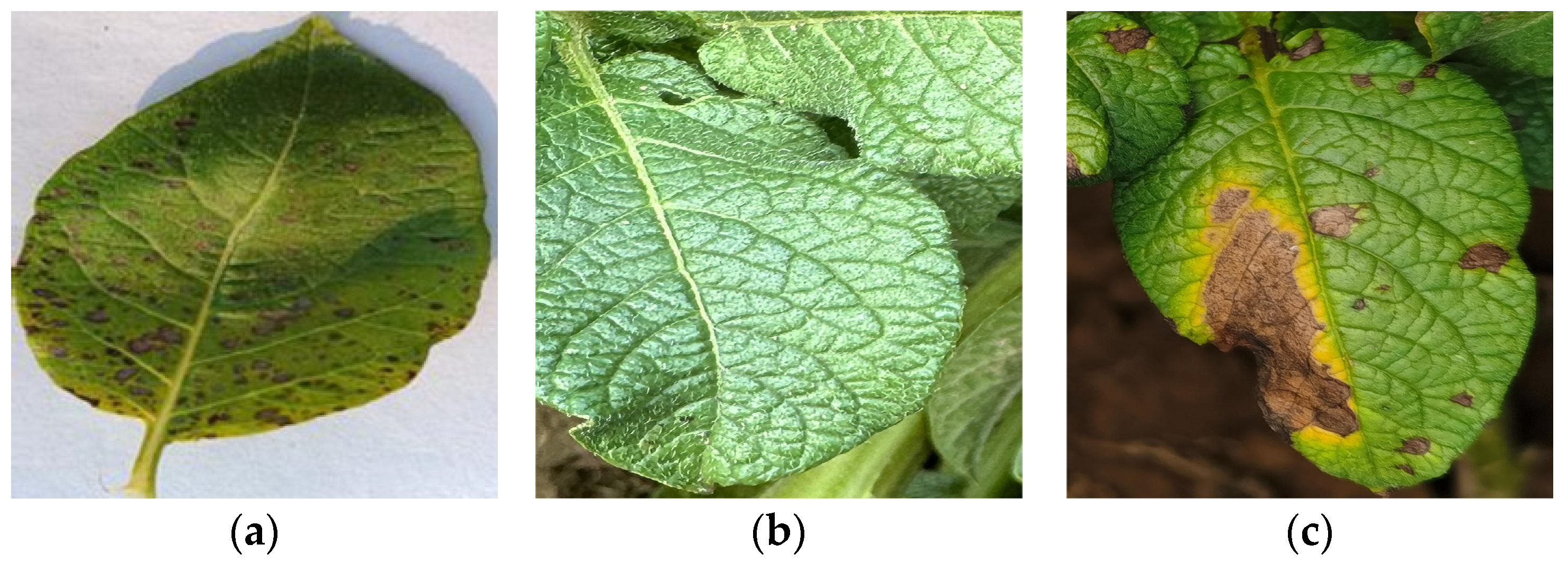




| Class Labels | Samples |
|---|---|
| Early_Blight | 1628 |
| Healthy | 1020 |
| Late_Blight | 1692 |
| CCMT Dataset | |||
|---|---|---|---|
| Class Labels | Samples | Class Labels | Samples |
| Cashew_anthracnose | 4940 | Maize_healthy | 1041 |
| Cashew_healthy | 7213 | Maize_leaf_blight | 5029 |
| Cashew_leaf_miner | 4953 | Maize_leaf_spot | 4285 |
| Cashew_red_rust | 6566 | Maize_streak_virus | 5047 |
| Cassava_bacterial_blight | 5864 | Tomato_healthy | 2500 |
| Cassava_brown_spot | 4733 | Tomato_leaf_blight | 6509 |
| Cassava_green_mite | 4266 | Tomato_leaf_curl | 2582 |
| Cassava_healthy | 3455 | Tomato_septoria_leaf_spot | 11,713 |
| Cassava_mosaic | 3450 | Tomato_verticulium_wilt | 3864 |
| Dataset | Added Modules | Accuracy | Recall | F1 |
|---|---|---|---|---|
| Our dataset | GRU | 98.81% | 98.16% | 98.17% |
| CBAM | 98.92% | 98.11% | 98.10% | |
| GRU + CBAM | 99.35% | 98.51% | 98.62% | |
| CCMT | GRU | 95.29% | 96.02% | 95.43% |
| CBAM | 95.17% | 95.81% | 95.1% | |
| GRU + CBAM | 95.61% | 96.20% | 95.52% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Yin, Y.; Li, Y.; Qu, T.; Guo, Z.; Peng, M.; Jia, S.; Wang, Q.; Zhang, W.; Li, F. Classification of Plant Leaf Disease Recognition Based on Self-Supervised Learning. Agronomy 2024, 14, 500. https://doi.org/10.3390/agronomy14030500
Wang Y, Yin Y, Li Y, Qu T, Guo Z, Peng M, Jia S, Wang Q, Zhang W, Li F. Classification of Plant Leaf Disease Recognition Based on Self-Supervised Learning. Agronomy. 2024; 14(3):500. https://doi.org/10.3390/agronomy14030500
Chicago/Turabian StyleWang, Yuzhi, Yunzhen Yin, Yaoyu Li, Tengteng Qu, Zhaodong Guo, Mingkang Peng, Shujie Jia, Qiang Wang, Wuping Zhang, and Fuzhong Li. 2024. "Classification of Plant Leaf Disease Recognition Based on Self-Supervised Learning" Agronomy 14, no. 3: 500. https://doi.org/10.3390/agronomy14030500
APA StyleWang, Y., Yin, Y., Li, Y., Qu, T., Guo, Z., Peng, M., Jia, S., Wang, Q., Zhang, W., & Li, F. (2024). Classification of Plant Leaf Disease Recognition Based on Self-Supervised Learning. Agronomy, 14(3), 500. https://doi.org/10.3390/agronomy14030500






