Spectroscopy Technologies to Screen Peanut Seeds with Superior Vigor Through “Chemical Fingerprinting”
Abstract
1. Introduction
2. Materials and Methods
2.1. Biomaterial
2.2. Seed Production in the Field
2.3. Seed Analysis
2.3.1. Seed Weight
2.3.2. Hydrogen Peroxide (H2O2)
2.3.3. Lipid Peroxidation: Malondialdehyde (MDA)
2.3.4. Seed Germination
2.3.5. Germination Rate
2.3.6. Normal Seedlings: The Strongest Ones
2.3.7. Post-Storage Seed Germination
2.3.8. Seedling Length Analysis
2.3.9. Plant Establishment and Shoot Dry Weight
2.4. Spectroscopy Technologies
2.4.1. Nuclear Magnetic Resonance (NMR)
2.4.2. Laser-Induced Breakdown Spectroscopy (LIBS)
2.4.3. Energy-Dispersive X-Ray Fluorescence (ED-XRF)
2.5. Statistical Analysis
2.5.1. ANOVA
2.5.2. Principal Component Analysis and Correlation
2.5.3. Quadratic Discriminant Analysis (QDA) and Confusion Matrix
3. Results
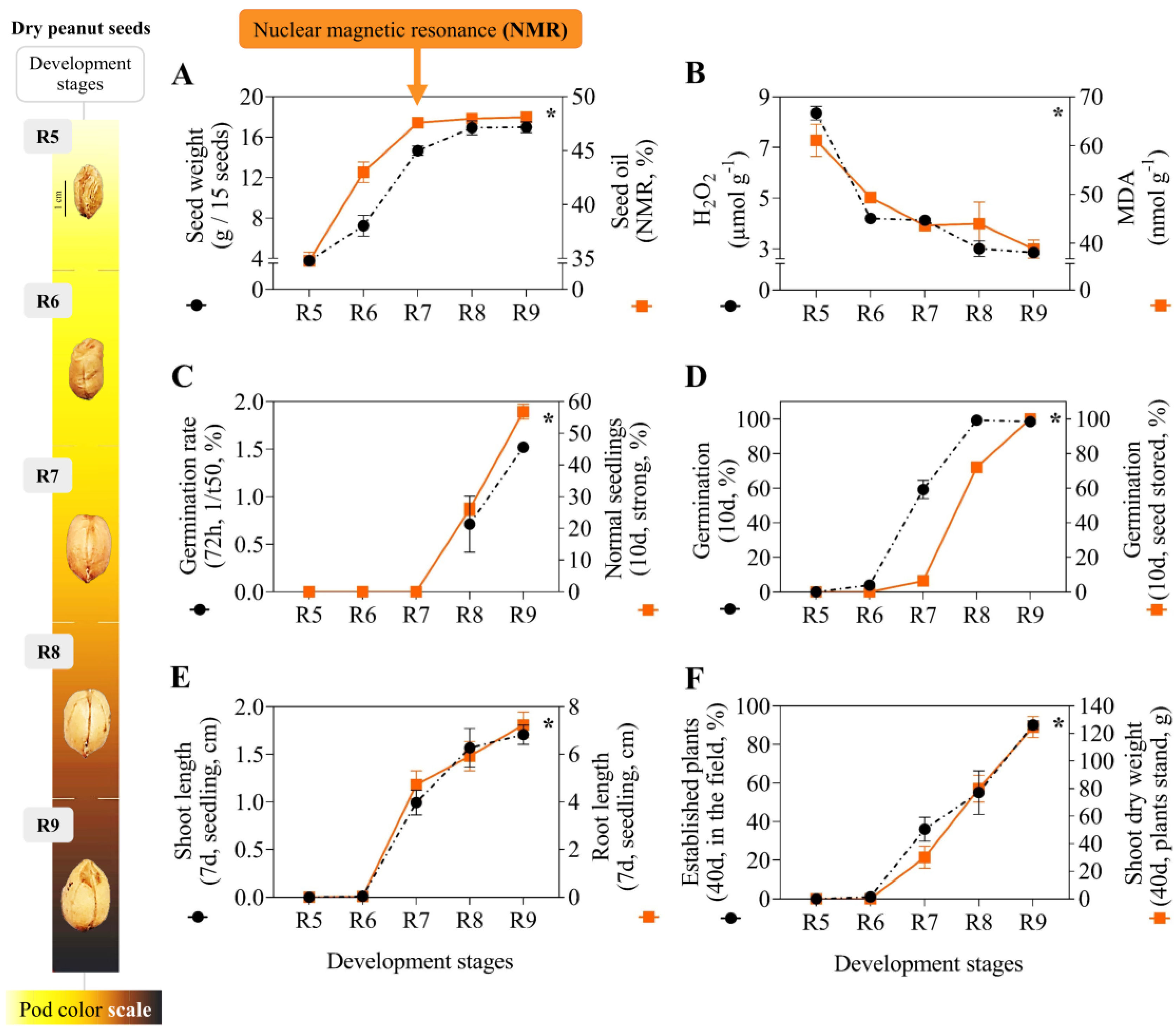
3.1. Physical, Chemical, Physiological Parameters and Oxidative Stress
3.2. LIBS and ED-XRF: Seed Minerals
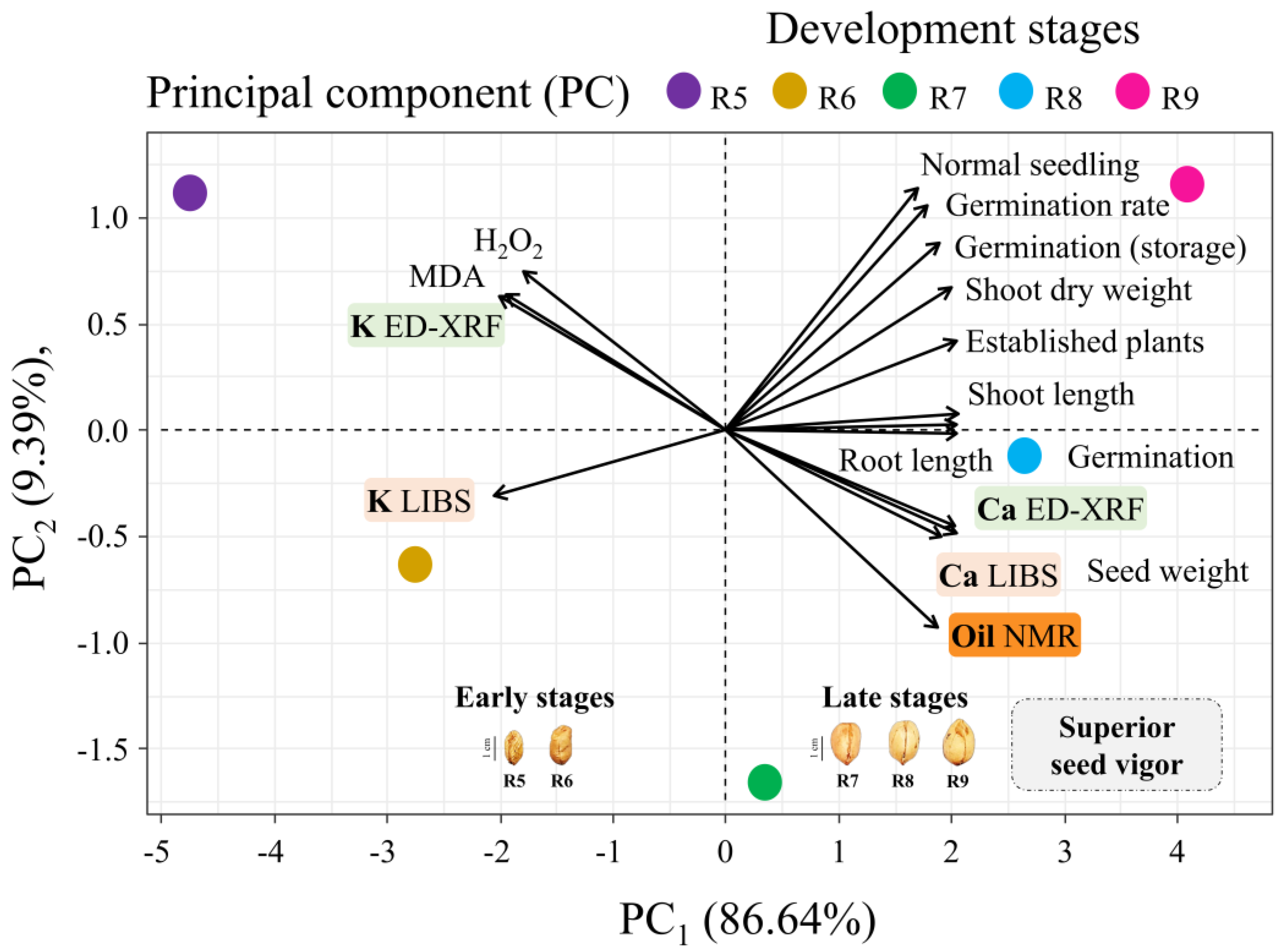
3.3. Principal Component Analysis
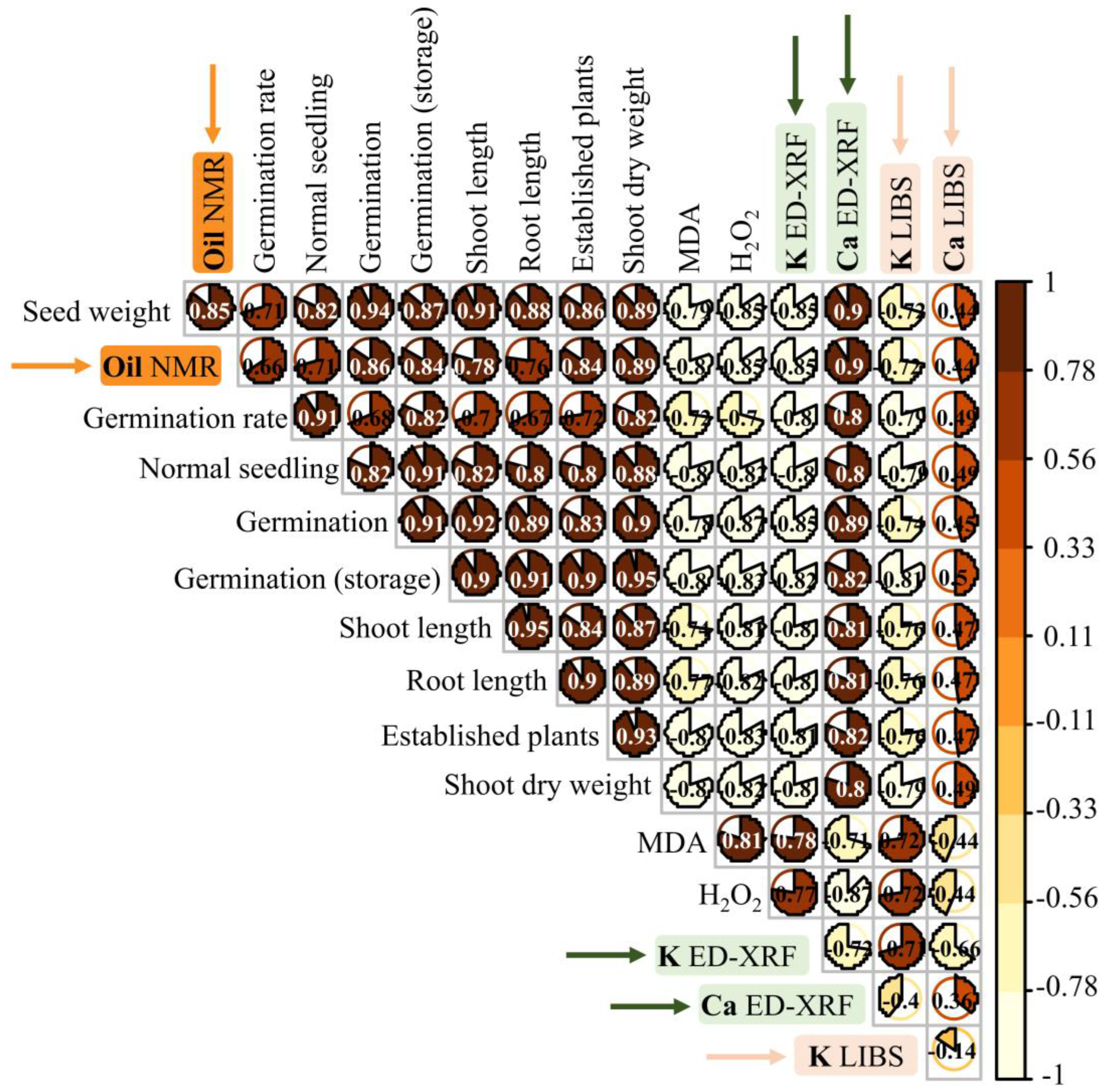
3.4. Correlation Analysis
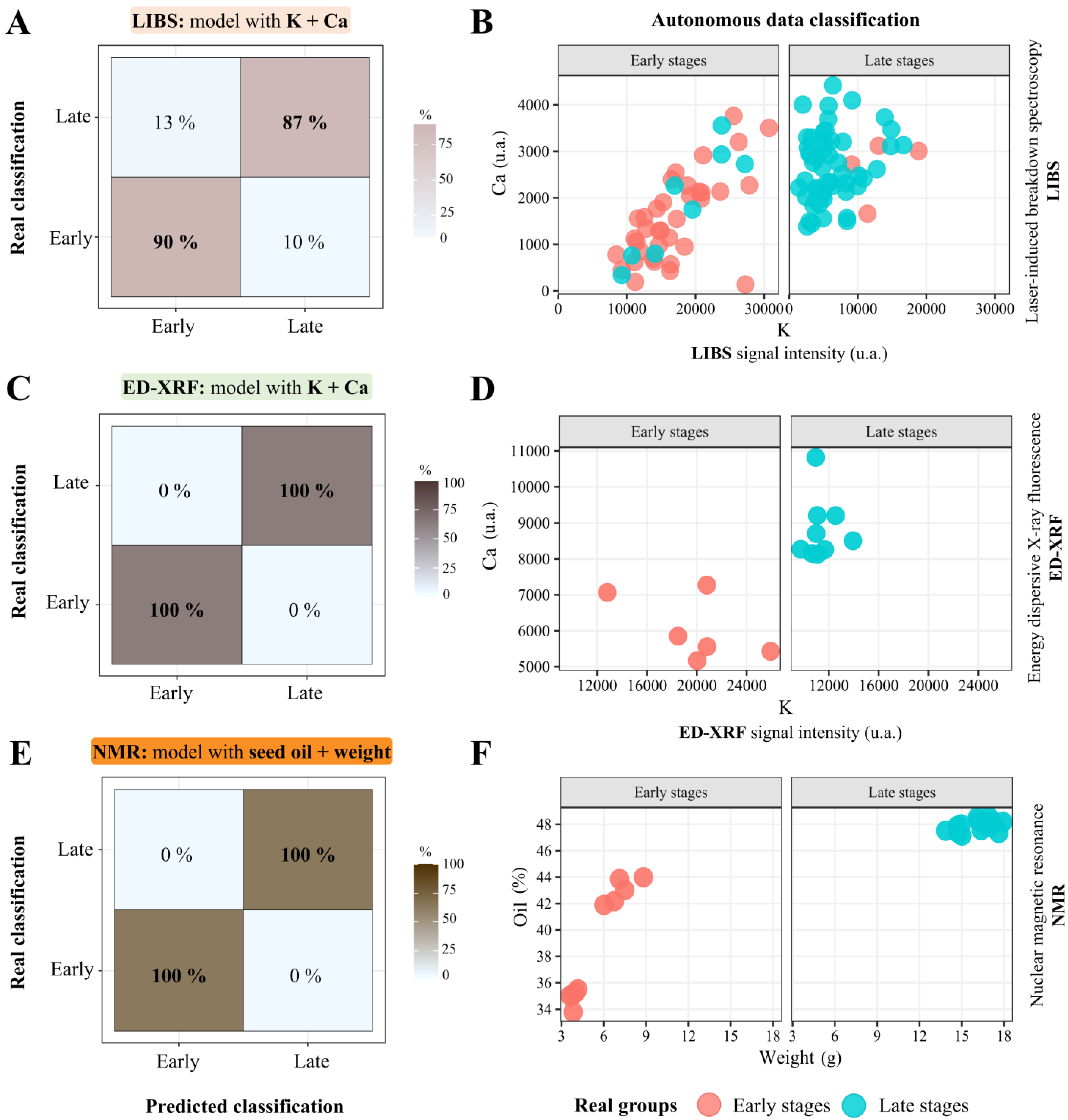
3.5. Predicting the Seed Chemical Autonomously
4. Discussion
4.1. Oil Through NMR: Biological Relationships with Seed Development and Applications
4.2. K Spectral Dynamics and Seed Oxidative Stability
4.3. Ca Chemical “Fingerprints”: Technological Insights into Seed Vigor
4.4. Screening Seed Vigor Through Spectral Chemical Patterns: Perspectives
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- U.S. Department of Agriculture. Peanut Explorer. Available online: https://ipad.fas.usda.gov/cropexplorer/cropview/commodityView.aspx?cropid=2221000 (accessed on 9 October 2024).
- Bonku, R.; Yu, J. Health Aspects of Peanuts as an Outcome of Its Chemical Composition. Food Sci. Hum. Wellness 2020, 9, 21–30. [Google Scholar] [CrossRef]
- Nakagawa, J.; Rosolem, C.A. O Amendoim; FEPAF: Botucatu, SP, Brazil, 2011; ISBN 978-85-98187-29-7. [Google Scholar]
- Qiao, M.; Sun, R.; Wang, Z.; Dumack, K.; Xie, X.; Dai, C.; Wang, E.; Zhou, J.; Sun, B.; Peng, X.; et al. Legume Rhizodeposition Promotes Nitrogen Fixation by Soil Microbiota under Crop Diversification. Nat. Commun. 2024, 15, 2924. [Google Scholar] [CrossRef] [PubMed]
- USDA Oilseeds: World Markets and Trade. Available online: https://fas.usda.gov/data/oilseeds-world-markets-and-trade-06122024 (accessed on 9 October 2024).
- FAO. Food and Agriculture Organization of the United Nations. Crops and Livestock Products. Available online: https://www.fao.org/faostat/en/#data/QCL/visualize (accessed on 7 October 2024).
- Stalker, H.T.; Wilson, R.F. Peanuts: Genetics, Processing, and Utilization; Academic Press: Cambridge, MA, USA; AOCS Press: Champaign, IL, USA, 2016; ISBN 9781630670382. [Google Scholar]
- Fonseca de Oliveira, G.R.; Amaral da Silva, E.A. Tropical Peanut Maturation Scale for Harvesting Seeds with Superior Quality. Front. Plant Sci. 2024, 15, 1376370. [Google Scholar] [CrossRef] [PubMed]
- Groot, S.P.C. Seed Maturation and Its Practical Implications. Seed Sci. Technol. 2022, 50, 141–151. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Bassel, G.W. Seed Vigour and Crop Establishment: Extending Performance beyond Adaptation. J. Exp. Bot. 2016, 67, 567–591. [Google Scholar] [CrossRef]
- Pattee, H.E.; Johns, E.B.; Singleton, J.A.; Sanders, T.H. Composition Changes of Peanut Fruit Parts During Maturation. Peanut Sci. 1974, 1, 57–62. [Google Scholar] [CrossRef]
- Moreno, L.; Santos, A.F.; Tubbs, R.S.; Grey, T.L.; Monfort, W.S.; Lamb, M.C.; Pilon, C. Physiological Components of Seed Quality in Runner-type Peanut during Seed Formation. Agron. J. 2023, 116, 189–201. [Google Scholar] [CrossRef]
- de Aguila Moreno, L.; Fonseca de Oliveira, G.R.; Batista, T.B.; Bossolani, J.W.; Ducatti, K.R.; Guimarães, C.C.; da Silva, E.A.A. Quality of Cowpea Seeds: A Food Security Strategy in the Tropical Environment. PLoS ONE 2022, 17, e0276136. [Google Scholar] [CrossRef]
- Song, Y.; Rowland, D.L.; Tillman, B.L.; Wilson, C.H.; Sarnoski, P.J.; Zurweller, B.A. Impact of Seed Maturity on Season-Long Physiological Performance and Offspring Seed Quality in Peanut (Arachis hypogaea L.). F. Crop Res. 2022, 288, 108674. [Google Scholar] [CrossRef]
- Leprince, O.; Pellizzaro, A.; Berriri, S.; Buitink, J. Late Seed Maturation: Drying without Dying. J. Exp. Bot. 2017, 68, 827–841. [Google Scholar] [CrossRef]
- Chatelain, E.; Hundertmark, M.; Leprince, O.; Gall, S.L.; Satour, P.; Deligny-Penninck, S.; Rogniaux, H.; Buitink, J. Temporal Profiling of the Heat-Stable Proteome during Late Maturation of Medicago truncatula Seeds Identifies a Restricted Subset of Late Embryogenesis Abundant Proteins Associated with Longevity. Plant Cell Environ. 2012, 35, 1440–1455. [Google Scholar] [CrossRef] [PubMed]
- Bewley, J.D.; Bradford, K.J.; Hilhorst, H.W.M.; Nonogaki, H. Seeds: Physiology of Development, Germination and Dormancy, 3rd ed.; Springer: New York, NY, USA, 2013; Volume 4, ISBN 9781461446927. [Google Scholar]
- Cao, D.; Ma, Y.; Cao, Z.; Hu, S.; Li, Z.; Li, Y.; Wang, K.; Wang, X.; Wang, J.; Zhao, K.; et al. Coordinated Lipid Mobilization during Seed Development and Germination in Peanut (Arachis hypogaea L.). J. Agric. Food Chem. 2024, 72, 3218–3230. [Google Scholar] [CrossRef] [PubMed]
- Sattler, S.E.; Gilliland, L.U.; Magallanes-Lundback, M.; Pollard, M.; DellaPenna, D. Vitamin E Is Essential for Seed Longevity and for Preventing Lipid Peroxidation during Germination. Plant Cell 2004, 16, 1419–1432. [Google Scholar] [CrossRef] [PubMed]
- Che, Y.; Yao, T.; Wang, H.; Wang, Z.; Zhang, H.; Sun, G.; Zhang, H. Potassium Ion Regulates Hormone, Ca2+ and H2O2 Signal Transduction and Antioxidant Activities to Improve Salt Stress Resistance in Tobacco. Plant Physiol. Biochem. 2022, 186, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Fatnani, D.; Parida, A.K. Potassium Deficiency Stress Tolerance in Peanut (Arachis hypogaea) through Ion Homeostasis, Activation of Antioxidant Defense, and Metabolic Dynamics: Alleviatory Role of Silicon Supplementation. Plant Physiol. Biochem. 2022, 182, 55–75. [Google Scholar] [CrossRef]
- Kadirimangalam, S.R.; Sawargaonkar, G.; Choudhari, P. Morphological and Molecular Insights of Calcium in Peanut Pod Development. J. Agric. Food Res. 2022, 9, 100320. [Google Scholar] [CrossRef]
- Li, Y.; Meng, J.; Yang, S.; Guo, F.; Zhang, J.; Geng, Y.; Cui, L.; Wan, S.; Li, X. Transcriptome Analysis of Calcium-and Hormone-Related Gene Expressions during Different Stages of Peanut Pod Development. Front. Plant Sci. 2017, 8, 1241. [Google Scholar] [CrossRef]
- de Carvalho, L.C.; Pereira, F.M.V.; de Morais, C.D.L.M.; de Lima, K.M.G.; de Almeida Teixeira, G.H. Assessment of Macadamia Kernel Quality Defects by Means of near Infrared Spectroscopy (NIRS) and Nuclear Magnetic Resonance (NMR). Food Control 2019, 106, 106695. [Google Scholar] [CrossRef]
- Santos, J.F.; Godoy, I.J.; Moraes, R.A.; Doniseti Michelotto, M.; Mello Martins, A.L.; Bolonhezi, D. Métodos De Avaliação De Maturação De Linhagens De Amendoim Em Colheita Antecipada. Encontro Sobre Cult. Amend. 2017, 1, 4–8. [Google Scholar] [CrossRef]
- Borisjuk, L.; Rolletschek, H.; Fuchs, J.; Melkus, G.; Neuberger, T. Low and High Field Magnetic Resonance for In Vivo Analysis of Seeds. Materials 2011, 4, 1426–1439. [Google Scholar] [CrossRef]
- Song, C.; Xiang, D.B.; Yan, L.; Song, Y.; Zhao, G.; Wang, Y.H.; Zhangb, B.L. Changes in Seed Growth, Levels and Distribution of Flavonoids during Tartary Buckwheat Seed Development. Plant Prod. Sci. 2016, 19, 518–527. [Google Scholar] [CrossRef]
- Mikkelsted, F.N.; Adén, D.; Nikolajsen, T.; Laursen, K.H. A Novel LIBS Method for Quantitative and High-Throughput Analysis of Macro and Micronutrients in Plants. J. Anal. At. Spectrom. 2024, 39, 2008–2020. [Google Scholar] [CrossRef]
- Gamela; Costa, V.C.; Babos, D.V.; Araújo, A.S.; Pereira-Filho, E.R. Direct Determination of Ca, K, and Mg in Cocoa Beans by Laser-Induced Breakdown Spectroscopy (LIBS): Evaluation of Three Univariate Calibration Strategies for Matrix Matching. Food Anal. Methods 2020, 13, 1017–1026. [Google Scholar] [CrossRef]
- Gamela; Costa, V.C.; Sperança, M.A.; Pereira-Filho, E.R. Laser-Induced Breakdown Spectroscopy (LIBS) and Wavelength Dispersive X-Ray Fluorescence (WDXRF) Data Fusion to Predict the Concentration of K, Mg and P in Bean Seed Samples. Food Res. Int. 2020, 132, 109037. [Google Scholar] [CrossRef]
- Li, F.; Meng, L.; Ding, W.; Wang, J.; Ge, L. Review of Energy-Dispersive X-Ray Fluorescence on Food Elements Detection. X-Ray Spectrom. 2022, 51, 346–364. [Google Scholar] [CrossRef]
- Guild, G.E.; Stangoulis, J.C.R. Screening Ca Concentration in Staple Food Crops with Energy Dispersive X-Ray Fluorescence (EDXRF). Plant Soil 2022, 473, 659–667. [Google Scholar] [CrossRef]
- Guild, G.E.; Stangoulis, J.C.R. EDXRF for Screening Micronutrients in Lentil and Sorghum Biofortification Breeding Programs. Plant Soil 2021, 463, 461–469. [Google Scholar] [CrossRef]
- Barboza da Silva, C.; Bianchini, V.J.M.; Medeiros, A.D.; Moraes, M.H.D.; Marassi, A.G.; Tannús, A. A Novel Approach for Jatropha Curcas Seed Health Analysis Based on Multispectral and Resonance Imaging Techniques. Ind. Crops Prod. 2021, 161, 113186. [Google Scholar] [CrossRef]
- Cioccia, G.; Pereira de Morais, C.; Babos, D.V.; Milori, D.M.B.P.; Alves, C.Z.; Cena, C.; Nicolodelli, G.; Marangoni, B.S. Laser-Induced Breakdown Spectroscopy Associated with the Design of Experiments and Machine Learning for Discrimination of Brachiaria Brizantha Seed Vigor. Sensors 2022, 22, 5067. [Google Scholar] [CrossRef]
- USDA. Illustrated Guide to Soil Taxonomy; Version 2; US Department of Agriculture, Natural Resources Conservation Service, National Soil Survey Center: Lincoln, NE, USA, 2015. [Google Scholar]
- Quaggio, J.A.; Zambrosi, F.C.B.; Cantarella, H.; Godoy, I.J.; Crusciol, C.A.C.; Bolonhezi, D. Amendoim (Arachis hypogaea). In Boletim 100: Recomendaçõesde Adubação e Calagem Para o Estado de São Paulo; IAC: Campinas, Brazil, 2022; pp. 243–244. [Google Scholar]
- Prela, A.; Ribeiro, A.M.A. Soma de Graus-Dia Para o Sub-Período Semeadura-Maturação Do Amendoinzeiro. Rev. Bras. Agrometeorol. 2000, 8, 321–324. [Google Scholar]
- Alexieva, V.; Sergiev, I.; Mapelli, S.; Karanov, E. The Effect of Drought and Ultraviolet Radiation on Growth and Stress Markers in Pea and Wheat. Plant Cell Environ. 2001, 24, 1337–1344. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in Isolated Chloroplasts. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Joosen, R.V.L.; Kodde, J.; Willems, L.A.J.; Ligterink, W.; Van Der Plas, L.H.W.; Hilhorst, H.W.M. Germinator: A Software Package for High-Throughput Scoring and Curve Fitting of Arabidopsis Seed Germination. Plant J. 2010, 62, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Krzyzanowski, F.C.; França-Neto, J.B.; Gomes-Junior, F.G.; Nakagawa, J. Testes de Vigor Baseado Em Desempenho de Plântulas. In Vigor de Sementes: Conceitos e Testes; Abrates: Londrina, PR, Brazil, 2020; pp. 79–140. [Google Scholar]
- Chinachoti, P.; Peter, H. Krygsman Application of Low-Resolution Nmr for Simultaneous Moisture and Oil Determination in Food (Oilseeds). In Current Protocols in Food Analytical Chemistry; John Wiley & Sons: New York, NY, USA, 2001. [Google Scholar]
- Moraes, T.B.; Colnago, L.A. Noninvasive Analyses of Food Products Using Low-Field Time-Domain NMR: A Review of Relaxometry Methods. Braz. J. Phys. 2022, 52, 43. [Google Scholar] [CrossRef]
- Castro, J.P.; Pereira-Filho, E.R. Twelve Different Types of Data Normalization for the Proposition of Classification, Univariate and Multivariate Regression Models for the Direct Analyses of Alloys by Laser-Induced Breakdown Spectroscopy (LIBS). J. Anal. At. Spectrom. 2016, 31, 2005–2014. [Google Scholar] [CrossRef]
- Rodrigues, L.; Pereira-Filho, E.R.; Pereira, F.M.V. Analytical Chemistry Nutritional Insights: Exploring ED-XRF, LIBS, and Chemometric Techniques for Macronutrient Determination in Non-Conventional Food Plants (PANC). Food Anal. Methods 2024, 17, 358–365. [Google Scholar] [CrossRef]
- R Core Team. A Language and Environment for Statistical Computing: R Foundation for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 3 August 2024).
- Hastie, T.; Tibshirani, R.; Friedman, J.H. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning with Applications in R; Springer: New York, NY, USA, 2021. [Google Scholar] [CrossRef]
- Clarke, W.R.; Lachenbruch, P.A.; Broffitt, B. How Non-Normality Affects the Quadratic Discriminant Function. Commun. Stat. Theory Methods 1979, 8, 1285–1301. [Google Scholar] [CrossRef]
- Ellis, R.H. Temporal Patterns of Seed Quality Development, Decline, and Timing of Maximum Quality during Seed Development and Maturation. Seed Sci. Res. 2019, 29, 135–142. [Google Scholar] [CrossRef]
- Zhou, W.; Branch, W.D.; Gilliam, L.; Marshall, J.A. Phytosterol Composition of Arachis hypogaea Seeds from Different Maturity Classes. Molecules 2019, 24, 106. [Google Scholar] [CrossRef]
- Ramtekey, V.; Cherukuri, S.; Kumar, S.; V, S.K.; Sheoran, S.; K, U.B.; K, B.N.; Kumar, S.; Singh, A.N.; Singh, H.V. Seed Longevity in Legumes: Deeper Insights into Mechanisms and Molecular Perspectives. Front. Plant Sci. 2022, 13, 918206. [Google Scholar] [CrossRef]
- Rehmani, M.S.; Xian, B.S.; Wei, S.; He, J.; Feng, Z.; Huang, H.; Shu, K. Seedling Establishment: The Neglected Trait in the Seed Longevity Field. Plant Physiol. Biochem. 2023, 200, 107765. [Google Scholar] [CrossRef] [PubMed]
- Sano, N.; Rajjou, L.; North, H.M.; Debeaujon, I.; Marion-Poll, A.; Seo, M. Staying Alive: Molecular Aspects of Seed Longevity. Plant Cell Physiol. 2016, 57, 660–674. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.P.J.; Rajendra Prasad, S.; Banerjee, R.; Thammineni, C. Seed Birth to Death: Dual Functions of Reactive Oxygen Species in Seed Physiology. Ann. Bot. 2015, 116, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Sardans, J.; Peñuelas, J. Potassium Control of Plant Functions: Ecological and Agricultural Implications. Plants 2021, 10, 419. [Google Scholar] [CrossRef]
- Ragel, P.; Raddatz, N.; Leidi, E.O.; Quintero, F.J.; Pardo, J.M. Regulation of K + Nutrition in Plants. Front. Plant Sci. 2019, 10, 281. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Nahar, K.; Hossain, M.S.; Al Mahmud, J.; Hossen, M.S.; Masud, A.A.C.; Moumita; Fujita, M. Potassium: A Vital Regulator of Plant Responses and Tolerance to Abiotic Stresses. Agronomy 2018, 8, 31. [Google Scholar] [CrossRef]
- Liu, C.; Liao, W. Potassium Signaling in Plant Abiotic Responses: Crosstalk with Calcium and Reactive Oxygen Species/Reactive Nitrogen Species. Plant Physiol. Biochem. 2022, 173, 110–121. [Google Scholar] [CrossRef]
- Buitink, J.; Leprince, O. Glass Formation in Plant Anhydrobiotes: Survival in the Dry State. Cryobiology 2004, 48, 215–228. [Google Scholar] [CrossRef]
- Buitink, J.; Leprince, O. Intracellular Glasses and Seed Survival in the Dry State. Comptes Rendus Biol. 2008, 331, 788–795. [Google Scholar] [CrossRef]
- Li, W.; Niu, Y.; Zheng, Y.; Wang, Z. Advances in the Understanding of Reactive Oxygen Species-Dependent Regulation on Seed Dormancy, Germination, and Deterioration in Crops. Front. Plant Sci. 2022, 13, 826809. [Google Scholar] [CrossRef]
- Kurek, K.; Plitta-Michalak, B.; Ratajczak, E. Reactive Oxygen Species as Potential Drivers of the Seed Aging Process. Plants 2019, 8, 174. [Google Scholar] [CrossRef] [PubMed]
- Larios, G.S.; Nicolodelli, G.; Senesi, G.S.; Ribeiro, M.C.S.; Xavier, A.A.P.; Milori, D.M.B.P.; Alves, C.Z.; Marangoni, B.S.; Cena, C. Laser-Induced Breakdown Spectroscopy as a Powerful Tool for Distinguishing High- and Low-Vigor Soybean Seed Lots. Food Anal. Methods 2020, 13, 1691–1698. [Google Scholar] [CrossRef]
- Crusciol, C.A.C.; Portugal, J.R.; Bossolani, J.W.; Moretti, L.G.; Fernandes, A.M.; Garcia, J.L.N.; Garcia, G.L.d.B.; Pilon, C.; Cantarella, H. Dynamics of Macronutrient Uptake and Removal by Modern Peanut Cultivars. Plants 2021, 10, 2167. [Google Scholar] [CrossRef] [PubMed]
- Thor, K. Calcium—Nutrient and Messenger. Front. Plant Sci. 2019, 10, 440. [Google Scholar] [CrossRef]
- Yang, S.; Wang, J.; Tang, Z.; Guo, F.; Zhang, Y.; Zhang, J.; Meng, J.; Zheng, L.; Wan, S.; Li, X. Transcriptome of Peanut Kernel and Shell Reveals the Mechanism of Calcium on Peanut Pod Development. Sci. Rep. 2020, 10, 15723. [Google Scholar] [CrossRef]
- Verma, G.; Khan, S.; Agarwal, S.K.; Sharma, S. Role of Apoplastic Calcium during Germination and Initial Stages of Seedling Establishment in Vigna radiata Seeds. J. Plant Physiol. 2019, 236, 66–73. [Google Scholar] [CrossRef]
- Ebone, L.A.; Caverzan, A.; Tagliari, A.; Chiomento, J.L.T.; Silveira, D.C.; Chavarria, G. Soybean Seed Vigor: Uniformity and Growth as Key Factors to Improve Yield. Agronomy 2020, 10, 545. [Google Scholar] [CrossRef]
- Bagateli, J.R.; Dörr, C.S.; Schuch, L.O.B.; Meneghello, G.E. Productive Performance of Soybean Plants Originated from Seed Lots with Increasing Vigor Levels. J. Seed Sci. 2019, 41, 151–159. [Google Scholar] [CrossRef]
- Reed, R.C.; Bradford, K.J.; Khanday, I. Seed Germination and Vigor: Ensuring Crop Sustainability in a Changing Climate. Heredity 2022, 128, 450–459. [Google Scholar] [CrossRef]
- Galletti, P.A.; Carvalho, M.E.A.; Hirai, W.Y.; Brancaglioni, V.A.; Arthur, V.; Barboza da Silva, C. Integrating Optical Imaging Tools for Rapid and Non-Invasive Characterization of Seed Quality: Tomato (Solanum lycopersicum L.) and Carrot (Daucus carota L.) as Study Cases. Front. Plant Sci. 2020, 11, 577851. [Google Scholar] [CrossRef]
- Elmasry, G.; Mandour, N.; Wagner, M.H.; Demilly, D.; Verdier, J.; Belin, E.; Rousseau, D. Utilization of Computer Vision and Multispectral Imaging Techniques for Classification of Cowpea (Vigna unguiculata) Seeds. Plant Methods 2019, 15, 24. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Yang, L.; Zhang, Z. Non-Destructive Identification of Single Hard Seed via Multispectral Imaging Analysis in Six Legume Species. Plant Methods 2020, 16, 116. [Google Scholar] [CrossRef] [PubMed]
- Arya, S.S.; Salve, A.R.; Chauhan, S. Peanuts as Functional Food: A Review. J. Food Sci. Technol. 2016, 53, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Batista, T.B.; Mastrangelo, C.B.; de Medeiros, A.D.; Petronilio, A.C.P.; Fonseca de Oliveira, G.R.; Santos, I.L.; Crusciol, C.A.C.; Amaral da Silva, E.A. A Reliable Method to Recognize Soybean Seed Maturation Stages Based on Autofluorescence-Spectral Imaging Combined with Machine Learning Algorithms. Front. Plant Sci. 2022, 13, 914287. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fonseca de Oliveira, G.R.; Hirai, W.Y.; Ferreira, D.S.; Silva, K.P.O.M.d.; Silva, G.C.; Moraes, T.B.; Mastrangelo, C.B.; Pereira, F.M.V.; Pereira-Filho, E.R.; Amaral da Silva, E.A. Spectroscopy Technologies to Screen Peanut Seeds with Superior Vigor Through “Chemical Fingerprinting”. Agronomy 2024, 14, 2529. https://doi.org/10.3390/agronomy14112529
Fonseca de Oliveira GR, Hirai WY, Ferreira DS, Silva KPOMd, Silva GC, Moraes TB, Mastrangelo CB, Pereira FMV, Pereira-Filho ER, Amaral da Silva EA. Spectroscopy Technologies to Screen Peanut Seeds with Superior Vigor Through “Chemical Fingerprinting”. Agronomy. 2024; 14(11):2529. https://doi.org/10.3390/agronomy14112529
Chicago/Turabian StyleFonseca de Oliveira, Gustavo Roberto, Welinton Yoshio Hirai, Dennis Silva Ferreira, Karolyne Priscila Oliveira Mota da Silva, Giovani Chaves Silva, Tiago Bueno Moraes, Clissia Barboza Mastrangelo, Fabiola Manhas Verbi Pereira, Edenir Rodrigues Pereira-Filho, and Edvaldo Aparecido Amaral da Silva. 2024. "Spectroscopy Technologies to Screen Peanut Seeds with Superior Vigor Through “Chemical Fingerprinting”" Agronomy 14, no. 11: 2529. https://doi.org/10.3390/agronomy14112529
APA StyleFonseca de Oliveira, G. R., Hirai, W. Y., Ferreira, D. S., Silva, K. P. O. M. d., Silva, G. C., Moraes, T. B., Mastrangelo, C. B., Pereira, F. M. V., Pereira-Filho, E. R., & Amaral da Silva, E. A. (2024). Spectroscopy Technologies to Screen Peanut Seeds with Superior Vigor Through “Chemical Fingerprinting”. Agronomy, 14(11), 2529. https://doi.org/10.3390/agronomy14112529








