The Relationship Between Cover Crop Species and Soil Fungal Communities in Irrigated Vineyards in the Okanagan Valley, Canada
Abstract
1. Introduction
2. Methods
2.1. Study Area
2.2. Soil Sampling DNA Extractions and Sequencing
2.3. Data Processing
2.4. Statistical Analyses
2.4.1. Does Site Affect Soil Fungal Communities?
2.4.2. Does Cover Crop Identity Affect Soil Fungal Communities?
2.4.3. Community Structure
2.4.4. Role of Cover Crop/Site on Occurrence of Putative Pathogens/Beneficials
3. Results
3.1. Overall Soil Properties and Fungal Communities
3.2. Cover Crop Choice and Soil Fungal Community Structure
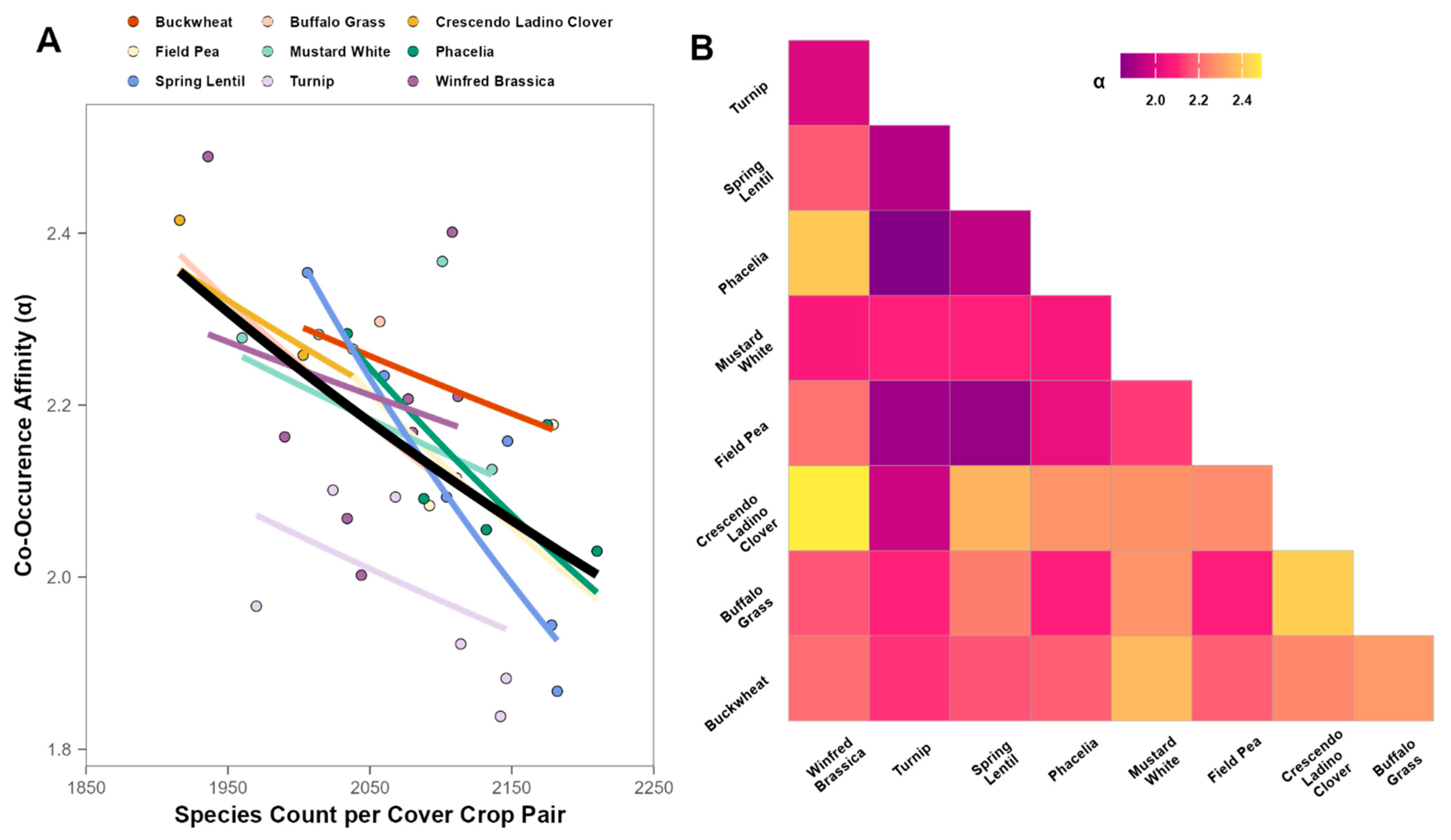
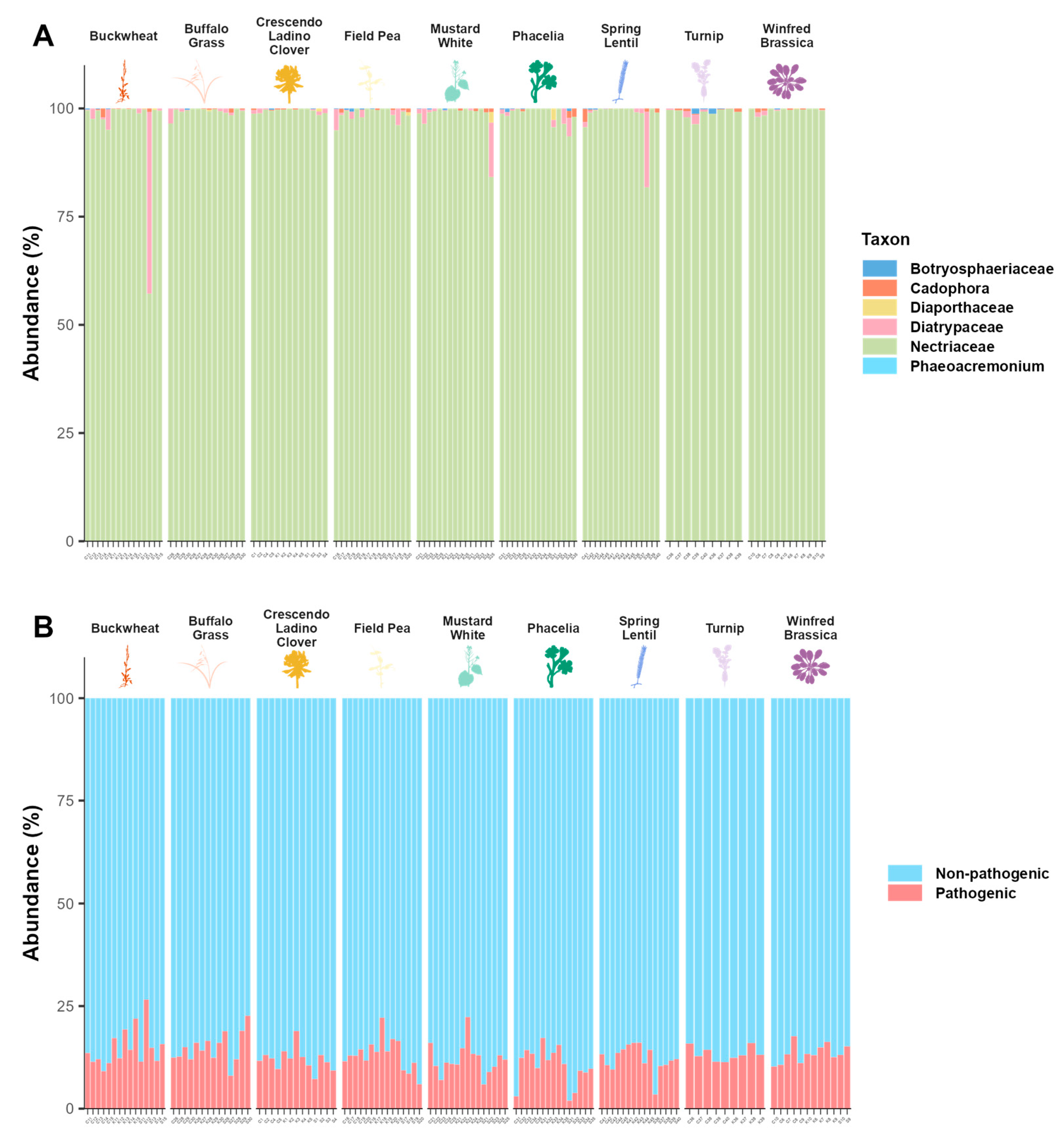
3.3. Pathogens and Beneficials
4. Discussion
4.1. Cover Crop Host Effects
4.2. Site Effects
4.3. Fungal Community Composition and Key Fungal Taxa
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zohry, A.; Ouda, S. Crop Rotation Defeats Pests and Weeds. In Crop Rotation; Springer International Publishing: Cham, Switzerland, 2018; pp. 77–88. ISBN 978-3-030-05350-5. [Google Scholar]
- Dzvene, A.R.; Tesfuhuney, W.A.; Walker, S.; Ceronio, G. Management of Cover Crop Intercropping for Live Mulch on Plant Productivity and Growth Resources: A Review. Air Soil Water Res. 2023, 16, 11786221231180079. [Google Scholar] [CrossRef]
- Jian, J.; Du, X.; Reiter, M.S.; Stewart, R.D. A Meta-Analysis of Global Cropland Soil Carbon Changes Due to Cover Cropping. Soil Biol. Biochem. 2020, 143, 107735. [Google Scholar] [CrossRef]
- McClelland, S.C.; Paustian, K.; Schipanski, M.E. Management of Cover Crops in Temperate Climates Influences Soil Organic Carbon Stocks: A Meta-analysis. Ecol. Appl. 2021, 31, e02278. [Google Scholar] [CrossRef] [PubMed]
- Helgason, B.L.; Janzen, H.H.; Chantigny, M.H.; Drury, C.F.; Ellert, B.H.; Gregorich, E.G.; Lemke, R.L.; Pattey, E.; Rochette, P.; Wagner-Riddle, C. Toward Improved Coefficients for Predicting Direct N2O Emissions from Soil in Canadian Agroecosystems. Nutr. Cycl. Agroecosyst. 2005, 72, 87–99. [Google Scholar] [CrossRef]
- Wang, H.; Beule, L.; Zang, H.; Pfeiffer, B.; Ma, S.; Karlovsky, P.; Dittert, K. The Potential of Ryegrass as Cover Crop to Reduce Soil N2O Emissions and Increase the Population Size of Denitrifying Bacteria. Eur. J. Soil Sci. 2021, 72, 1447–1461. [Google Scholar] [CrossRef]
- Zhang, H.; Ghahramani, A.; Ali, A.; Erbacher, A. Cover Cropping Impacts on Soil Water and Carbon in Dryland Cropping System. PLoS ONE 2023, 18, e0286748. [Google Scholar] [CrossRef]
- Wang, Q.; Li, Y.; Klassen, W. Influence of Summer Cover Crops on Conservation of Soil Water and Nutrients in a Subtropical Area. J. Soil Water Conserv. 2005, 60, 58–63. [Google Scholar]
- Murrell, E.G.; Ray, S.; Lemmon, M.E.; Luthe, D.S.; Kaye, J.P. Cover Crop Species Affect Mycorrhizae-Mediated Nutrient Uptake and Pest Resistance in Maize. Renew. Agric. Food Syst. 2020, 35, 467–474. [Google Scholar] [CrossRef]
- Rosado, M.D.C.; Araújo, G.J.D.; Pallini, A.; Venzon, M. Cover Crop Intercropping Increases Biological Control in Coffee Crops. Biol. Control 2021, 160, 104675. [Google Scholar] [CrossRef]
- Berlanas, C.; Andrés-Sodupe, M.; López-Manzanares, B.; Maldonado-González, M.M.; Gramaje, D. Effect of White Mustard Cover Crop Residue, Soil Chemical Fumigation and Trichoderma spp. Root Treatment on Black-foot Disease Control in Grapevine. Pest. Manag. Sci. 2018, 74, 2864–2873. [Google Scholar] [CrossRef]
- Mazzola, M.; Manici, L.M. Apple Replant Disease: Role of Microbial Ecology in Cause and Control. Annu. Rev. Phytopathol. 2012, 50, 45–65. [Google Scholar] [CrossRef] [PubMed]
- Latz, E.; Eisenhauer, N.; Rall, B.C.; Scheu, S.; Jousset, A. Unravelling Linkages between Plant Community Composition and the Pathogen-Suppressive Potential of Soils. Sci. Rep. 2016, 6, 23584. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Smalla, K. Plant Species and Soil Type Cooperatively Shape the Structure and Function of Microbial Communities in the Rhizosphere: Plant Species, Soil Type and Rhizosphere Communities. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Aleklett, K.; Hart, M. The Root Microbiota—A Fingerprint in the Soil? Plant Soil 2013, 370, 671–686. [Google Scholar] [CrossRef]
- Rosa, D.; Sharifi, M.; Hart, M.M. Cover Crops as Reservoirs for Young Vine Decline Pathogens. Agronomy 2022, 12, 2422. [Google Scholar] [CrossRef]
- Bleach, C.M. Management of Cylindrocarpon Black Foot Disease in New Zealand Nurseries and Vineyards. Ph.D. Thesis, Lincoln University, Christchurch, New Zealand, 2013. [Google Scholar]
- Richards, A.; Estaki, M.; Úrbez-Torres, J.R.; Bowen, P.; Lowery, T.; Hart, M. Cover Crop Diversity as a Tool to Mitigate Vine Decline and Reduce Pathogens in Vineyard Soils. Diversity 2020, 12, 128. [Google Scholar] [CrossRef]
- Shen, B.-B.; Yang, Y.-P.; Yasamin, S.; Liang, N.; Su, W.; Chen, S.-H.; Wang, X.-J.; Wang, W. Analysis of the Phytochemistry and Bioactivity of the Genus Polygonum of Polygonaceae. Digit. Chin. Med. 2018, 1, 19–36. [Google Scholar] [CrossRef]
- Sparg, S.G.; Light, M.E.; Van Staden, J. Biological Activities and Distribution of Plant Saponins. J. Ethnopharmacol. 2004, 94, 219–243. [Google Scholar] [CrossRef]
- Mohammadi, A.; Hashemi, M.; Hosseini, S.M. Comparison of Antifungal Activities of Various Essential Oils on the Phytophthora Drechsleri, the Causal Agent of Fruit Decay. Iran. J. Microbiol. 2015, 7, 31–37. [Google Scholar]
- O’Farrell, C.; Forge, T.; Hart, M.M. Using Brassica Cover Crops as Living Mulch in a Vineyard, Changes over One Growing Season. Int. J. Plant Biol. 2023, 14, 1105–1116. [Google Scholar] [CrossRef]
- Jung, S.C.; Martinez-Medina, A.; Lopez-Raez, J.A.; Pozo, M.J. Mycorrhiza-Induced Resistance and Priming of Plant Defenses. J. Chem. Ecol. 2012, 38, 651–664. [Google Scholar] [CrossRef]
- Nader, A.A.; Hauka, F.I.A.; Afify, A.H.; El-Sawah, A.M. Drought-Tolerant Bacteria and Arbuscular Mycorrhizal Fungi Mitigate the Detrimental Effects of Drought Stress Induced by Withholding Irrigation at Critical Growth Stages of Soybean (Glycine max, L.). Microorganisms 2024, 12, 1123. [Google Scholar] [CrossRef]
- Guerra, B.; Steenwerth, K. Influence of Floor Management Technique on Grapevine Growth, Disease Pressure, and Juice and Wine Composition: A Review. Am. J. Enol. Vitic. 2012, 63, 149–164. [Google Scholar] [CrossRef]
- White, C.A.; Holmes, H.F.; Morris, N.L.; Stobart, R.M. A Review of the Benefits, Optimal Crop Management Practices and Knowledge Gaps Associated with Different Cover Crop Species; AHDB Cereals & Oilseeds; Agriculture and Horticulture Development Board (AHDB): Coventry, UK, 2016; pp. 1–92. [Google Scholar]
- Ettema, C.; Wardle, D.A. Spatial Soil Ecology. Trends Ecol. Evol. 2002, 17, 177–183. [Google Scholar] [CrossRef]
- Philippot, L.; Chenu, C.; Kappler, A.; Rillig, M.C.; Fierer, N. The Interplay between Microbial Communities and Soil Properties. Nat. Rev. Microbiol. 2024, 22, 226–239. [Google Scholar] [CrossRef]
- Mehlich, A. Mehlich 3 Soil Test Extractant: A Modification of Mehlich 2 Extractant. Commun. Soil Sci. Plant Anal. 1984, 15, 1409–1416. [Google Scholar] [CrossRef]
- Sharifi, M.; Salimi, K.; Rosa, D.; Hart, M. Screening Cover Crops for Utilization in Irrigated Vineyards: A Greenhouse Study on Species’ Nitrogen Uptake and Carbon Sequestration Potential. Plants 2024, 13, 1959. [Google Scholar] [CrossRef]
- Tompkins, J.M. Ecosystem Services Provided by Native New Zealand Plants in Vineyards. Ph.D. Thesis, Lincoln University, Christchurch, New Zealand, 2010. [Google Scholar]
- Olmstead, M.A.; Wample, R.L.; Greene, S.L.; Tarara, J.M. Evaluation of Potential Cover Crops for Inland Pacific Northwest Vineyards. Am. J. Enol. Vitic. 2001, 52, 292–303. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Workflow for Microbiome Data Analysis: From Raw Reads to Community Analyses. Available online: https://bioconductor.org/help/course-materials/2017/BioC2017/Day1/Workshops/Microbiome/MicrobiomeWorkflowII.html (accessed on 11 May 2023).
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact Sequence Variants Should Replace Operational Taxonomic Units in Marker-Gene Data Analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef]
- Lin, E.Y.; Noonan, M.J. Mga: User-Friendly R Package for Microbial Genetic Analysis for Amplicon Data; Canadian Journal of Undergraduate Research: Vancouver, BC, Canada, 2024; Volume MURCxCJUR. [Google Scholar]
- Abarenkov, K.; Zirk, A.; Piirmann, T.; Pöhönen, R.; Ivanov, F.; Nilsson, R.H.; Kõljalg, U. UNITE General FASTA Release for Fungi; UNITE Community. 2022. Available online: https://cir.nii.ac.jp/crid/1370013198746680974 (accessed on 29 October 2024). [CrossRef]
- Wright, E. Using DECIPHER v2.0 to Analyze Big Biological Sequence Data in R. R J. 2016, 8, 352–359. [Google Scholar] [CrossRef]
- Schliep, K.P. Phangorn: Phylogenetic Analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Yan, L. Ggvenn: Draw Venn Diagram by “Ggplot2”, version 0.1.10; MIT License: Cambridge, MA, USA, 2023. [Google Scholar]
- Fedor, P.; Zvaríková, M. Biodiversity Indices. In Encyclopedia of Ecology; Earth Systems and Environmental Sciences; Elsevier: Amsterdam, The Netherlands, 2019; Volume 1, pp. 337–346. ISBN 978-0-444-64130-4. [Google Scholar]
- Safi, K.; Cianciaruso, M.V.; Loyola, R.D.; Brito, D.; Armour-Marshall, K.; Diniz-Filho, J.A.F. Understanding Global Patterns of Mammalian Functional and Phylogenetic Diversity. Phil. Trans. R. Soc. B 2011, 366, 2536–2544. [Google Scholar] [CrossRef]
- Csárdi, G.; Nepusz, T.; Müller, K.; Horvát, S.; Traag, V.; Zanini, F.; Noom, D. Igraph for R: R Interface of the igraph Library for Graph Theory and Network Analysis, Version 1.4.2. 2024. Available online: https://zenodo.org/records/10369053 (accessed on 29 October 2024). [CrossRef]
- Lurgi, M. Networks as Graphs. Available online: https://mlurgi.github.io/networks_for_r/lesson-4.html (accessed on 12 October 2024).
- Mainali, K.P.; Slud, E.; Singer, M.C.; Fagan, W.F. A Better Index for Analysis of Co-Occurrence and Similarity. Sci. Adv. 2022, 8, eabj9204. [Google Scholar] [CrossRef]
- Pinheiro, J.; Bates, D. R Core Team Nlme: Linear and Nonlinear Mixed Effects Models, version 3.1-162; Free Software Foundation, Inc.: Boston, MA, USA, 2012. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Soft. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Wilke, C.O. Ggridges: Ridgeline Plots in Ggplot2, version 0.5.4; Free Software Foundation, Inc.: Boston, MA, USA, 2022. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Bekris, F.; Vasileiadis, S.; Papadopoulou, E.; Samaras, A.; Testempasis, S.; Gkizi, D.; Tavlaki, G.; Tzima, A.; Paplomatas, E.; Markakis, E.; et al. Grapevine Wood Microbiome Analysis Identifies Key Fungal Pathogens and Potential Interactions with the Bacterial Community Implicated in Grapevine Trunk Disease Appearance. Environ. Microbiome 2021, 16, 23. [Google Scholar] [CrossRef]
- Travadon, R.; Lawrence, D.P.; Moyer, M.M.; Fujiyoshi, P.T.; Baumgartner, K. Fungal Species Associated with Grapevine Trunk Diseases in Washington Wine Grapes and California Table Grapes, with Novelties in the Genera Cadophora, Cytospora, and Sporocadus. Front. Fungal Biol. 2022, 3, 1018140. [Google Scholar] [CrossRef]
- Úrbez-Torres, J.R.; Haag, P.; Bowen, P.; O’Gorman, D.T. Grapevine Trunk Diseases in British Columbia: Incidence and Characterization of the Fungal Pathogens Associated with Black Foot Disease of Grapevine. Plant Dis. 2014, 98, 456–468. [Google Scholar] [CrossRef]
- Chen, M.; Arato, M.; Borghi, L.; Nouri, E.; Reinhardt, D. Beneficial Services of Arbuscular Mycorrhizal Fungi–From Ecology to Application. Front. Plant Sci. 2018, 9, 1270. [Google Scholar] [CrossRef]
- Van Herk, W.G.; Vernon, R.S. Wireworm Damage to Wheat Seedlings: Effect of Temperature and Wireworm State. J. Pest. Sci. 2013, 86, 63–75. [Google Scholar] [CrossRef]
- Poggi, S.; Le Cointe, R.; Riou, J.-B.; Larroudé, P.; Thibord, J.-B.; Plantegenest, M. Relative Influence of Climate and Agroenvironmental Factors on Wireworm Damage Risk in Maize Crops. J. Pest. Sci. 2018, 91, 585–599. [Google Scholar] [CrossRef]
- Hermann, A.; Brunner, N.; Hann, P.; Wrbka, T.; Kromp, B. Correlations between Wireworm Damages in Potato Fields and Landscape Structure at Different Scales. J. Pest. Sci. 2013, 86, 41–51. [Google Scholar] [CrossRef]
- Toom, M.; Talgre, L.; Mäe, A.; Tamm, S.; Narits, L.; Edesi, L.; Haljak, M.; Lauringson, E. Selecting Winter Cover Crop Species for Northern Climatic Conditions. Biol. Agric. Hortic. 2019, 35, 263–274. [Google Scholar] [CrossRef]
- Mäkelä, P.S.A.; Tuulos, A.; Turakainen, M.; Santanen, A.; Stoddard, F.L. Revitalizing the Winter Turnip Rape Crop in the Northern Latitudes. Acta Agric. Scand. Sect B 2011, 61, 195–201. [Google Scholar] [CrossRef][Green Version]
- Gupta, S.C.; Larson, W.E. Estimating Soil Water Retention Characteristics from Particle Size Distribution, Organic Matter Percent, and Bulk Density. Water Resour. Res. 1979, 15, 1633–1635. [Google Scholar] [CrossRef]
- Saxton, K.E.; Rawls, W.J. Soil Water Characteristic Estimates by Texture and Organic Matter for Hydrologic Solutions. Soil Sci. Soc. Am. J. 2006, 70, 1569–1578. [Google Scholar] [CrossRef]
- Ferreira, C.S.; Veiga, A.; Caetano, A.; Gonzalez-Pelayo, O.; Karine-Boulet, A.; Abrantes, N.; Keizer, J.; Ferreira, A.J. Assessment of the Impact of Distinct Vineyard Management Practices on Soil Physico-Chemical Properties. Air Soil Water Res. 2020, 13, 1178622120944847. [Google Scholar] [CrossRef]
- Hannam, K.D.; Neilsen, G.H.; Forge, T.A.; Neilsen, D.; Losso, I.; Jones, M.D.; Nichol, C.; Fentabil, M.M. Irrigation Practices, Nutrient Applications, and Mulches Affect Soil Nutrient Dynamics in a Young Merlot (Vitis vinifera L.) Vineyard. Can. J. Soil. Sci. 2016, 96, 23–36. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tendersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.V.; Saxena, R.K.; et al. Outline of Fungi and Fungus-like Taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
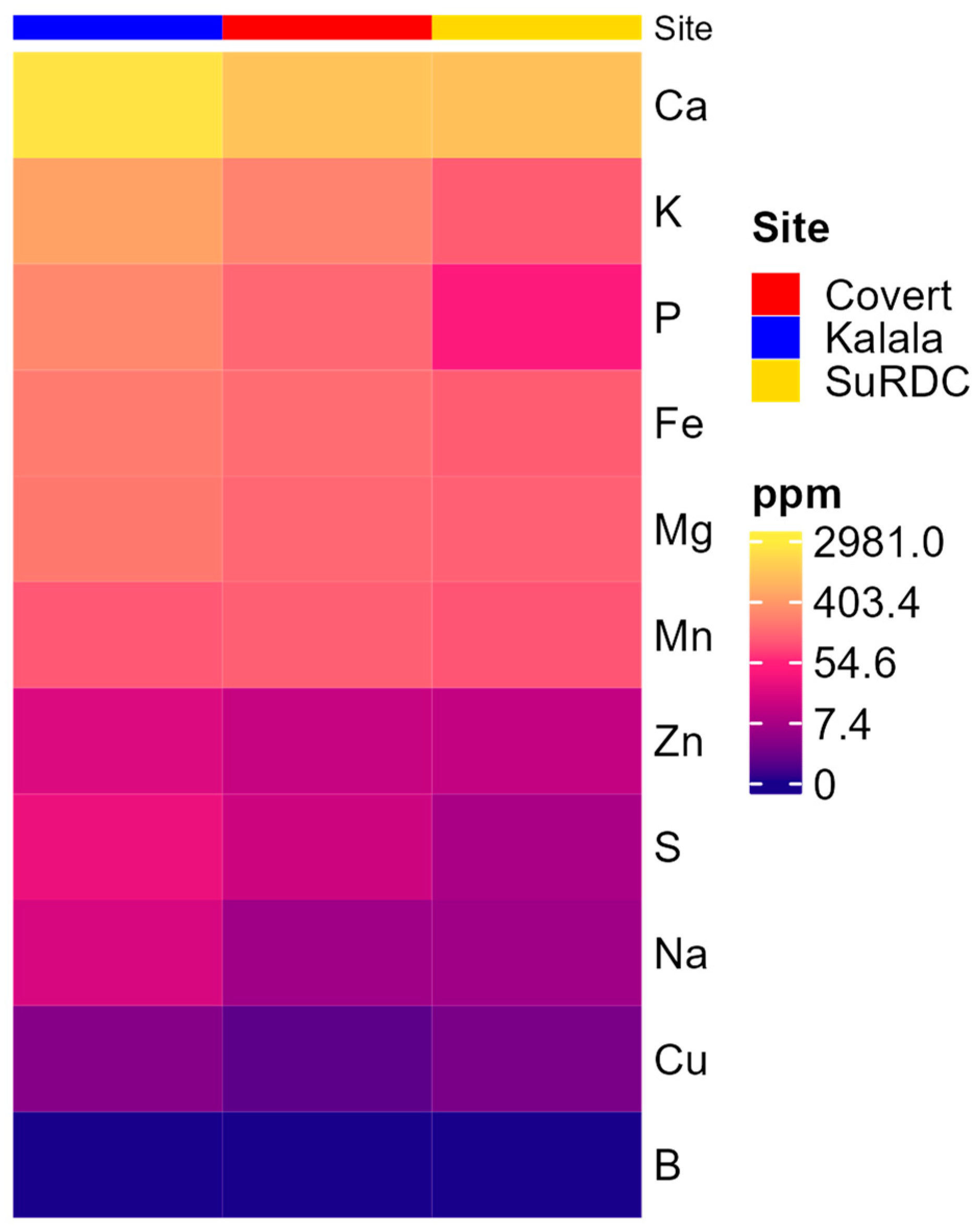
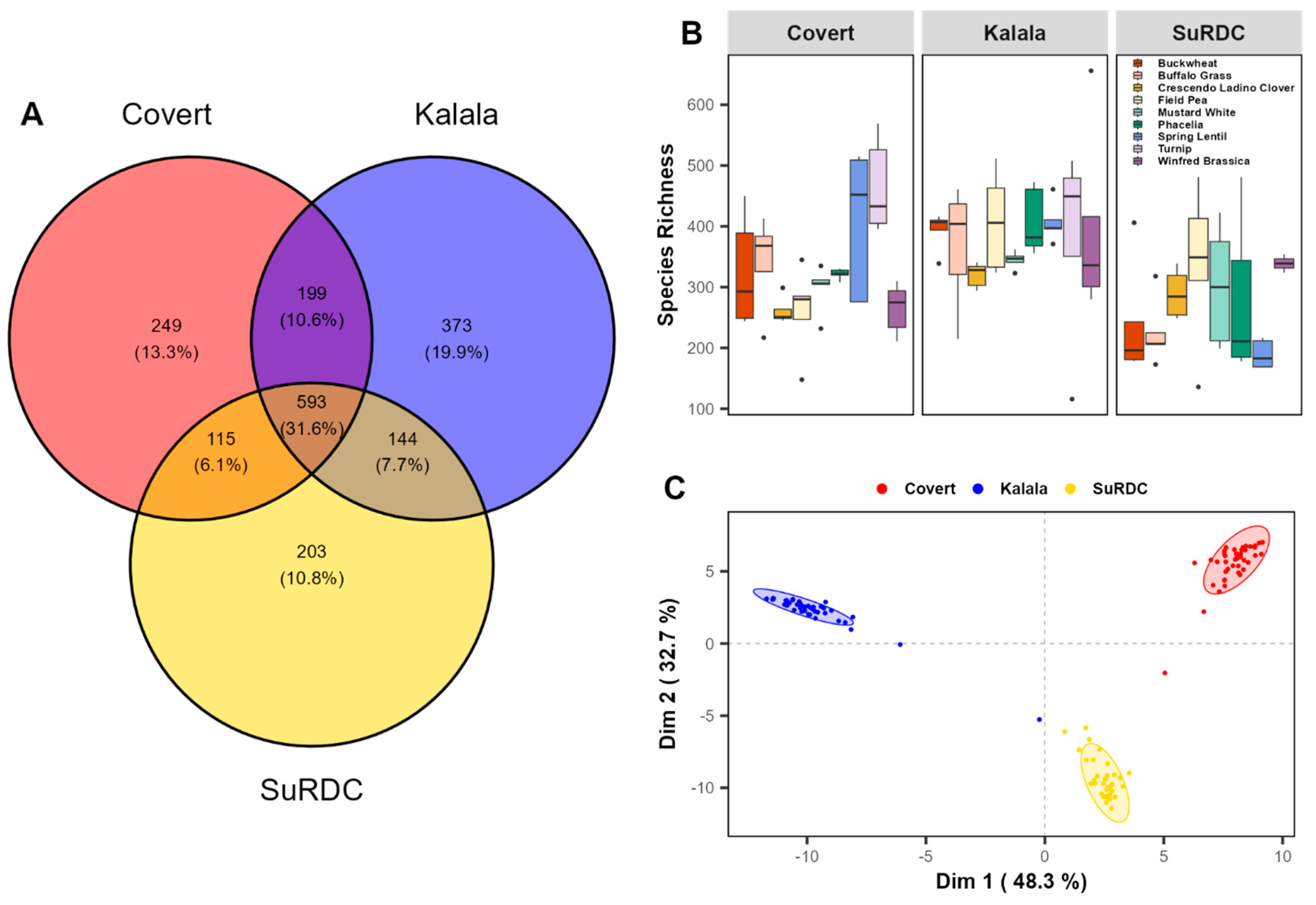
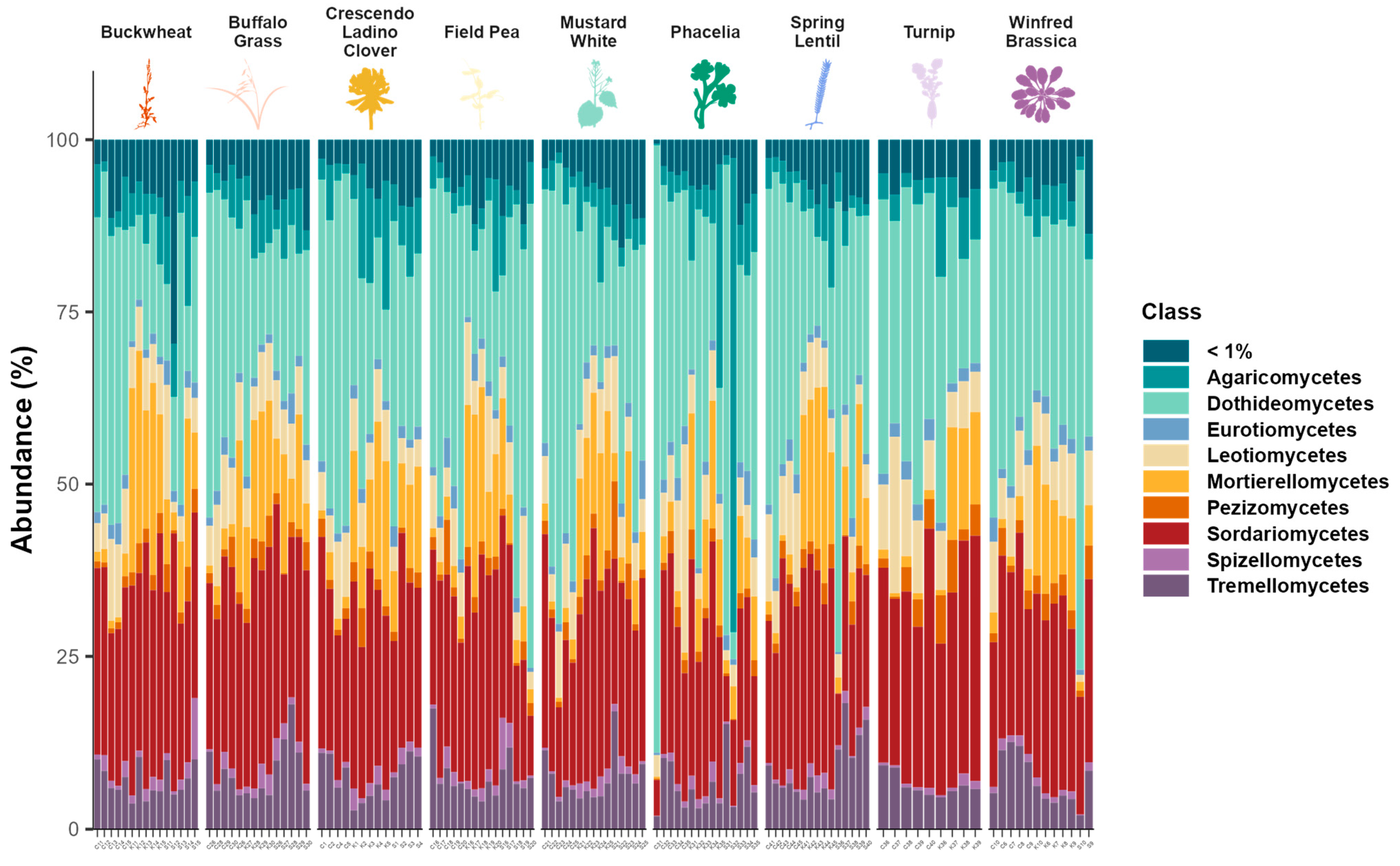
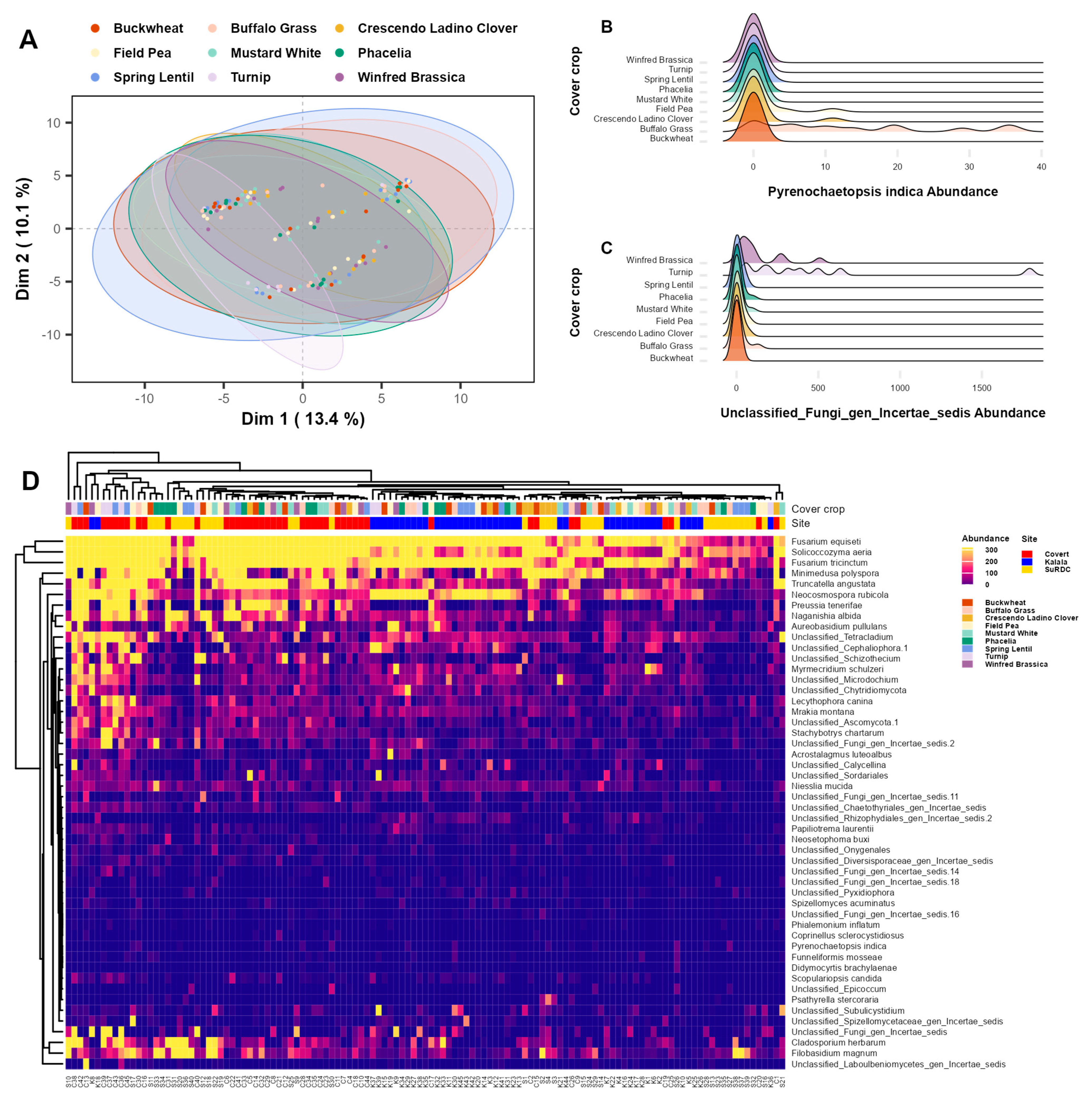
| Site | Latitude and Longitude (°N, °W) | Growing Season Rainfall † (mm) | Mean Growing Season Air Temperature † (°C) | Fertilizer Rates Applied (kg ha−1) | Starter N Applied (kg N ha−1) | Grape Variety | Soil Type | Management Practices |
|---|---|---|---|---|---|---|---|---|
| Covert | 49°14′39.8″ 119°32′42.7″ | 134 | 10.7 | None applied (organic) | None applied | 13-yr old Merlot | Loamy sand | Drip irrigation, Overhead sprinkler |
| SuRDC | 49°33′49.2″ 119°38′19.0″ | 225 | 15.6 | 107 N 8.00 P 54 K | 2-yr old Riesling | Loamy sand | Drip irrigation | |
| Kalala | 49°50′31.2″ 119°38′42″ | 143 | 18.6 | None applied (organic) | 10-yr old Zweigelt | Sandy loam | Drip irrigation, Overhead sprinkler |
| Site | Electerdial Conductivity (EC) (μS/cm) | pH | %C | %N | C/N | % Clay | % Silt | % Sand | Classification |
|---|---|---|---|---|---|---|---|---|---|
| Covert | 148 (115, 167) | 6.34 (6.30, 6.40) | 0.813 (0.510, 1.356) | 0.0716 (0.0446, 0.1199) | 11.5 (10.4, 13.2) | 6.00 (4.67, 6.87) | 9.72 (6.30, 12.39) | 84.27 (80.73, 89.03) | Loamy sand |
| Kalala | 262 (70, 91) | 6.31 (6.03, 6.63) | 1.492 (0.958, 2.075) | 0.1258 (0.0865, 0.1678) | 11.9 (11.1, 12. 7) | 14.00 (11.72, 16.12) | 32.64 (27.39, 37.33) | 53.36 (46.55, 60.89) | Sandy loam |
| SuRDC | 228 (188, 272) | 6.32 (6.22, 6.39) | 0.7843 (0.586,1.050) | 0.0604 (0.0452, 0.0782) | 13.0 (11.1, 16.2) | 5.86 (4.90, 7.73) | 13.67 (11.51, 17.85) | 80.47 (74.43, 83.58) | Loamy sand |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, E.Y.; Rosa, D.; Sharifi, M.; Noonan, M.J.; Hart, M. The Relationship Between Cover Crop Species and Soil Fungal Communities in Irrigated Vineyards in the Okanagan Valley, Canada. Agronomy 2024, 14, 2835. https://doi.org/10.3390/agronomy14122835
Lin EY, Rosa D, Sharifi M, Noonan MJ, Hart M. The Relationship Between Cover Crop Species and Soil Fungal Communities in Irrigated Vineyards in the Okanagan Valley, Canada. Agronomy. 2024; 14(12):2835. https://doi.org/10.3390/agronomy14122835
Chicago/Turabian StyleLin, Erika Y., Daniel Rosa, Mehdi Sharifi, Michael J. Noonan, and Miranda Hart. 2024. "The Relationship Between Cover Crop Species and Soil Fungal Communities in Irrigated Vineyards in the Okanagan Valley, Canada" Agronomy 14, no. 12: 2835. https://doi.org/10.3390/agronomy14122835
APA StyleLin, E. Y., Rosa, D., Sharifi, M., Noonan, M. J., & Hart, M. (2024). The Relationship Between Cover Crop Species and Soil Fungal Communities in Irrigated Vineyards in the Okanagan Valley, Canada. Agronomy, 14(12), 2835. https://doi.org/10.3390/agronomy14122835







