Automated Seedling Contour Determination and Segmentation Using Support Vector Machine and Image Features
Abstract
1. Introduction
- Combined the color and texture features with the SVM to improve boundary contour determination, with higher segmentation accuracy;
- Enhanced the segmentation performance under different lighting conditions;
- Enabled automated contour-based annotation for real-time monitoring models;
- Captured intricate contours to support the precise morphological analysis and monitoring of seedlings.
2. Materials and Methods
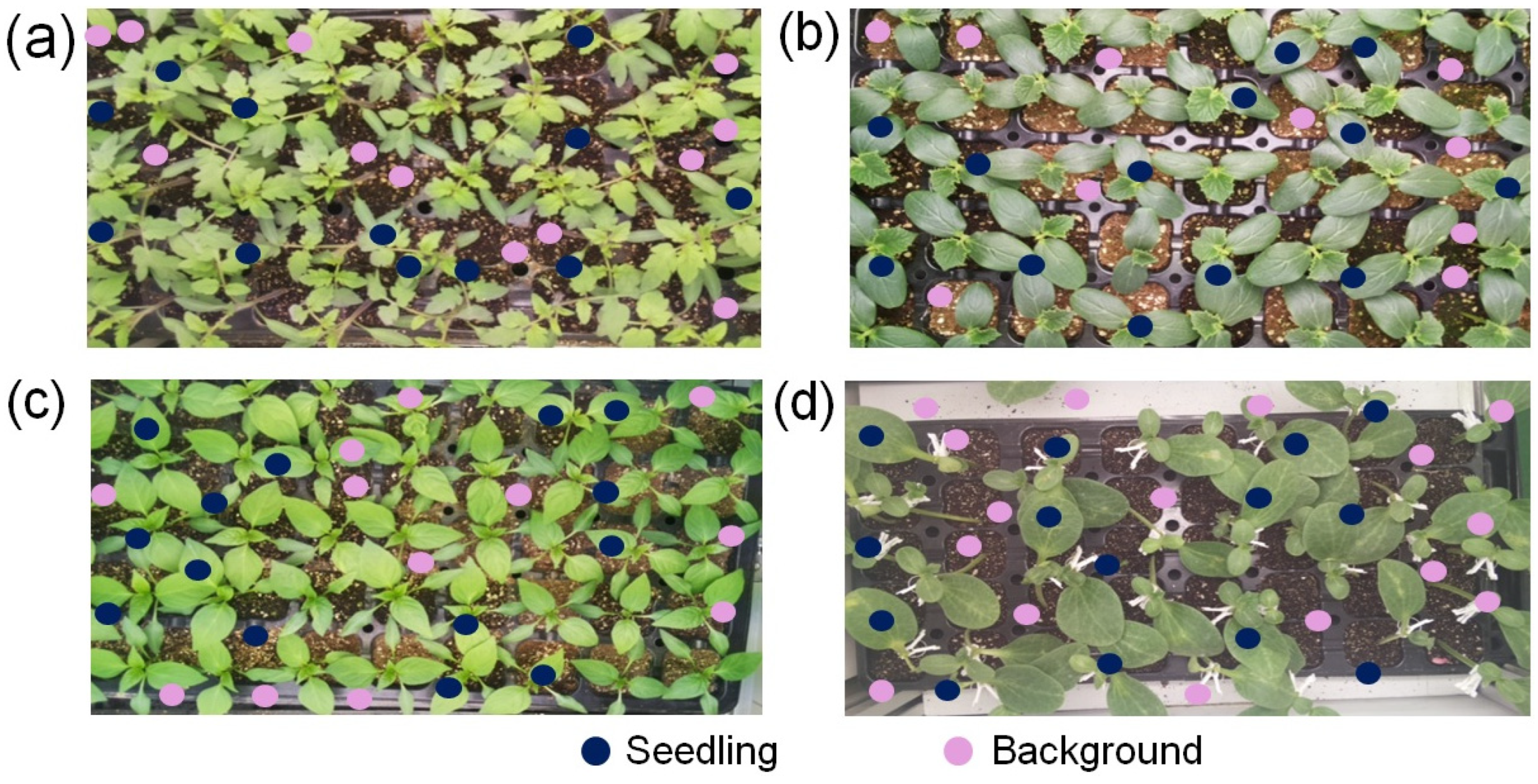
2.1. Image Acquisition Setup
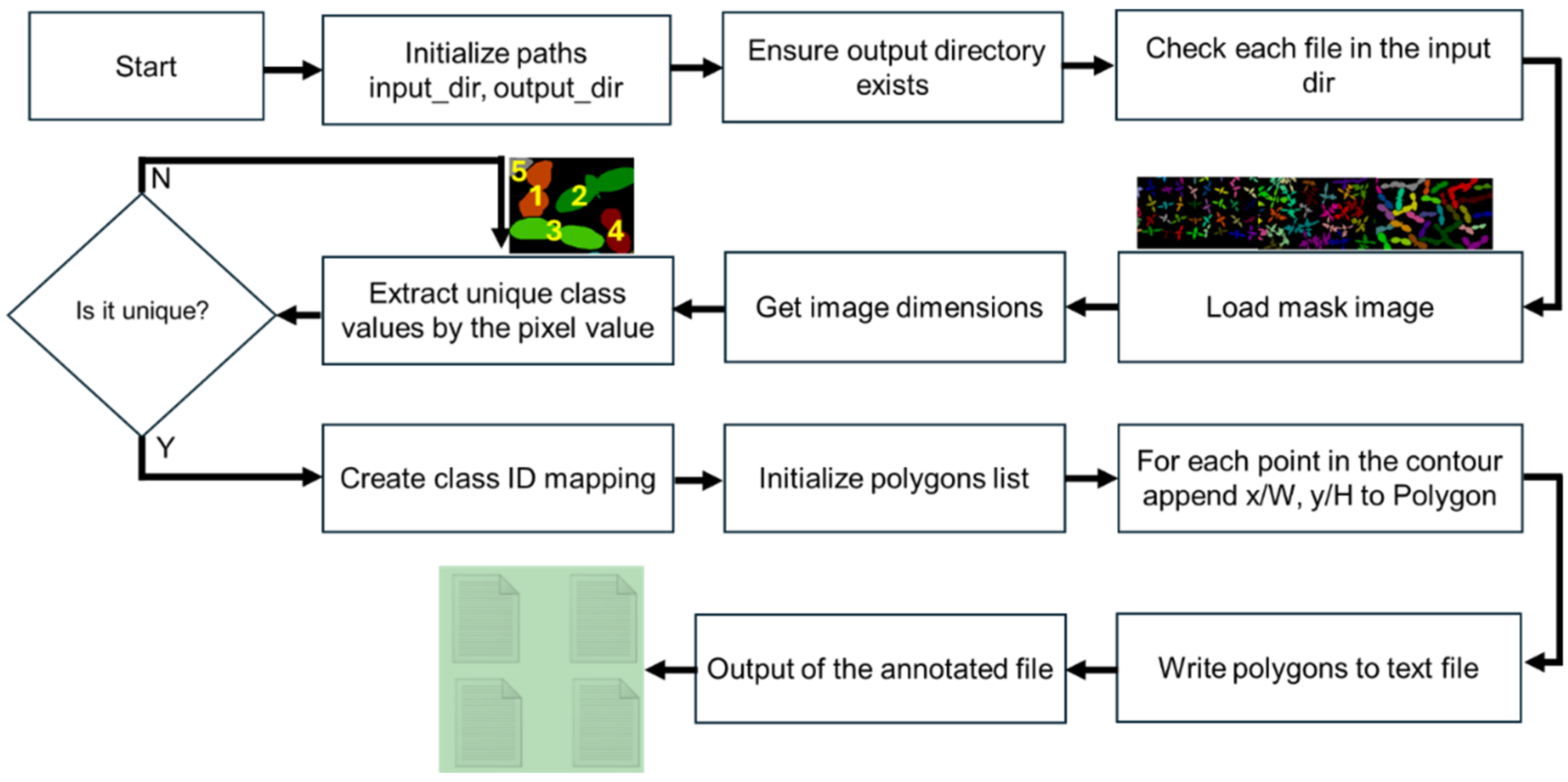
2.2. Dataset Preparation
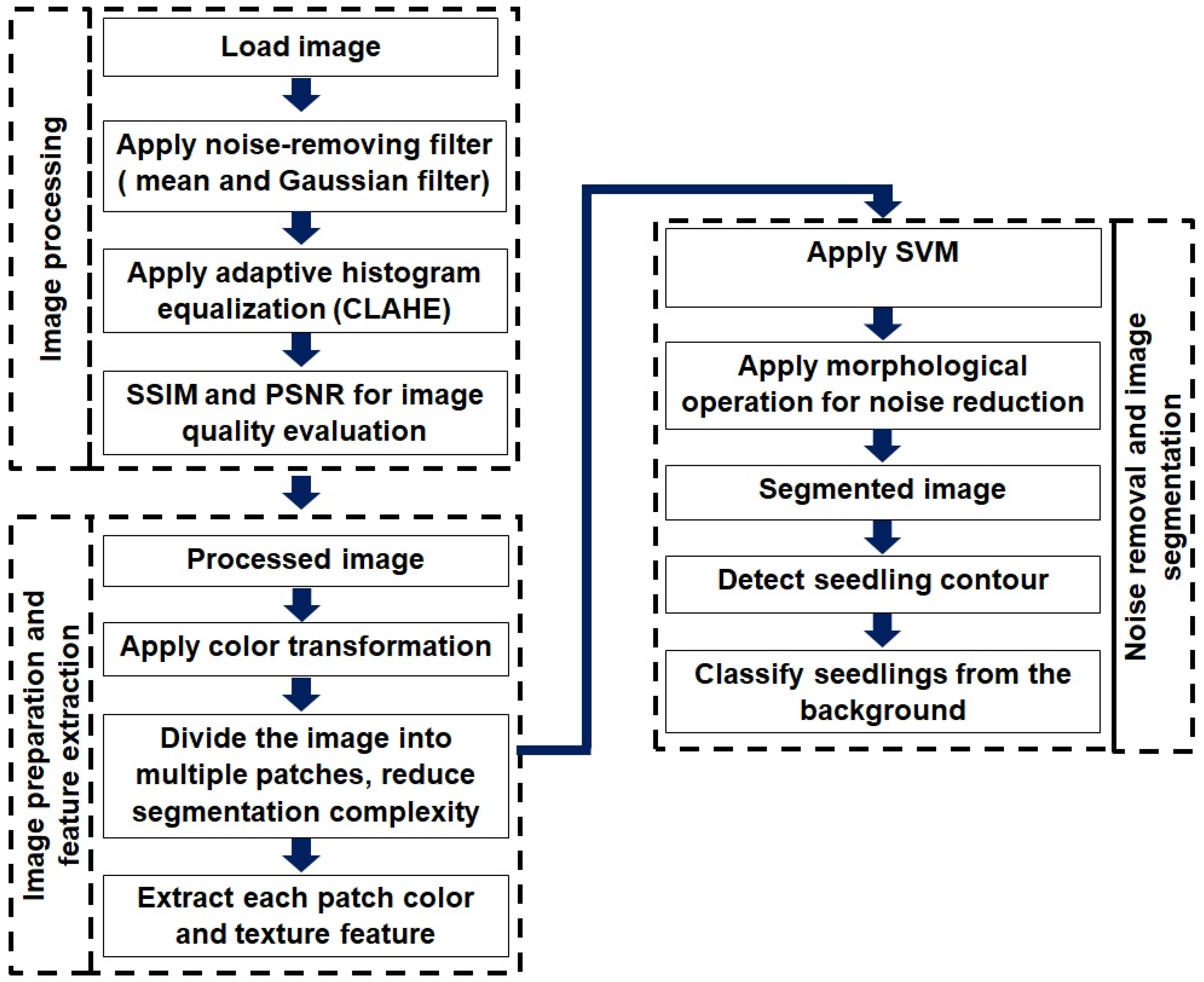
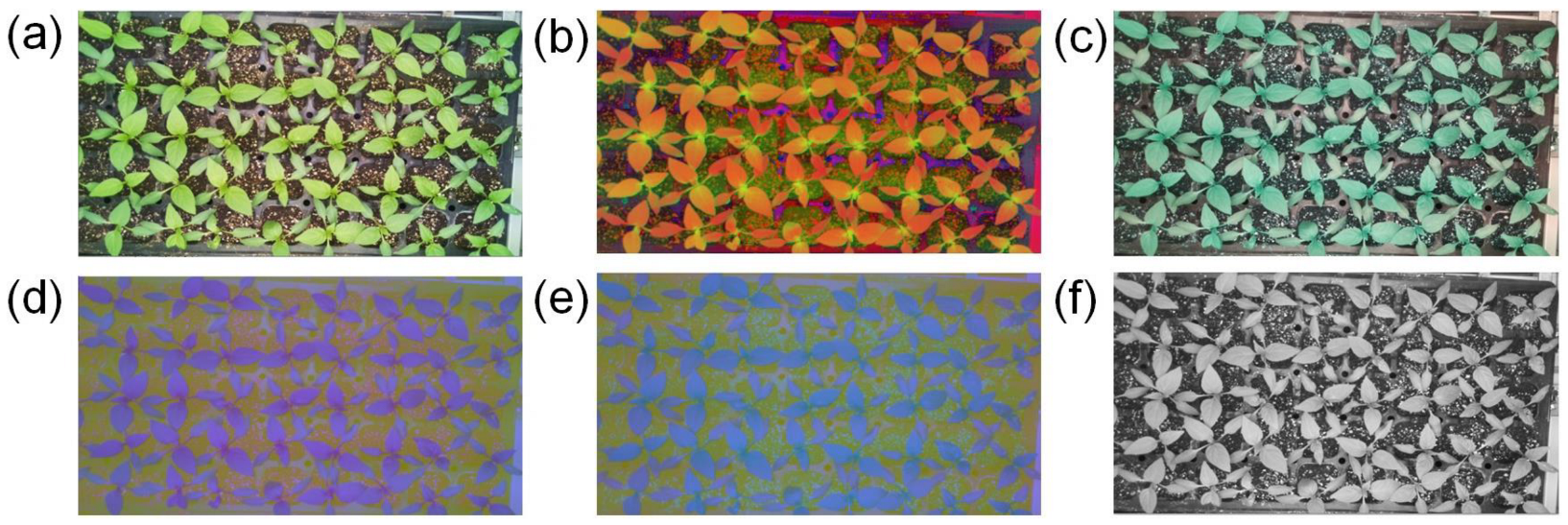
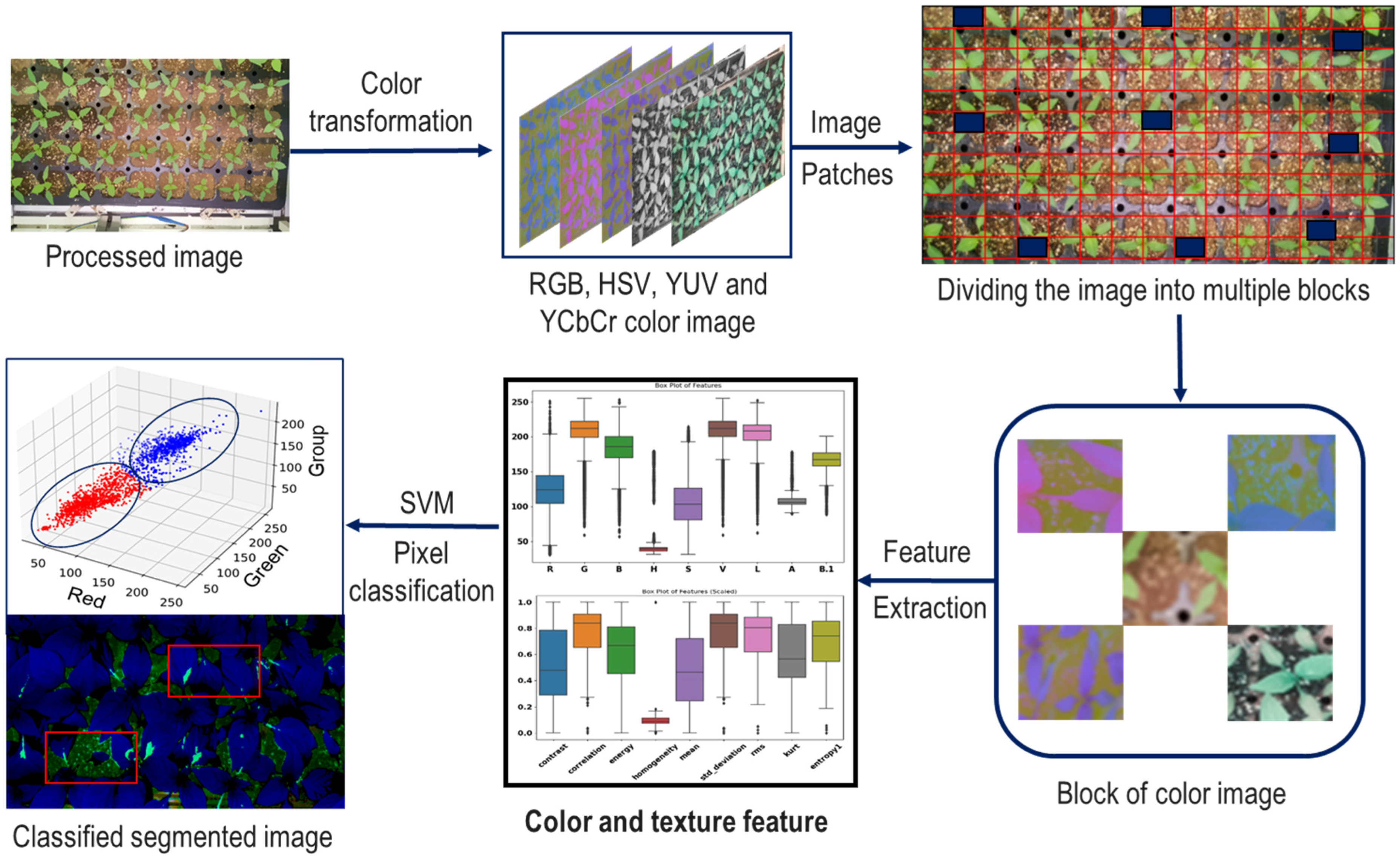
2.3. Image Processing Procedure
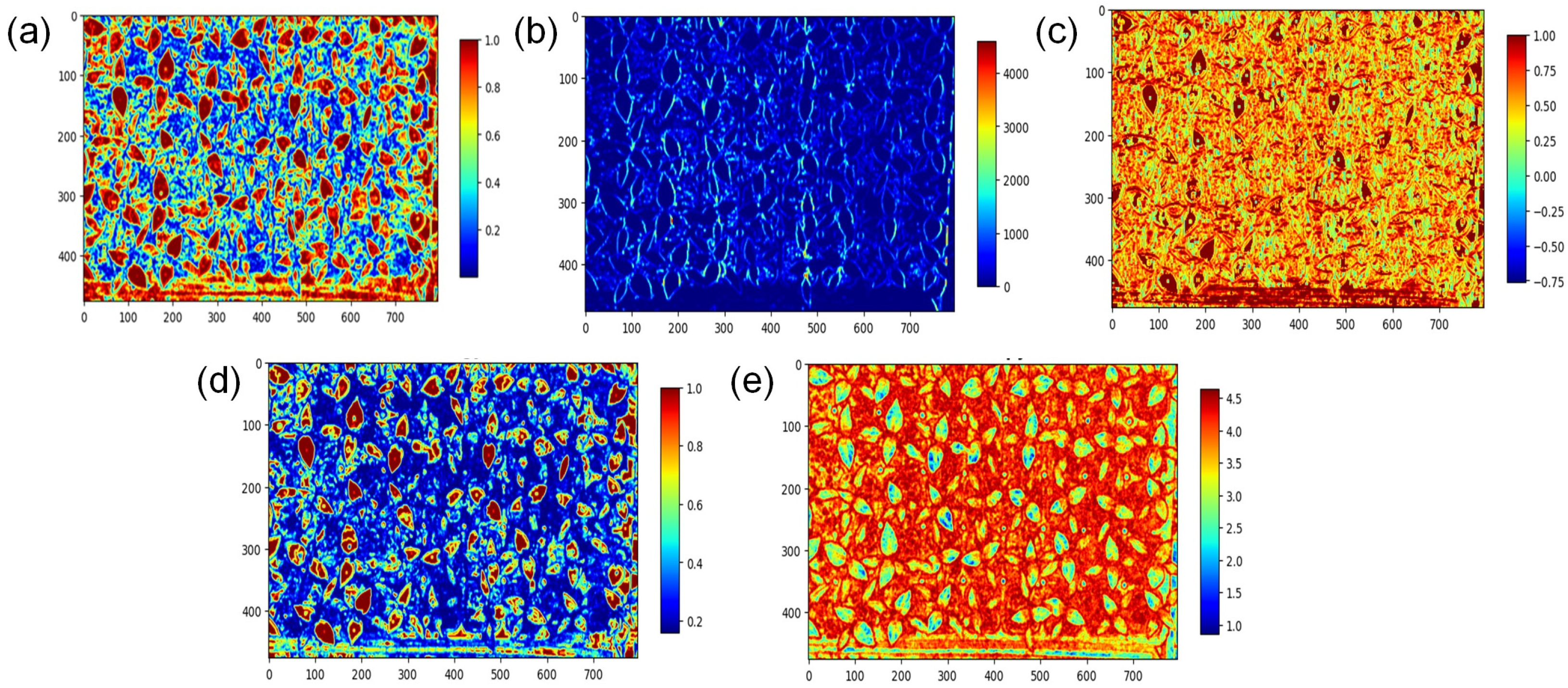
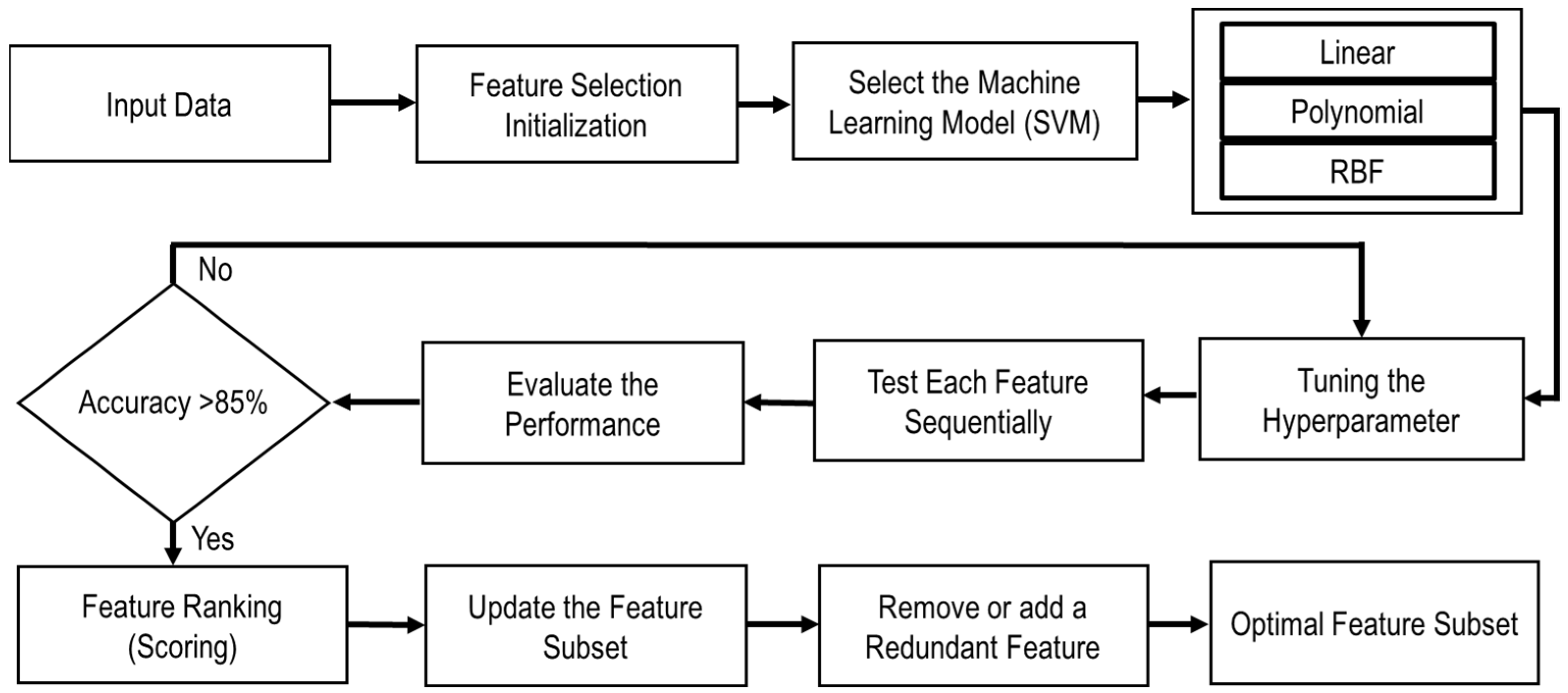
2.4. Feature Pattern and Feature Selection
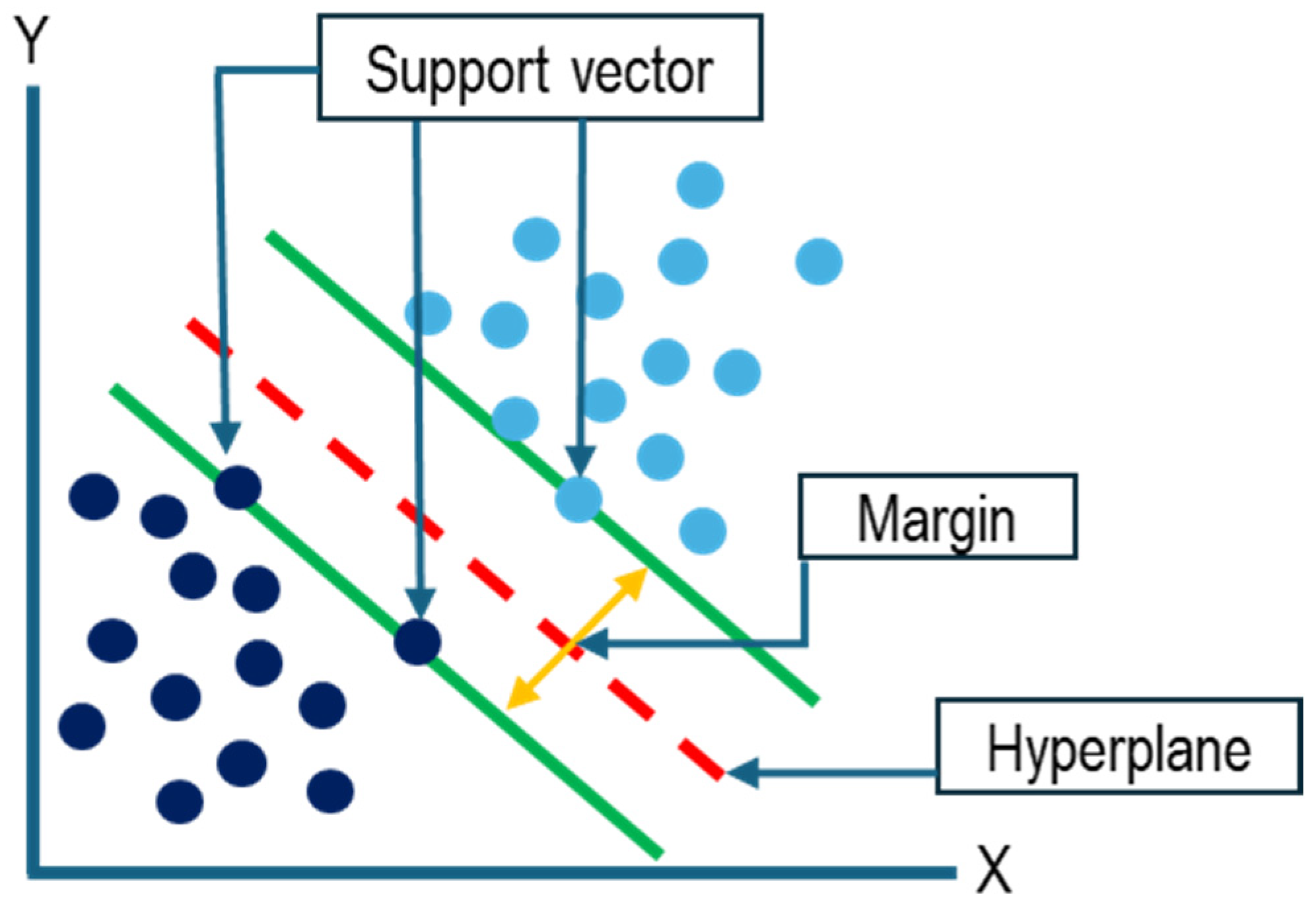
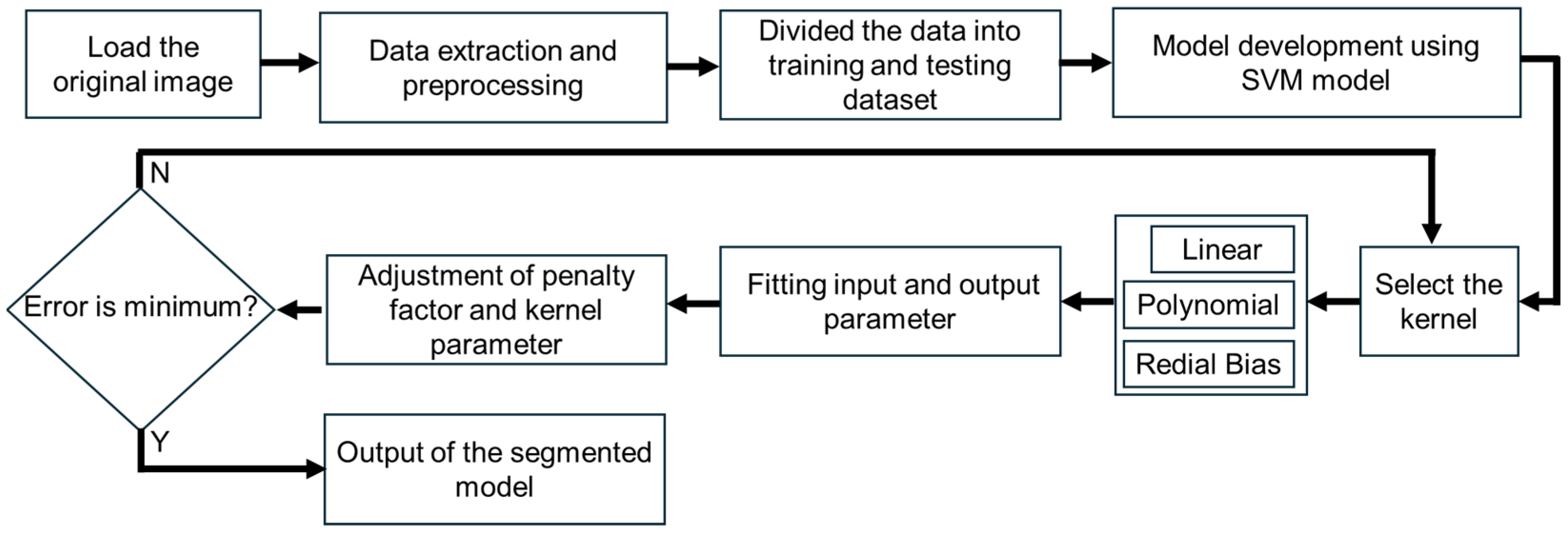
2.5. SVM Segmentation Model
2.6. Overall Image Segmentation Process
2.7. Performance Evaluation for Boundary Contour Determination
3. Results
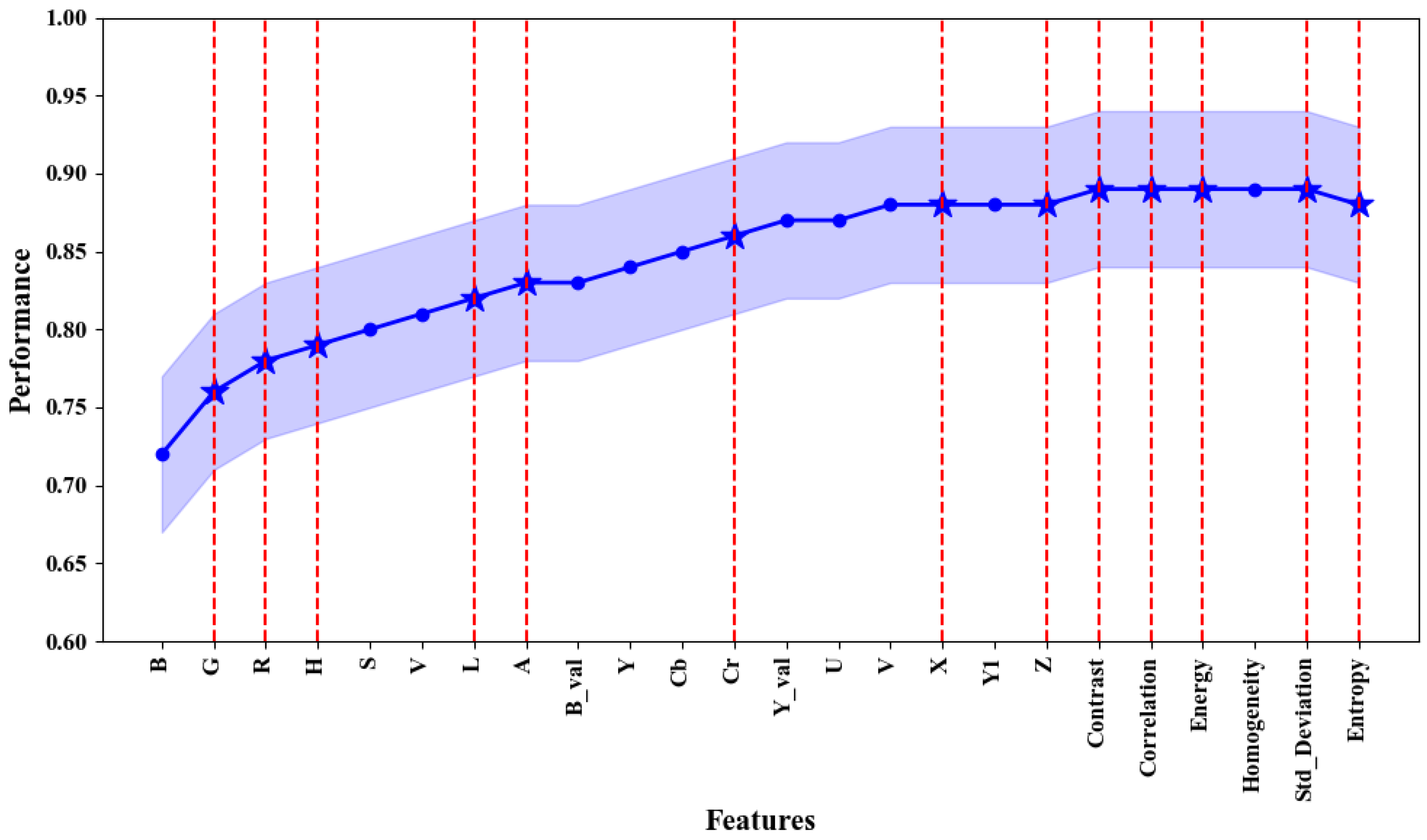
3.1. Selected Features
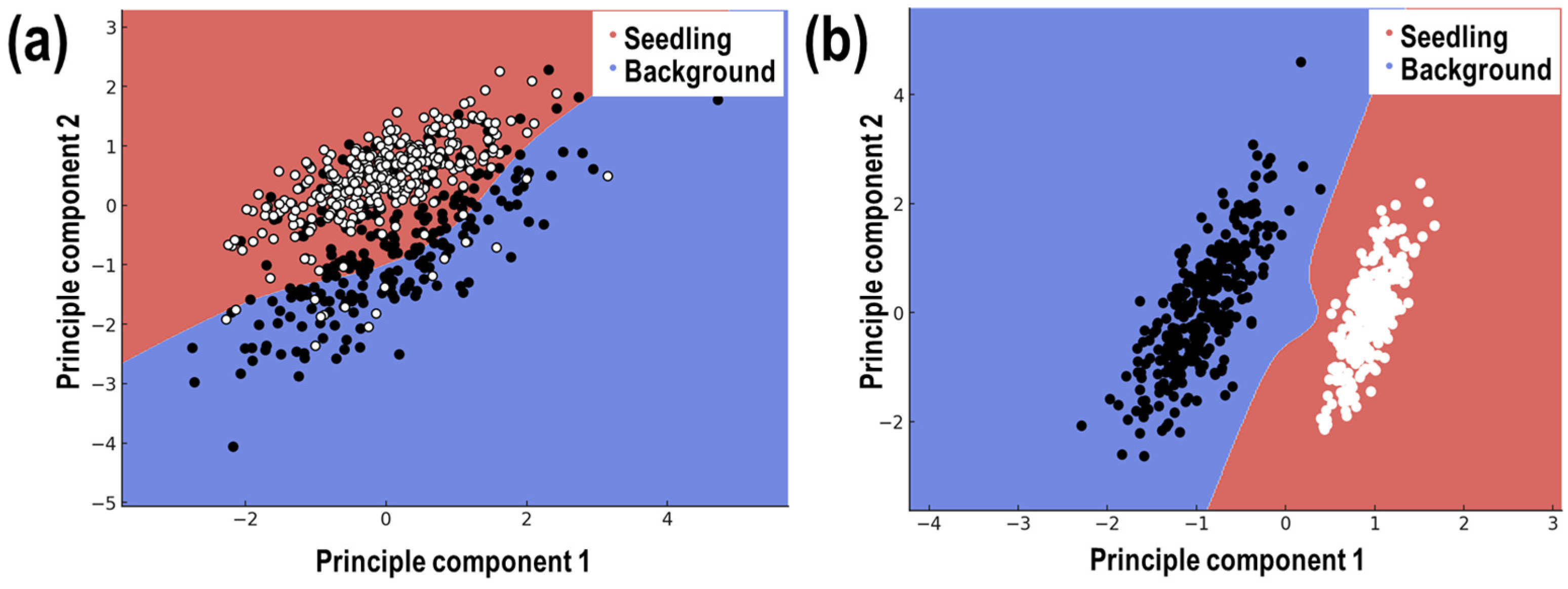
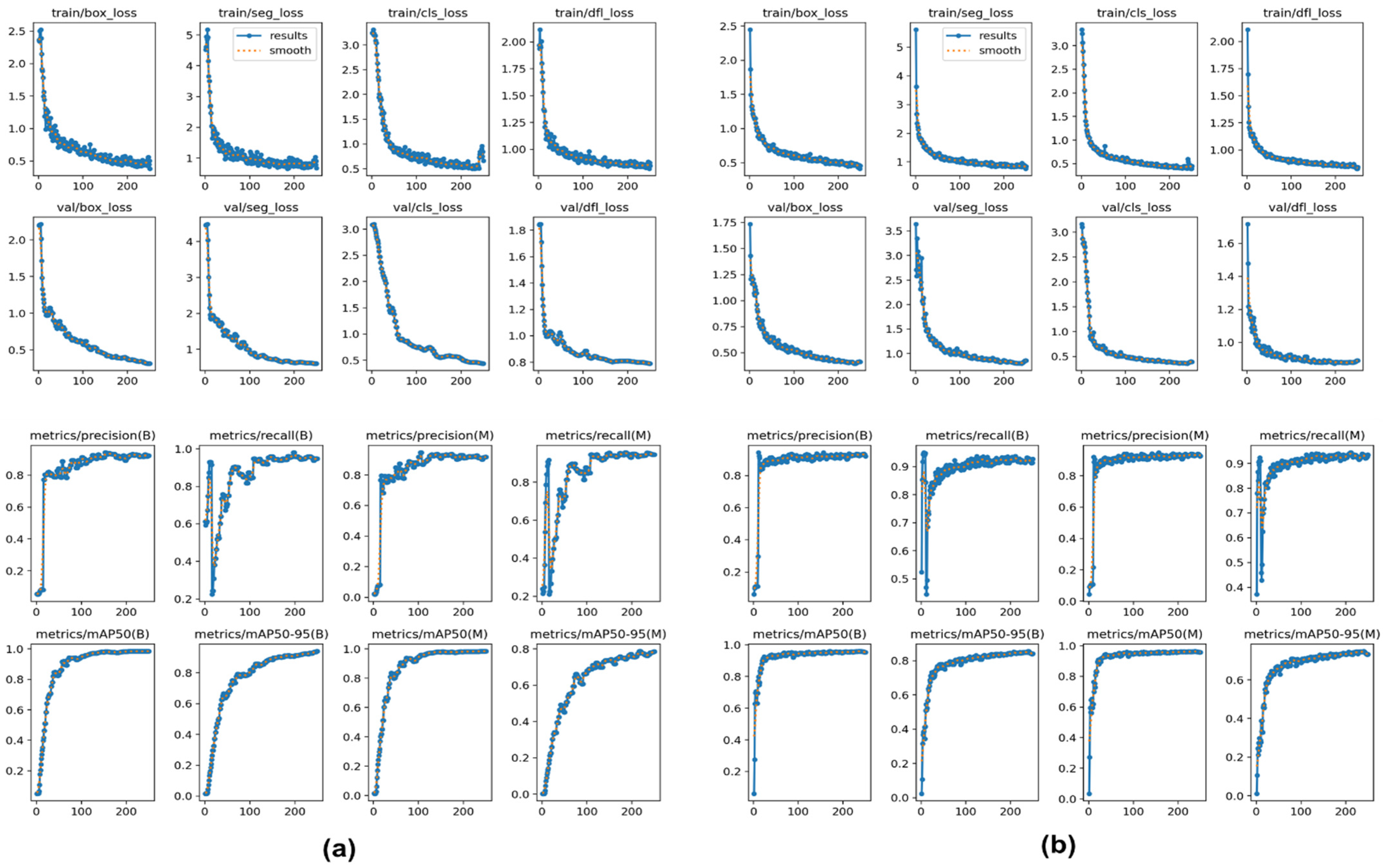
3.2. Performance of the SVM Segmentation Model
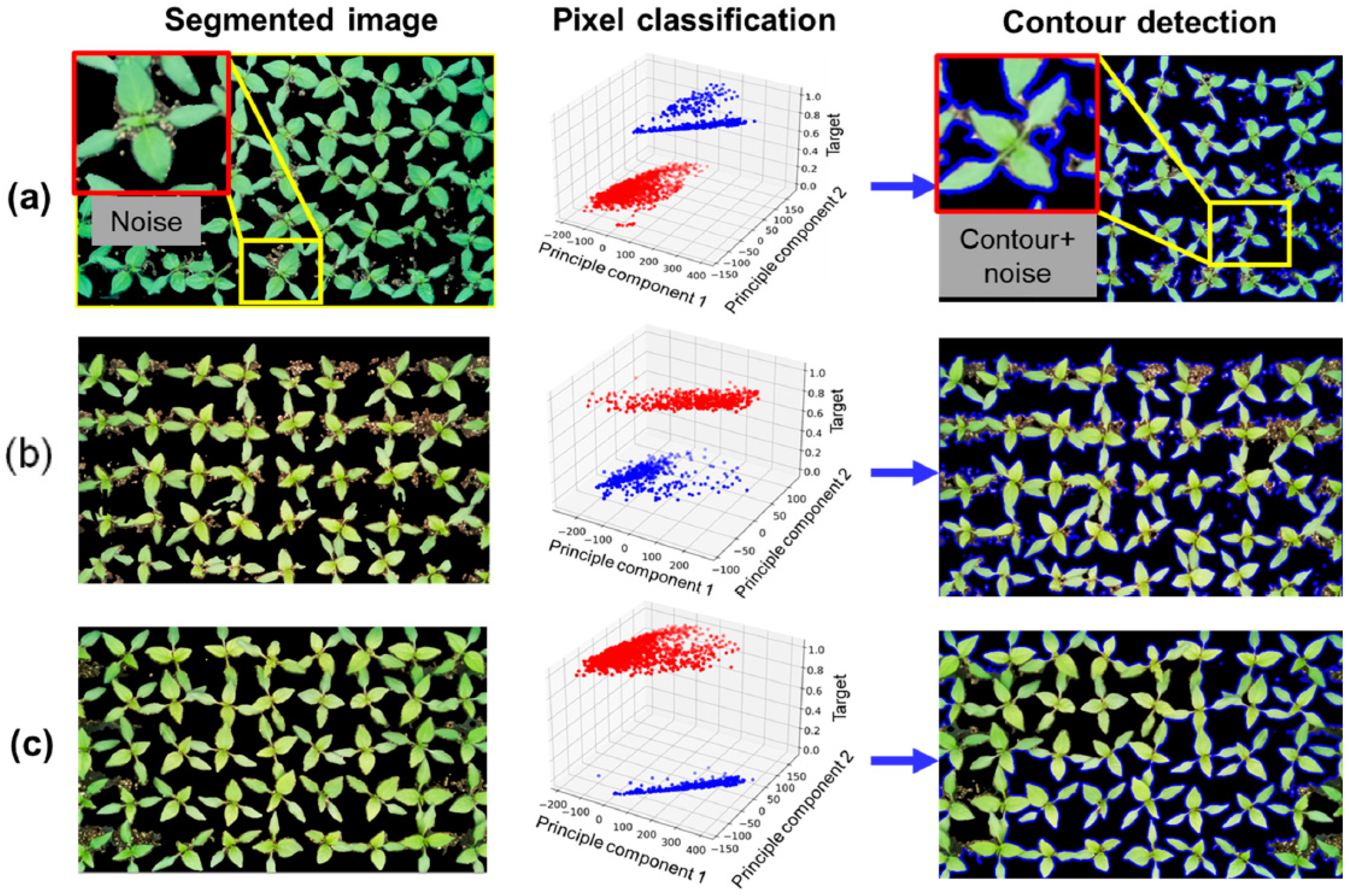
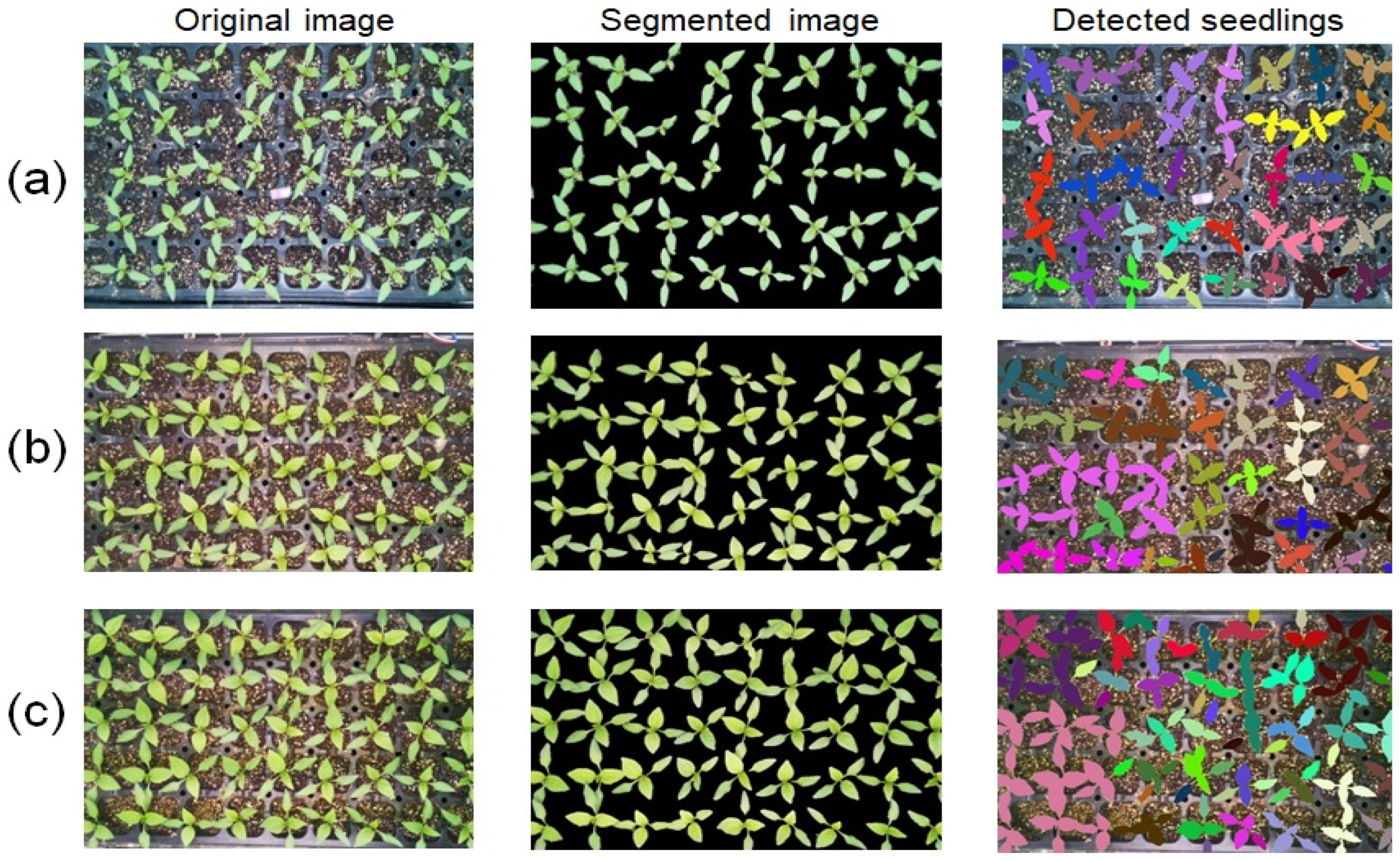
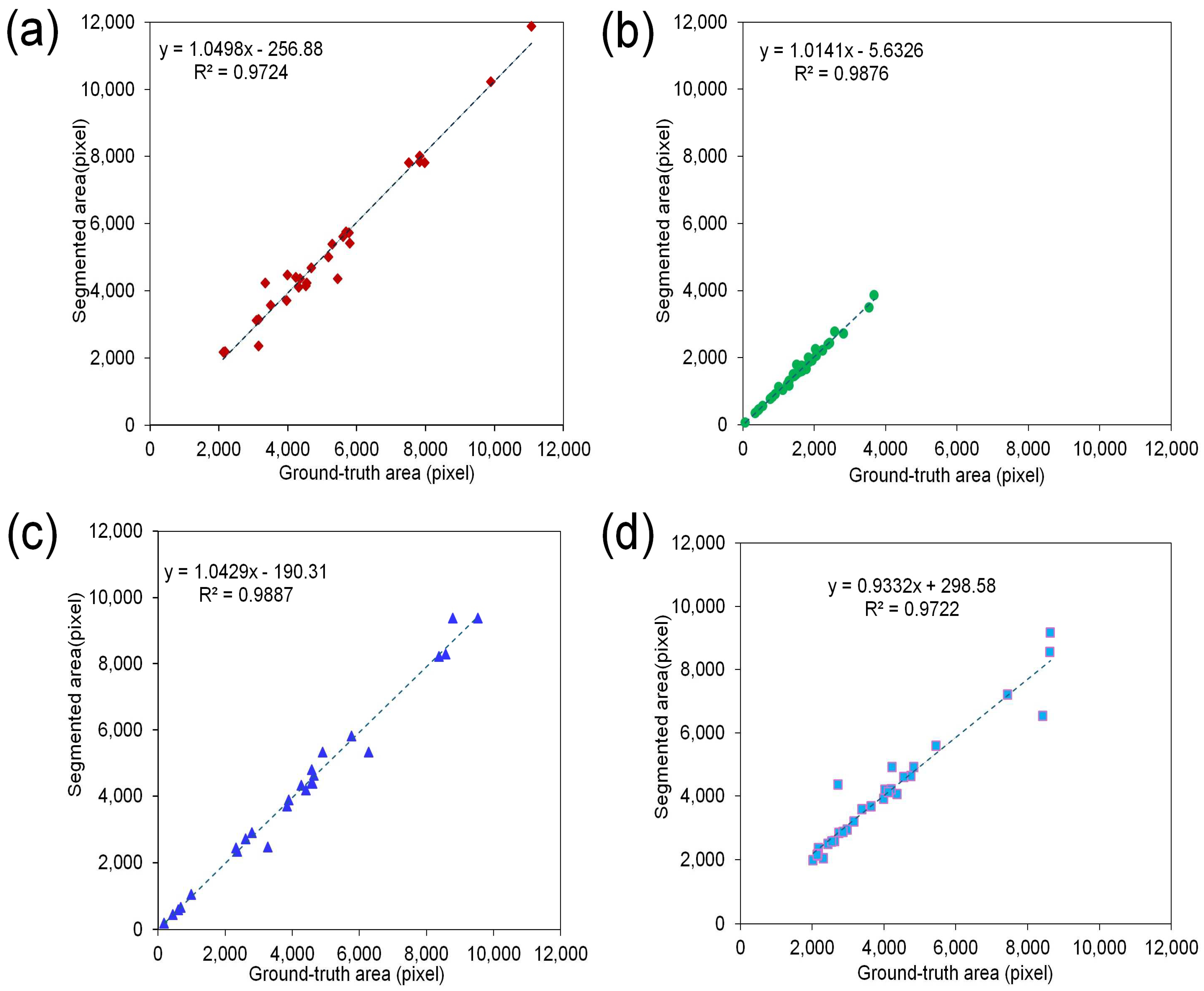
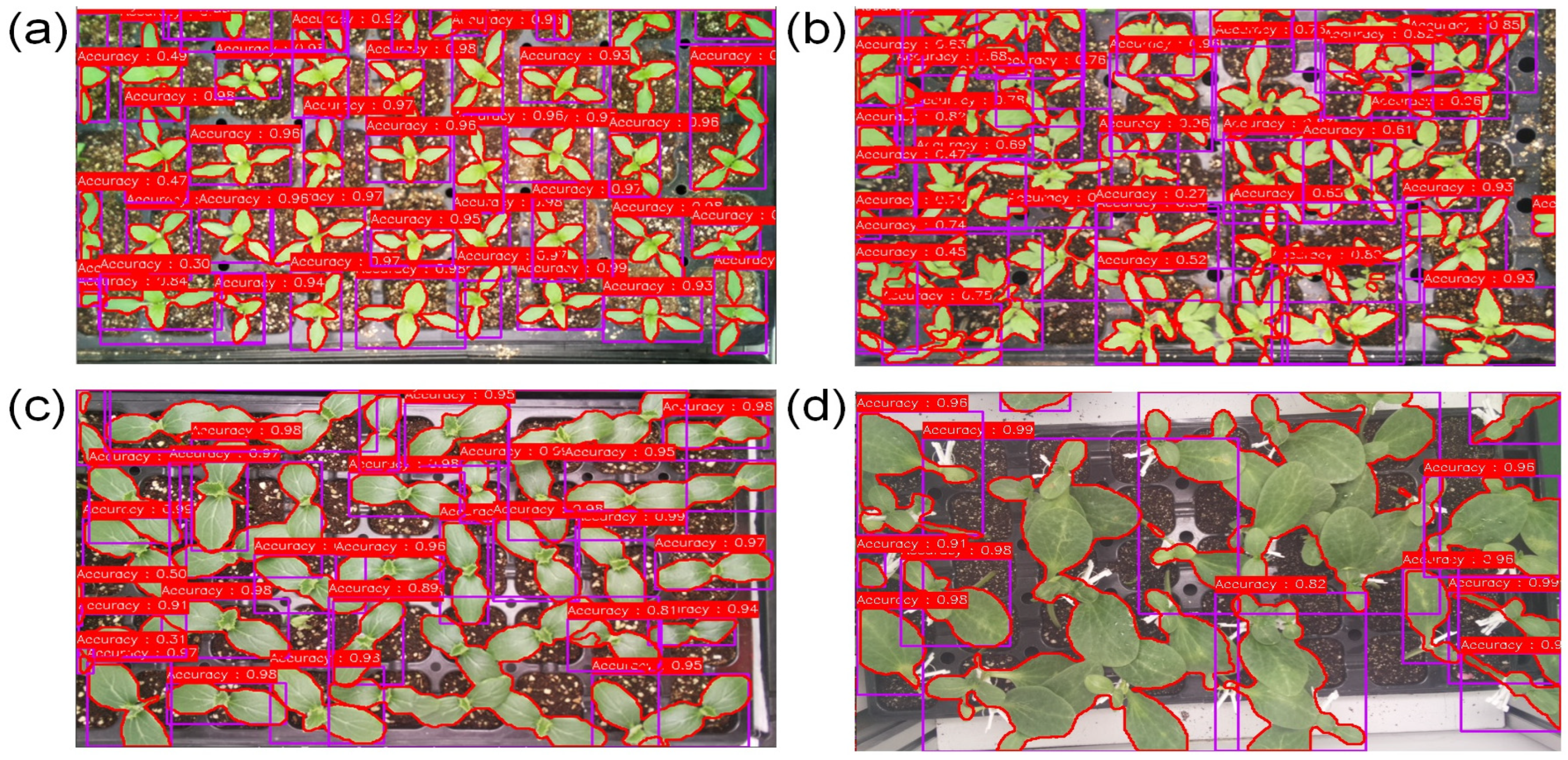
3.3. Segmentation Performance Evaluation
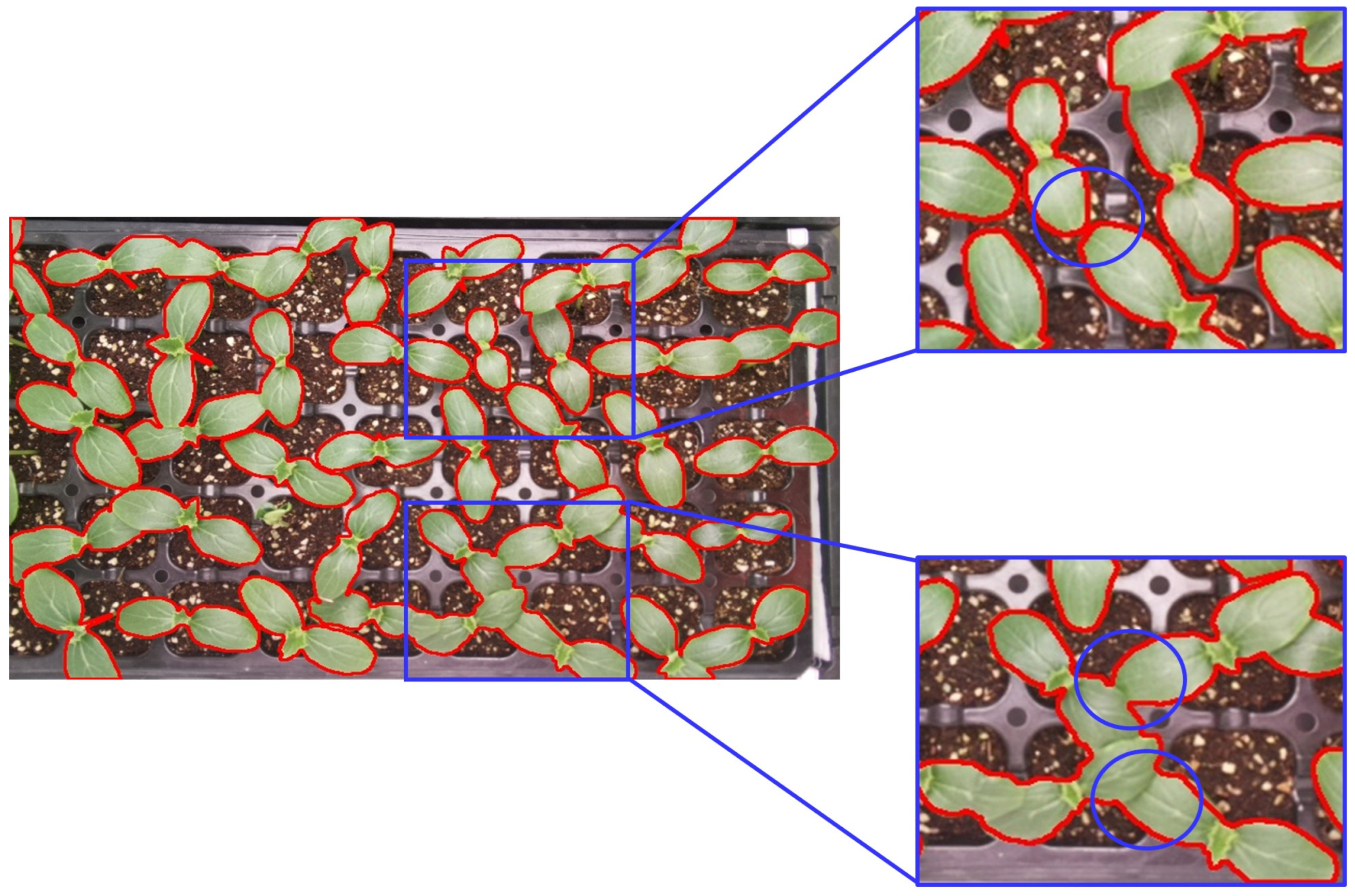
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Geng, T.; Yu, H.; Yuan, X.; Ma, R.; Li, P. Research on Segmentation Method of Maize Seedling Plant Instances Based on UAV Multispectral Remote Sensing Images. Plants 2024, 13, 1842. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Wang, T.; Liu, Y.; Qiao, X.; Li, Y. Computer Vision Technology in Agricultural Automation—A Review. Inf. Process. Agric. 2020, 7, 1–19. [Google Scholar] [CrossRef]
- Gupta, M.K.; Samuel, D.V.K.; Sirohi, N.P.S. Decision Support System for Greenhouse Seedling Production. Comput. Electron. Agric. 2010, 73, 133–145. [Google Scholar] [CrossRef]
- Ahmed, H.A.; Tong, Y.-X.; Yang, Q.C. Optimal Control of Environmental Conditions Affecting Lettuce Plant Growth in a Controlled Environment with Artificial Lighting: A Review. S. Afr. J. Bot. 2020, 130, 75–89. [Google Scholar] [CrossRef]
- Proietti, S.; Moscatello, S.; Riccio, F.; Downey, P.; Battistelli, A. Continuous Lighting Promotes Plant Growth, Light Conversion Efficiency, and Nutritional Quality of Eruca vesicaria (L.) Cav. in Controlled Environment with Minor Effects Due to Light Quality. Front. Plant Sci. 2021, 12, 730119. [Google Scholar] [CrossRef]
- Goto, E. Effects of Light Quality on Growth of Crop Plants under Artificial Lighting. Environ. Control Biol. 2003, 41, 121–132. [Google Scholar] [CrossRef]
- Islam, S.; Reza, M.N.; Ahmed, S.; Samsuzzaman; Cho, Y.J.; Noh, D.H.; Chung, S.O. Image Processing and Support Vector Machine (SVM) for Classifying Environmental Stress Symptoms of Pepper Seedlings Grown in a Plant Factory. Agronomy 2024, 14, 2043. [Google Scholar] [CrossRef]
- Blessing, E. Utilizing Deep Learning, Computer Vision, and Robotics for Crop Monitoring without Human Intervention: Showcasing Advancements like PATHoBot. February 2024. Available online: https://www.researchgate.net/publication/378498623 (accessed on 8 October 2024).
- Ruby, E.D.K.; Amirthayogam, G.; Sasi, G.; Chitra, T.; Choubey, A.; Gopalakrishnan, S. Advanced Image Processing Techniques for Automated Detection of Healthy and Infected Leaves in Agricultural Systems. Mesopotamian J. Comput. Sci. 2024, 2024, 62–70. [Google Scholar] [CrossRef]
- Lee, D.Y.; Na, D.Y.; Góngora-Canul, C.; Baireddy, S.; Lane, B.; Cruz, A.P.; Fernández-Campos, M.; Kleczewski, N.M.; Telenko, D.E.P.; Goodwin, S.B.; et al. Contour-Based Detection and Quantification of Tar Spot Stromata Using Red-Green-Blue (RGB) Imagery. Front. Plant Sci. 2021, 12, 675975. [Google Scholar] [CrossRef]
- Arbeláez, P.; Maire, M.; Fowlkes, C.; Malik, J. Contour Detection and Hierarchical Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2011, 33, 898–916. [Google Scholar] [CrossRef]
- Gwo, C.; Wei, C. Plant Identification through Images: Using Feature Extraction of Key Points on Leaf Contours. Appl. Plant Sci. 2013, 1, 1200005. [Google Scholar] [CrossRef] [PubMed]
- Desclaux, D.; Huynh, T.T.; Roumet, P. Identification of Soybean Plant Characteristics That Indicate the Timing of Drought Stress. Crop Sci. 2000, 40, 716–722. [Google Scholar] [CrossRef]
- Zhuang, M.; Chen, Z.; Wang, H.; Tang, H.; He, J.; Qin, B.; Yang, Y.; Jin, X.; Yu, M.; Jin, B.; et al. Efficient Contour-Based Annotation by Iterative Deep Learning for Organ Segmentation from Volumetric Medical Images. Int. J. Comput. Assist. Radiol. Surg. 2023, 18, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Zhang, B.; Deng, L.; Bozdar, B.; Li, J.; Chachar, S.; Chachar, Z.; Jahan, I.; Talpur, A.; Gishkori, M.S.; et al. Advancing Horizons in Vegetable Cultivation: A Journey from Age-Old Practices to High-Tech Greenhouse Cultivation—A Review. Front. Plant Sci. 2024, 15, 1357153. [Google Scholar] [CrossRef]
- Shamshiri, R.R.; Kalantari, F.; Ting, K.C.; Thorp, K.R.; Hameed, I.A.; Weltzien, C.; Ahmad, D.; Shad, Z. Advances in Greenhouse Automation and Controlled Environment Agriculture: A Transition to Plant Factories and Urban Agriculture. Int. J. Agric. Biol. Eng. 2018, 11, 1–22. [Google Scholar] [CrossRef]
- Chowdhury, M.; Reza, M.N.; Jin, H.; Islam, S.; Lee, G.J.; Chung, S.O. Defective Pennywort Leaf Detection Using Machine Vision and Mask R-CNN Model. Agronomy 2024, 14, 2313. [Google Scholar] [CrossRef]
- Wang, Q.; Du, W.; Ma, C.; Gu, Z. Gradient Color Leaf Image Segmentation Algorithm Based on Meanshift and Kmeans. In Proceedings of the 2021 IEEE 5th Advanced Information Technology, Electronic and Automation Control Conference (IAEAC), Chongqing, China, 12–14 March 2021; pp. 1609–1614. [Google Scholar] [CrossRef]
- Li, M.; Liao, J.J. Texture Image Segmentation Based on GLCM. Appl. Mech. Mater. 2012, 220–223, 1398–1401. [Google Scholar] [CrossRef]
- Samiei, S.; Rasti, P.; Vu, J.L.; Buitink, J.; Rousseau, D. Deep Learning-Based Detection of Seedling Development. Plant Methods 2020, 16, 103. [Google Scholar] [CrossRef]
- Hsu, R.C.; Chan, D.Y.; Liu, C.T.; Lai, W.C. Contour Extraction in Medical Images Using Initial Boundary Pixel Selection and Segmental Contour Following. Multidimens. Syst. Signal Process. 2012, 23, 469–498. [Google Scholar] [CrossRef]
- Yang, X.; Chen, A.; Zhou, G.; Wang, J.; Chen, W.; Gao, Y.; Jiang, R. Instance Segmentation and Classification Method for Plant Leaf Images Based on ISC-MRCNN and APS-DCCNN. IEEE Access 2020, 8, 151555–151573. [Google Scholar] [CrossRef]
- Oudah, M.; Al-Naji, A.; AL-Janabi, T.Y.; Namaa, D.S.; Chahl, J. Automatic Irrigation System Based on Computer Vision and an Artificial Intelligence Technique Using Raspberry Pi. Automation 2024, 5, 90–105. [Google Scholar] [CrossRef]
- Gai, J.; Tang, L.; Steward, B.L. Automated Crop Plant Detection Based on the Fusion of Color and Depth Images for Robotic Weed Control. J. Field Robot. 2020, 37, 35–52. [Google Scholar] [CrossRef]
- Sanou, I.W.; Baderot, J.; Bricq, S.; Benezeth, Y.; Marzani, F.; Martinez, S.; Foucher, J. Deep Learning Contour-Based Method for Semi-Automatic Annotation of Manufactured Objects in Electron Microscopy Images. J. Electron. Imaging 2024, 33, 031204. [Google Scholar] [CrossRef]
- Jasim, W.N.; Mohammed, R.J. A Survey on Segmentation Techniques for Image Processing. Iraqi J. Electr. Electron. Eng. 2021, 17, 73–93. [Google Scholar] [CrossRef]
- Kurugollu, F.; Sankur, B.; Harmanci, A.E. Color Image Segmentation Using Histogram Multithresholding and Fusion. Image Vis. Comput. 2001, 19, 915–928. [Google Scholar] [CrossRef]
- Muntarina, K.; Shorif, S.B.; Uddin, M.S. Notes on Edge Detection Approaches. Evol. Syst. 2022, 13, 169–182. [Google Scholar] [CrossRef]
- Xu, X.; Qiu, J.; Zhang, W.; Zhou, Z.; Kang, Y. Soybean Seedling Root Segmentation Using Improved U-Net Network. Sensors 2022, 22, 8904. [Google Scholar] [CrossRef]
- Sun, S.; Zhu, Y.; Liu, S.; Chen, Y.; Zhang, Y.; Li, S. An Integrated Method for Phenotypic Analysis of Wheat Based on Multi-View Image Sequences: From Seedling to Grain Filling Stages. Front. Plant Sci. 2024, 15, 1459968. [Google Scholar] [CrossRef]
- Wu, Y.; He, Y.; Wang, Y. Multi-Class Weed Recognition Using Hybrid CNN-SVM Classifier. Sensors 2023, 23, 7153. [Google Scholar] [CrossRef]
- Kumar, A.; Sachar, S. Deep Learning Techniques in Leaf Image Segmentation and Leaf Species Classification: A Survey. Wireless Pers. Commun. 2023, 133, 2379–2410. [Google Scholar] [CrossRef]
- Wang, S.; Li, C.; Wang, R.; Liu, Z.; Wang, M.; Tan, H.; Wu, Y.; Liu, X.; Sun, H.; Yang, R.; et al. Annotation-Efficient Deep Learning for Automatic Medical Image Segmentation. Nat. Commun. 2021, 12, 5915. [Google Scholar] [CrossRef] [PubMed]
- Thompson, N.; Greenewald, K.; Lee, K.; Manso, G.F. The Computational Limits of Deep Learning. arXiv 2021, arXiv:2007.05558. [Google Scholar]
- Kiss, A.; Moreau, T.; Mirabet, V.; Calugaru, C.I.; Boudaoud, A.; Das, P. Segmentation of 3D Images of Plant Tissues at Multiple Scales Using the Level Set Method. Plant Methods 2017, 13, 114. [Google Scholar] [CrossRef] [PubMed]
- Narisetti, N.; Henke, M.; Neumann, K.; Stolzenburg, F.; Altmann, T.; Gladilin, E. Deep Learning Based Greenhouse Image Segmentation and Shoot Phenotyping (DeepShoot). Front. Plant Sci. 2022, 13, 906410. [Google Scholar] [CrossRef]
- Yang, A.; Bai, Y.; Liu, H.; Jin, K.; Xue, T.; Ma, W. Application of SVM and Its Improved Model in Image Segmentation. Mob. Netw. Appl. 2022, 27, 851–861. [Google Scholar] [CrossRef]
- Attri, I.; Awasthi, L.K.; Sharma, T.P.; Rathee, P. A Review of Deep Learning Techniques Used in Agriculture. Ecol. Inform. 2023, 77, 102217. [Google Scholar] [CrossRef]
- Li, M.; Yu, X.; Ryu, K.H.; Lee, S.; Theera-Umpon, N. Face Recognition Technology Development with Gabor, PCA and SVM Methodology under Illumination Normalization Condition. Cluster Comput. 2018, 21, 1117–1126. [Google Scholar] [CrossRef]
- Islam, S.; Reza, M.N.; Ahmed, S.; Samsuzzaman; Cho, Y.J.; Noh, D.H.; Chung, S.O. Seedling Growth Stress Quantification Based on Environmental Factors Using Sensor Fusion and Image Processing. Horticulturae 2024, 10, 186. [Google Scholar] [CrossRef]
- Feng, L.; Raza, M.A.; Li, Z.; Chen, Y.; Khalid, M.H.B.; Du, J.; Liu, W.; Wu, X.; Song, C.; Yu, L.; et al. The Influence of Light Intensity and Leaf Movement on Photosynthesis Characteristics and Carbon Balance of Soybean. Front. Plant Sci. 2019, 9, 1952. [Google Scholar] [CrossRef]
- Javidan, S.M.; Banakar, A.; Rahnama, K.; Vakilian, K.A.; Ampatzidis, Y. Feature Engineering to Identify Plant Diseases Using Image Processing and Artificial Intelligence: A Comprehensive Review. Smart Agric. Technol. 2024, 8, 100480. [Google Scholar] [CrossRef]
- Yalman, Y. A Histogram Based Image Quality Index. Prz. Elektrotech. 2012, 88, 126–129. [Google Scholar]
- Guo, J.; Ma, J.; García-Fernández, Á.F.; Zhang, Y.; Liang, H. A Survey on Image Enhancement for Low-Light Images. Heliyon 2023, 9, e14558. [Google Scholar] [CrossRef] [PubMed]
- Pizer, S.M.; Amburn, E.P.; Austin, J.D.; Cromartie, R.; Geselowitz, A.; Greer, T.; Haar Romeny, B.t.; Zimmerman, J.B.; Zuiderveld, K. Adaptive Histogram Equalization and Its Variations. Comput. Vis. Graph. Image Process. 1987, 39, 355–368. [Google Scholar] [CrossRef]
- Dhal, K.G.; Das, A.; Ray, S.; Gálvez, J.; Das, S. Histogram Equalization Variants as Optimization Problems: A Review. Arch. Comput. Methods Eng. 2021, 28, 1471–1496. [Google Scholar] [CrossRef]
- Sepasian, M.; Balachandran, W.; Mares, C. Image Enhancement for Fingerprint Minutiae-Based Algorithms Using CLAHE, Standard Deviation Analysis and Sliding Neighborhood. Lect. Notes Eng. Comput. Sci. 2008, 2173, 1199–1203. Available online: https://www.researchgate.net/publication/44262481 (accessed on 5 October 2024).
- Azam, M.; Nouman, M. Evaluation of Image Support Resolution Deep Learning Technique Based on PSNR Value. KIET J. Comput. Inf. Sci. 2022, 6, 93–122. [Google Scholar] [CrossRef]
- Sridhar, S.; Kumar, P.R.; Ramanaiah, K.V. Wavelet Transform Techniques for Image Compression—An Evaluation. Int. J. Image Graph. Signal Process. 2014, 6, 54–67. [Google Scholar] [CrossRef]
- Sara, U.; Akter, M.; Uddin, M.S. Image Quality Assessment through FSIM, SSIM, MSE and PSNR—A Comparative Study. J. Comput. Commun. 2019, 7, 8–18. [Google Scholar] [CrossRef]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image Quality Assessment: From Error Visibility to Structural Similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef]
- Yuan, W.; Wijewardane, N.K.; Jenkins, S.; Bai, G.; Ge, Y.; Graef, G.L. Early Prediction of Soybean Traits through Color and Texture Features of Canopy RGB Imagery. Sci. Rep. 2019, 9, 14089. [Google Scholar] [CrossRef]
- Yue, J.; Li, Z.; Liu, L.; Fu, Z. Content-Based Image Retrieval Using Color and Texture Fused Features. Math. Comput. Model. 2011, 54, 1121–1127. [Google Scholar] [CrossRef]
- Shrivastava, V.K.; Pradhan, M.K. Rice Plant Disease Classification Using Color Features: A Machine Learning Paradigm. J. Plant Pathol. 2021, 103, 17–26. [Google Scholar] [CrossRef]
- Hu, X.; Ensor, A. Fourier Spectrum Image Texture Analysis. In Proceedings of the 2018 International Conference on Image and Vision Computing New Zealand (IVCNZ), Auckland, New Zealand, 19–21 November 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Szczypiński, P.M.; Strzelecki, M.; Materka, A.; Klepaczko, A. MaZda-A Software Package for Image Texture Analysis. Comput. Methods Programs Biomed. 2009, 94, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Zubair, A.R.; Alo, O.A. Grey Level Co-Occurrence Matrix (GLCM) Based Second Order Statistics for Image Texture Analysis. Int. J. Sci. Eng. Investig. 2019, 8, 93. Available online: www.IJSEI.com (accessed on 3 September 2024).
- Yao, Q.; Guan, Z.; Zhou, Y.; Tang, J.; Hu, Y.; Yang, B. Application of Support Vector Machine for Detecting Rice Diseases Using Shape and Color Texture Features. In Proceedings of the 2009 International Conference on Engineering Computation, Hong Kong, China, 2–3 May 2009; pp. 79–83. [Google Scholar] [CrossRef]
- Nadafzadeh, M.; Abdanan Mehdizadeh, S. Design and Fabrication of an Intelligent Control System for Determination of Watering Time for Turfgrass Plant Using Computer Vision System and Artificial Neural Network. Precis. Agric. 2019, 20, 857–879. [Google Scholar] [CrossRef]
- Cai, J.; Luo, J.; Wang, S.; Yang, S. Feature Selection in Machine Learning: A New Perspective. Neurocomputing 2018, 300, 70–79. [Google Scholar] [CrossRef]
- Al-Tashi, Q.; Abdulkadir, S.J.; Rais, H.M.; Mirjalili, S.; Alhussian, H. Approaches to Multi-Objective Feature Selection: A Systematic Literature Review. IEEE Access 2020, 8, 125076–125096. [Google Scholar] [CrossRef]
- Aggrawal, R.; Pal, S. Sequential Feature Selection and Machine Learning Algorithm-Based Patient’s Death Events Prediction and Diagnosis in Heart Disease. SN Comput. Sci. 2020, 1, 344. [Google Scholar] [CrossRef]
- Rückstieß, T.; Osendorfer, C.; Van Der Smagt, P. Sequential Feature Selection for Classification; Springer: Berlin/Heidelberg, Germany, 2011; pp. 132–141. [Google Scholar] [CrossRef]
- Dubey, S.R.; Dixit, P.; Singh, N.; Gupta, J.P. Infected Fruit Part Detection Using K-Means Clustering Segmentation Technique. Int. J. Interact. Multimed. Artif. Intell. 2013, 2, 65. [Google Scholar] [CrossRef][Green Version]
- Soliman, O.S.; Mahmoud, A.S. A Classification System for Remote Sensing Satellite Images Using Support Vector Machine with Non-Linear Kernel Functions. In Proceedings of the 2012 8th International Conference on Informatics and Systems (INFOS), Giza, Egypt, 14–16 May 2012. [Google Scholar]
- Hussain, M.; Wajid, S.K.; Elzaart, A.; Berbar, M. A Comparison of SVM Kernel Functions for Breast Cancer Detection. In Proceedings of the 2011 Eighth International Conference Computer Graphics, Imaging and Visualization, Singapore, 17–19 August 2011; pp. 145–150. [Google Scholar] [CrossRef]
- Wang, Z. Cell Segmentation for Image Cytometry: Advances, Insufficiencies, and Challenges. Cytom. Part A 2019, 95, 708–711. [Google Scholar] [CrossRef]
- Vădineanu, Ş.; Pelt, D.M.; Dzyubachyk, O.; Batenburg, K.J. Reducing Manual Annotation Costs for Cell Segmentation by Upgrading Low-Quality Annotations; Springer: Cham, Switzerland, 2023; pp. 3–13. [Google Scholar] [CrossRef]
- Lu, Y.; Zheng, K.; Li, W.; Wang, Y.; Harrison, A.P.; Lin, C.; Wang, S.; Xiao, J.; Lu, L.; Kuo, C.F.; et al. Contour Transformer Network for One-Shot Segmentation of Anatomical Structures. IEEE Trans. Med. Imaging 2021, 40, 2672–2684. [Google Scholar] [CrossRef] [PubMed]
- Tatsumi, K.; Tanino, T. Support Vector Machines Maximizing Geometric Margins for Multi-Class Classification. Top 2014, 22, 815–840. [Google Scholar] [CrossRef]
- Montesinos López, O.A.; Montesinos López, A.; Crossa, J. Overfitting, Model Tuning, and Evaluation of Prediction Performance; Springer: Cham, Switzerland, 2022. [Google Scholar] [CrossRef]
- Santos, M.S.; Soares, J.P.; Abreu, P.H.; Araujo, H.; Santos, J. Cross-Validation for Imbalanced Datasets: Avoiding Overoptimistic and Overfitting Approaches. IEEE Comput. Intell. Mag. 2018, 13, 59–76. [Google Scholar] [CrossRef]
- Hashemzadeh, K.; Hashemzadeh, S. Maximum Relative Margin and Data-Dependent Regularization Pannagadatta. Minerva Chir. 2012, 67, 327–335. [Google Scholar]
- Xue, H.; Chen, S.; Yang, Q. Discriminatively Regularized Least-Squares Classification. Pattern Recognit. 2009, 42, 93–104. [Google Scholar] [CrossRef]
- Wang, C.; Deng, C.; Yu, Z.; Hui, D.; Gong, X.; Luo, R. Adaptive Ensemble of Classifiers with Regularization for Imbalanced Data Classification. Inf. Fusion 2021, 69, 81–102. [Google Scholar] [CrossRef]
- Guo, Z.; Hu, X.; Zhao, B.; Wang, H.; Ma, X. StrawSnake: A Real-Time Strawberry Instance Segmentation Network Based on the Contour Learning Approach. Electronics 2024, 13, 3103. [Google Scholar] [CrossRef]
- Gao, L.; Lin, X. A Method for Accurately Segmenting Images of Medicinal Plant Leaves with Complex Backgrounds. Comput. Electron. Agric. 2018, 155, 426–445. [Google Scholar] [CrossRef]
- Sadeghi-Tehran, P.; Virlet, N.; Sabermanesh, K.; Hawkesford, M.J. Multi-Feature Machine Learning Model for Automatic Segmentation of Green Fractional Vegetation Cover for High-Throughput Field Phenotyping. Plant Methods 2017, 13, 103. [Google Scholar] [CrossRef]
- Ghosh, S.; Singh, A.; Kavita; Jhanjhi, N.Z.; Masud, M.; Aljahdali, S. SVM and KNN Based CNN Architectures for Plant Classification. Comput. Mater. Contin. 2022, 71, 4257–4274. [Google Scholar] [CrossRef]
- Hossain, E.; Hossain, M.F.; Rahaman, M.A. A Color and Texture-Based Approach for the Detection and Classification of Plant Leaf Disease Using KNN Classifier. In Proceedings of the 2019 International Conference on Electrical, Computer and Communication Engineering (ECCE), Cox’s Bazar, Bangladesh, 7–9 February 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, H.; Huang, W.; You, Z. Plant Diseased Leaf Segmentation and Recognition by Fusion of Superpixel, K-Means, and PHOG. Optik 2018, 157, 866–872. [Google Scholar] [CrossRef]
- Wang, W.; Chen, Z.; Yuan, X.; Wu, X. Adaptive Image Enhancement Method for Correcting Low-Illumination Images. Inf. Sci. 2019, 496, 25–41. [Google Scholar] [CrossRef]
- Ijaz, E.D.U.; Ijaz, E.A.; Ali Iqbal, D.F.G.; Hayat, M. Quantitative Analysis of Image Enhancement Algorithms for Diverse Applications. Int. J. Innov. Sci. Technol. 2023, 5, 694–707. [Google Scholar]
- Juneja, M.; Saini, S.K.; Gupta, J.; Garg, P.; Thakur, N.; Sharma, A.; Mehta, M.; Jindal, P. Survey of Denoising, Segmentation and Classification of Magnetic Resonance Imaging for Prostate Cancer. Multimed. Tools Appl. 2021, 80, 29199–29249. [Google Scholar] [CrossRef]
- Büyükarıkan, B.; Ülker, E. Convolutional Neural Network-Based Apple Images Classification and Image Quality Measurement by Light Colors Using the Color-Balancing Approach. Multimed. Syst. 2023, 29, 1651–1661. [Google Scholar] [CrossRef]
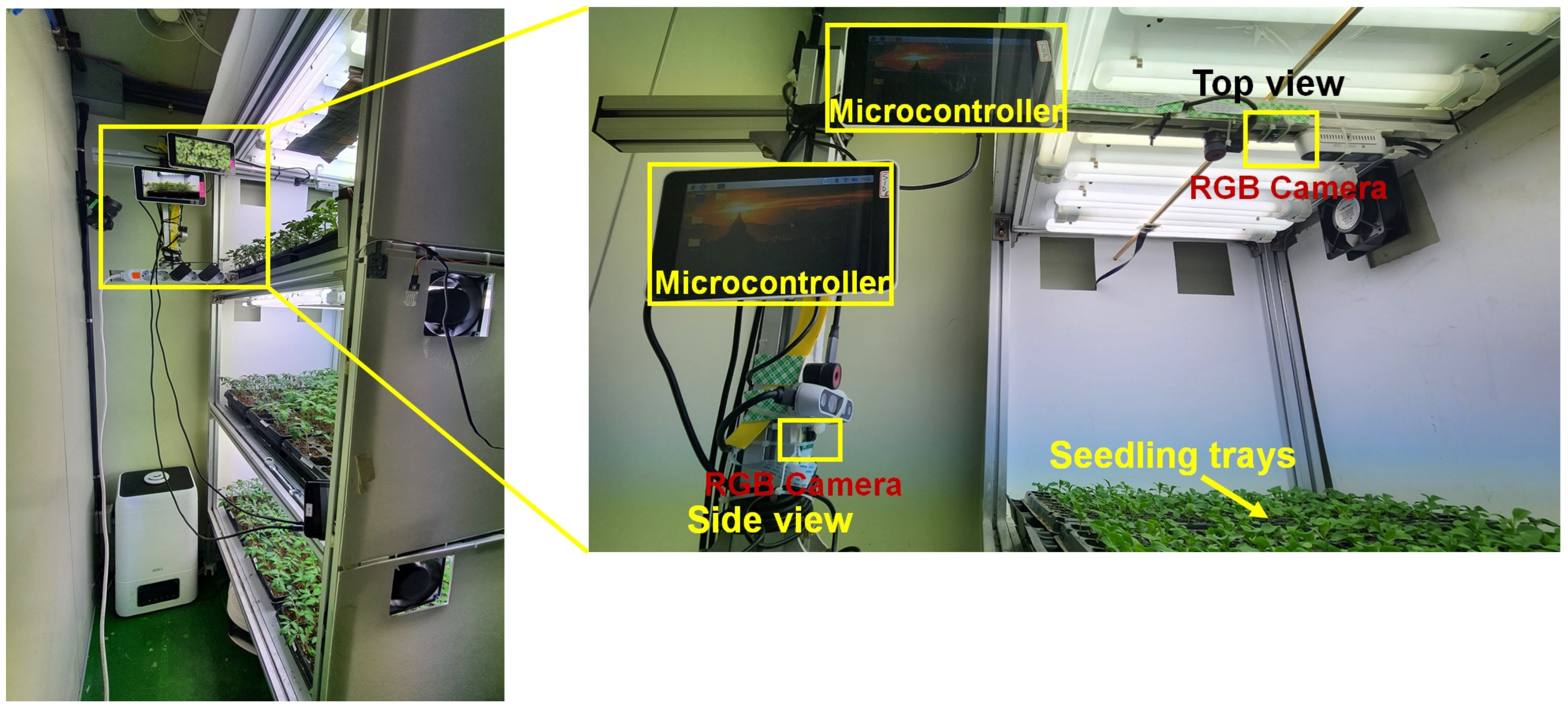
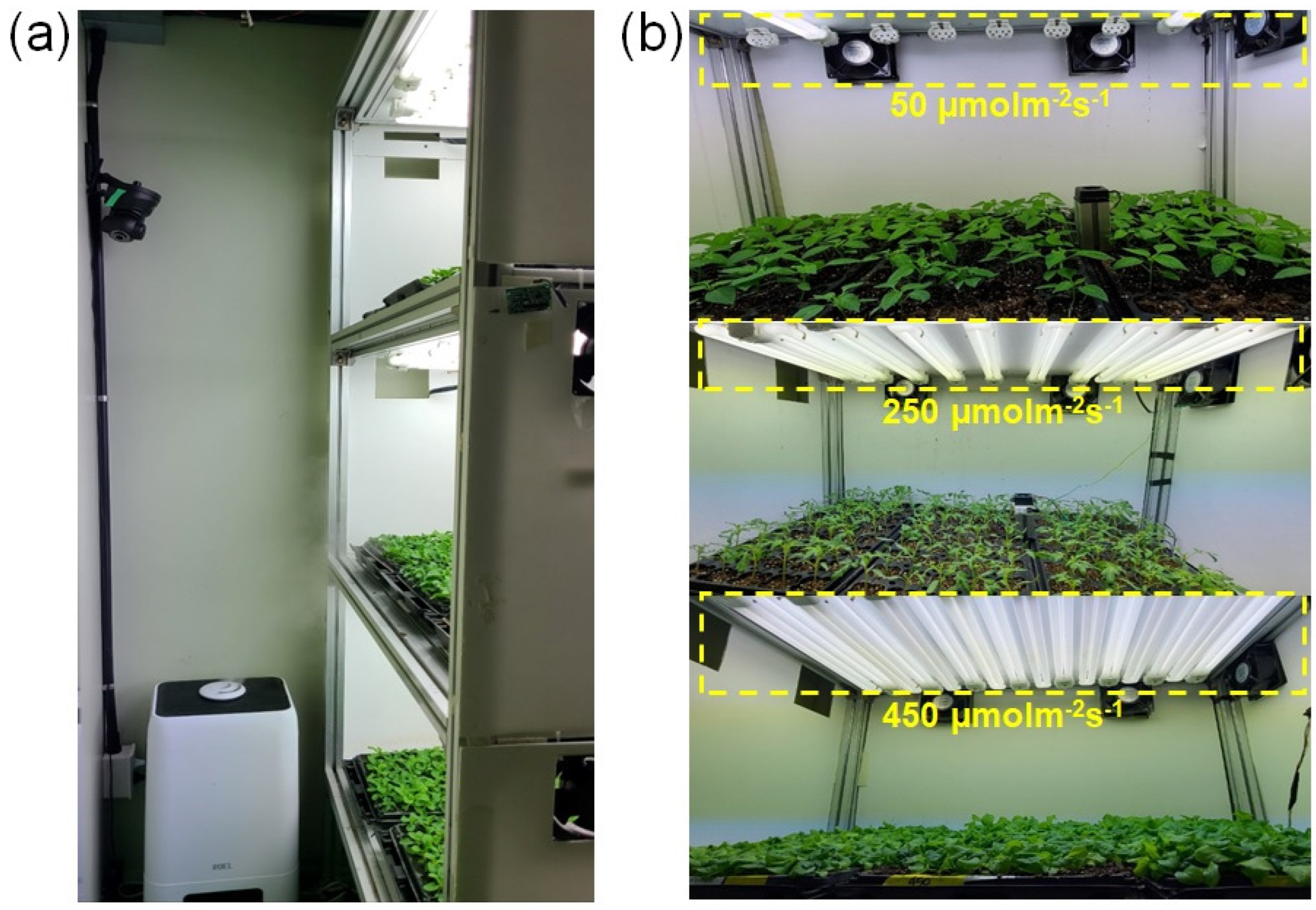
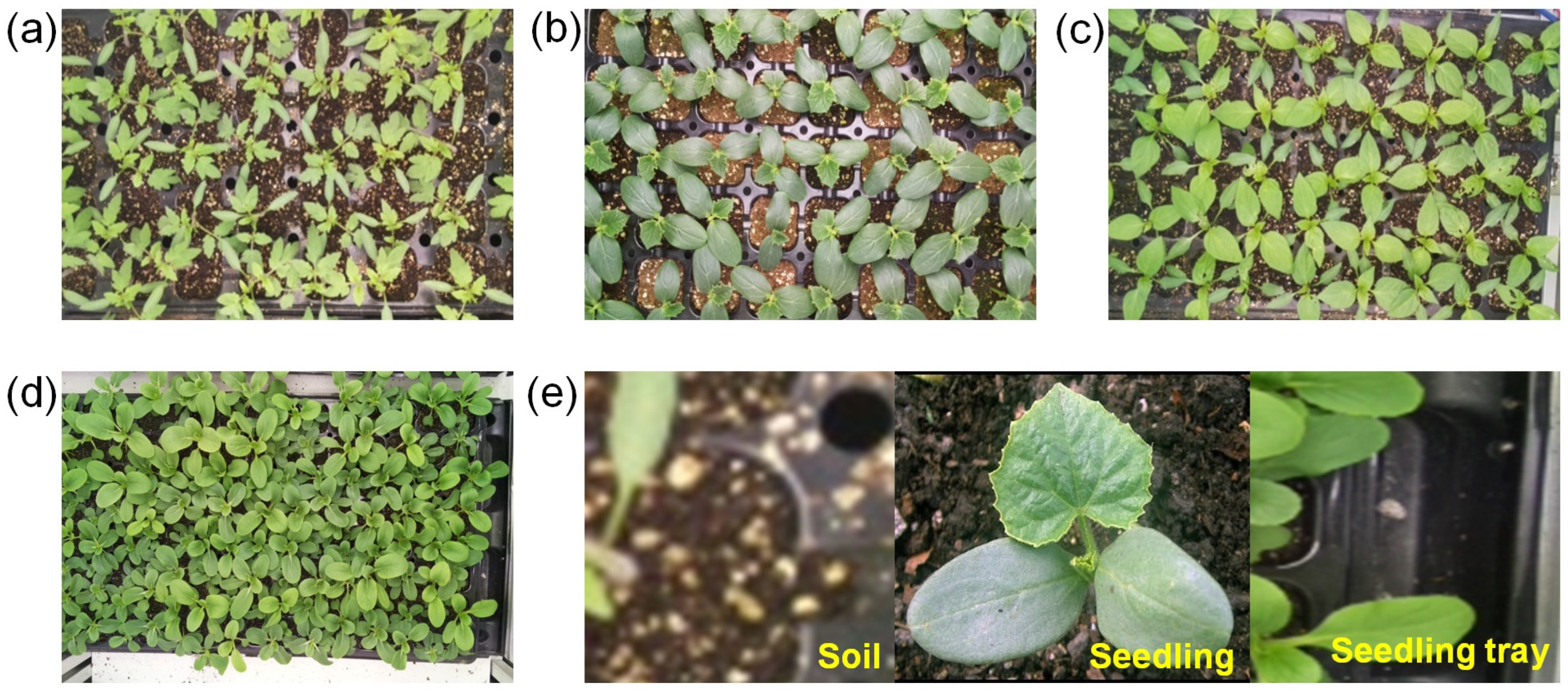







| Parameter | Microcontroller | Parameter | Camera |
|---|---|---|---|
| Name | Raspberry Pi 4B board | Name | Raspberry Pi Camera Module 2 |
| CPU | Quad-core Cortex-A72, 64-bit, 1.8 GHz | Sensor | Sony IMX 219 PQ CMOS |
| RAM | 8 GB LPDDR4-3200 | Resolution | 8 MP |
| Operating system | Linux based | FPS | 108p: 30; 720p: 60 |
| Connection | Standard 40-pin GPIO header | Resolution | 3280 × 2464 pixel |
| Power | 5 V DC | Connection | 15-pin MIPI CSI-2 |
| Operating temperature | 0 to 50 °C | Image control | Automatic |
| Parameter | SVM Classification with the SFS Method | SVM Classification Without the SFS Method | ||||||
|---|---|---|---|---|---|---|---|---|
| Precision | Recall | F1-Score | Support | Precision | Recall | F1-Score | Support | |
| Seedlings | 0.99 | 0.98 | 0.98 | 301 | 0.88 | 0.45 | 0.60 | 264 |
| Background | 0.98 | 0.99 | 0.98 | 299 | 0.69 | 0.95 | 0.80 | 336 |
| Accuracy | 0.98 | 600 | 0.73 | 600 | ||||
| Macro avg. | 0.98 | 0.98 | 0.98 | 600 | 0.79 | 0.70 | 0.70 | 600 |
| Weighted avg. | 0.98 | 0.97 | 0.98 | 600 | 0.77 | 0.73 | 0.71 | 600 |
| Parameter | SVM Classification with Linear Kernel | SVM Classification with Polynomial Kernel | SVM Classification with RBF Kernel | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Precision | Recall | F1-Score | Support | Precision | Recall | F1-Score | Support | Precision | Recall | F1-Score | Support | ||
| CV-0 | Seedling | 0.93 | 0.90 | 0.92 | 302 | 96 | 0.95 | 0.95 | 302 | 0.94 | 0.91 | 0.92 | 302 |
| Background | 0.91 | 0.94 | 0.92 | 298 | 0.95 | 0.96 | 0.95 | 298 | 0.92 | 0.91 | 0.92 | 298 | |
| Accuracy | 0.81 | 600 | 0.86 | 600 | 0.81 | 600 | |||||||
| Macro avg. | 0.80 | 0.77 | 0.78 | 600 | 0.91 | 0.81 | 0.83 | 600 | 0.79 | 0.78 | 0.79 | 600 | |
| Weighted avg. | 0.80 | 0.81 | 0.80 | 600 | 0.89 | 0.86 | 0.85 | 600 | 0.80 | 0.81 | 0.80 | 60 | |
| CV-5 | Seedling | 0.96 | 0.95 | 0.96 | 302 | 0.99 | 0.98 | 0.98 | 301 | 0.96 | 0.95 | 0.96 | 302 |
| Background | 0.95 | 0.96 | 0.96 | 298 | 0.98 | 0.99 | 0.98 | 299 | 0.95 | 0.96 | 0.96 | 298 | |
| Accuracy | 0.96 | 600 | 0.98 | 600 | 0.96 | 600 | |||||||
| Macro avg. | 0.96 | 0.96 | 0.96 | 600 | 0.98 | 0.98 | 0.98 | 600 | 0.96 | 0.95 | 0.96 | 600 | |
| Weighted avg. | 0.96 | 0.96 | 0.96 | 600 | 0.98 | 0.97 | 0.98 | 600 | 0.95 | 0.96 | 0.96 | 600 | |
| CV-10 | Seedling | 0.90 | 0.83 | 0.87 | 301 | 0.92 | 0.82 | 0.87 | 301 | 0.90 | 0.84 | 0.87 | 301 |
| Background | 0.84 | 0.91 | 0.87 | 299 | 0.84 | 0.93 | 0.89 | 299 | 0.85 | 0.91 | 0.88 | 299 | |
| Accuracy | 0.87 | 600 | 0.88 | 600 | 0.88 | 600 | |||||||
| Macro avg. | 0.87 | 0.87 | 0.87 | 600 | 0.88 | 0.88 | 0.88 | 600 | 0.88 | 0.88 | 0.87 | 600 | |
| Weighted avg. | 0.87 | 0.86 | 0.87 | 600 | 0.88 | 0.88 | 0.88 | 600 | 0.88 | 0.88 | 0.87 | 600 | |
| Kernel Type | CV | C, γ | MAE | Precision | Recall | F1-Score | Accuracy |
|---|---|---|---|---|---|---|---|
| Polynomial | 5 | C = 60 γ = 0 Degree = 3 | 0.27 | 0.77 | 0.73 | 0.71 | 73% |
| Linear | 0 | C = 10 | 0.19 | 0.80 | 0.81 | 0.80 | 81% |
| Linear | 5 | C = 10 | 0.04 | 0.96 | 0.96 | 0.96 | 96% |
| Linear | 10 | C = 30 | 0.13 | 0.87 | 0.86 | 0.87 | 87% |
| RBF | 0 | C = 128 γ = 128 | 0.19 | 0.80 | 0.81 | 0.80 | 81% |
| RBF | 5 | C = 100 γ = 512 | 0.04 | 0.95 | 0.96 | 0.96 | 96% |
| RBF | 10 | C = 128 γ = 128 | 0.13 | 0.88 | 0.88 | 0.87 | 87% |
| Polynomial | 0 | C = 60 γ = 0 Degree = 3 | 0.14 | 0.89 | 0.86 | 0.85 | 86% |
| Polynomial | 5 | C = 60 γ = 0 Degree = 3 | 0.02 | 0.98 | 0.97 | 0.98 | 98% |
| Polynomial | 10 | C = 60 γ = 0 Degree = 3 | 0.12 | 0.88 | 0.88 | 0.88 | 88% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samsuzzaman; Reza, M.N.; Islam, S.; Lee, K.-H.; Haque, M.A.; Ali, M.R.; Cho, Y.J.; Noh, D.H.; Chung, S.-O. Automated Seedling Contour Determination and Segmentation Using Support Vector Machine and Image Features. Agronomy 2024, 14, 2940. https://doi.org/10.3390/agronomy14122940
Samsuzzaman, Reza MN, Islam S, Lee K-H, Haque MA, Ali MR, Cho YJ, Noh DH, Chung S-O. Automated Seedling Contour Determination and Segmentation Using Support Vector Machine and Image Features. Agronomy. 2024; 14(12):2940. https://doi.org/10.3390/agronomy14122940
Chicago/Turabian StyleSamsuzzaman, Md Nasim Reza, Sumaiya Islam, Kyu-Ho Lee, Md Asrakul Haque, Md Razob Ali, Yeon Jin Cho, Dong Hee Noh, and Sun-Ok Chung. 2024. "Automated Seedling Contour Determination and Segmentation Using Support Vector Machine and Image Features" Agronomy 14, no. 12: 2940. https://doi.org/10.3390/agronomy14122940
APA StyleSamsuzzaman, Reza, M. N., Islam, S., Lee, K.-H., Haque, M. A., Ali, M. R., Cho, Y. J., Noh, D. H., & Chung, S.-O. (2024). Automated Seedling Contour Determination and Segmentation Using Support Vector Machine and Image Features. Agronomy, 14(12), 2940. https://doi.org/10.3390/agronomy14122940









