QTL Mapping and Functional Identification of Candidate Genes Regulated by Sinorhizobium fredii HH103 and Associated with Nodulation Traits in Soybean
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, and Soybean Genetic Materials
2.2. Nodulation Tests and Detection of Nitrogenase Activity
2.3. Whole-Genome Sequencing of the CSSL Population and Natural Varieties of Soybean
2.4. QTL Mapping and Screening of Candidate Genes
2.5. Verification of Candidate Genes Using qRT-PCR
2.6. Nodulation and Hairy Root Transformation in Soybean
2.7. Subcellular Localization of Candidate Genes
2.8. Phylogenetic Analysis of the Candidate Gene
2.9. Transcriptome Analysis
2.10. Haplotype Analysis of Candidate Gene
3. Results
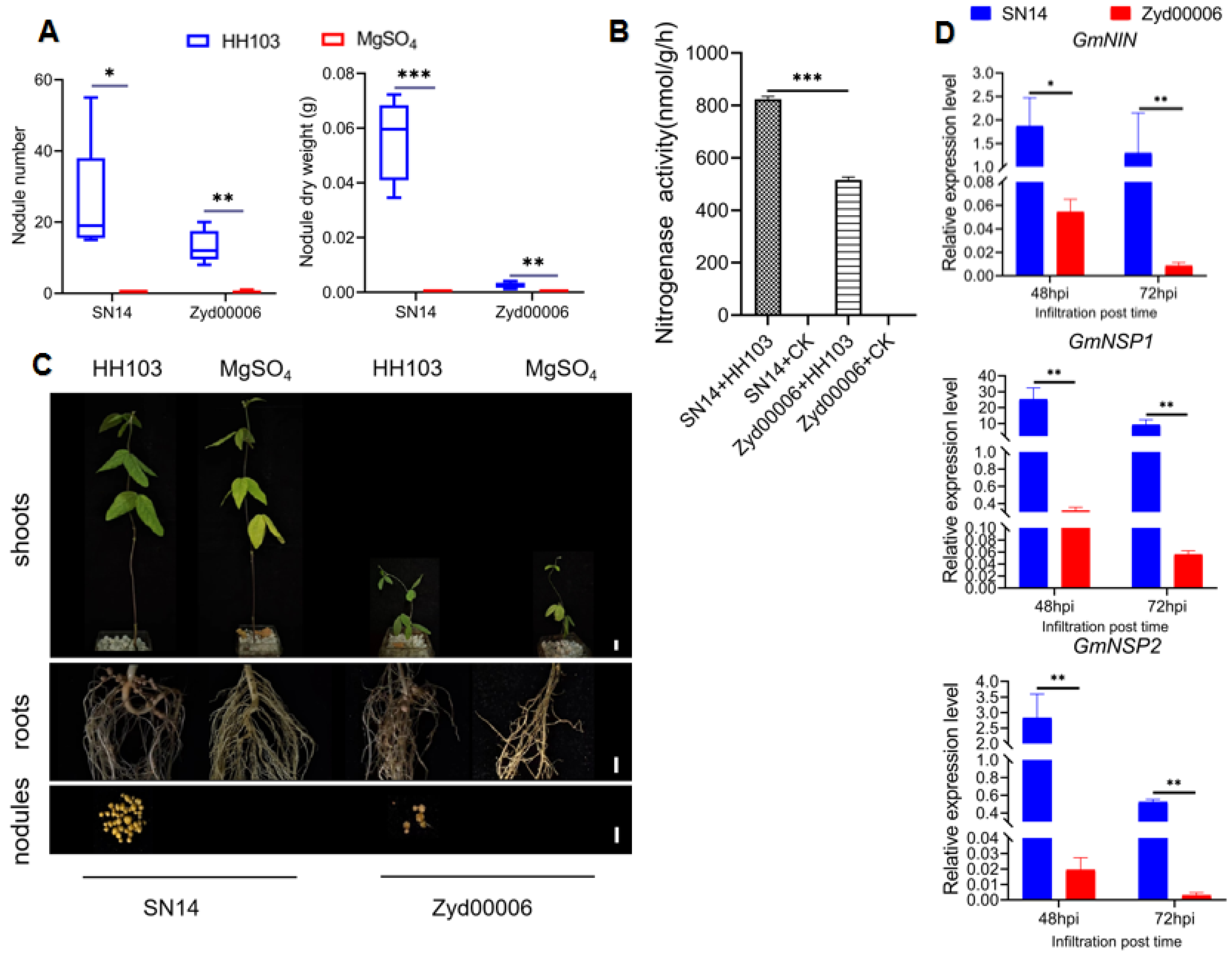
3.1. Improved Cultivar SN14 and Wild Soybean Zyd00006 Exhibited Different Phenotypes after HH103 Infection
3.2. Identification of Genomic Regions Associated with Different Nodulation Phenotypes in Soybean CSSL Population
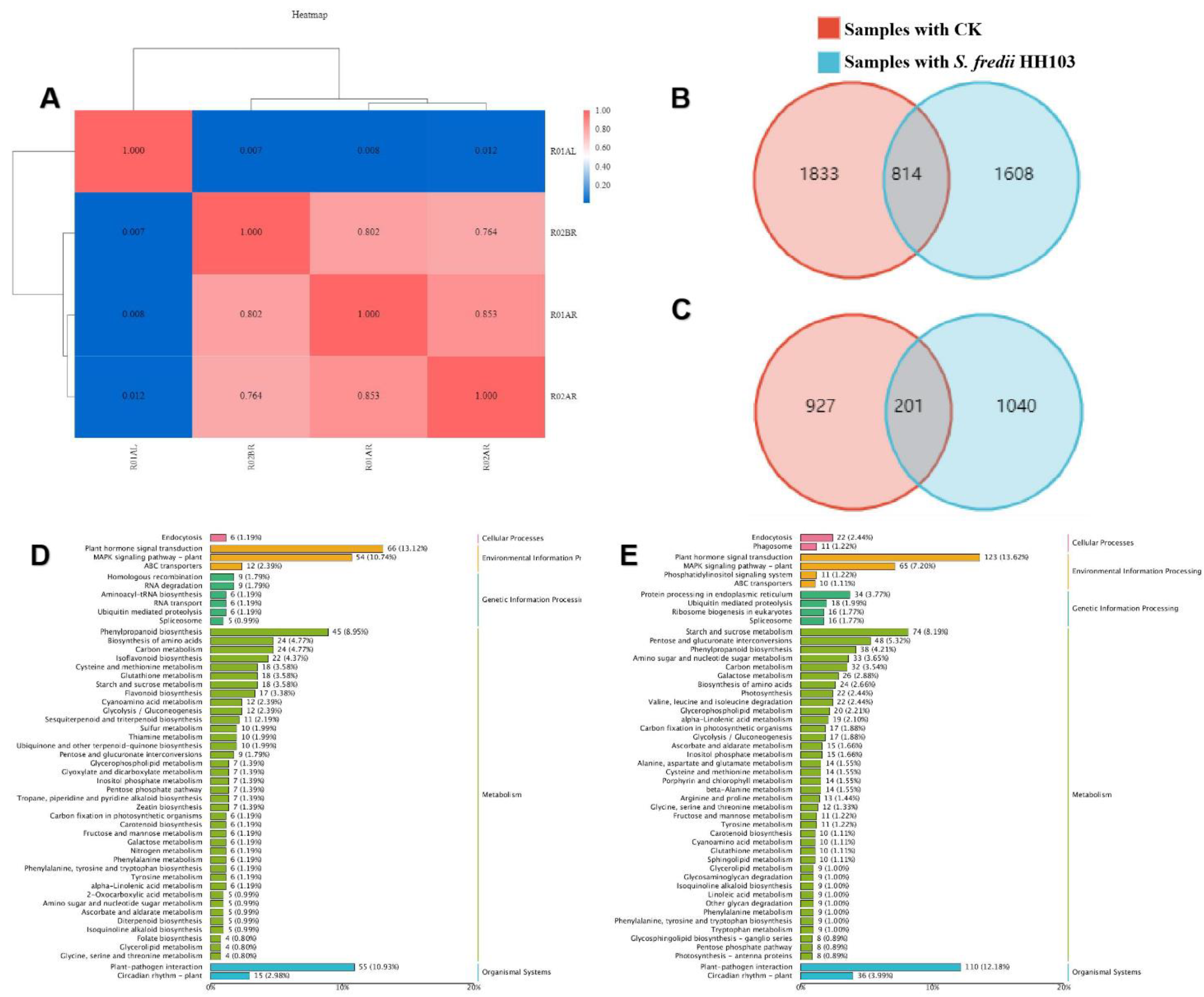
3.3. Analysis of the Response of SN14 to S. fredii HH103 Infection Using RNA-Seq
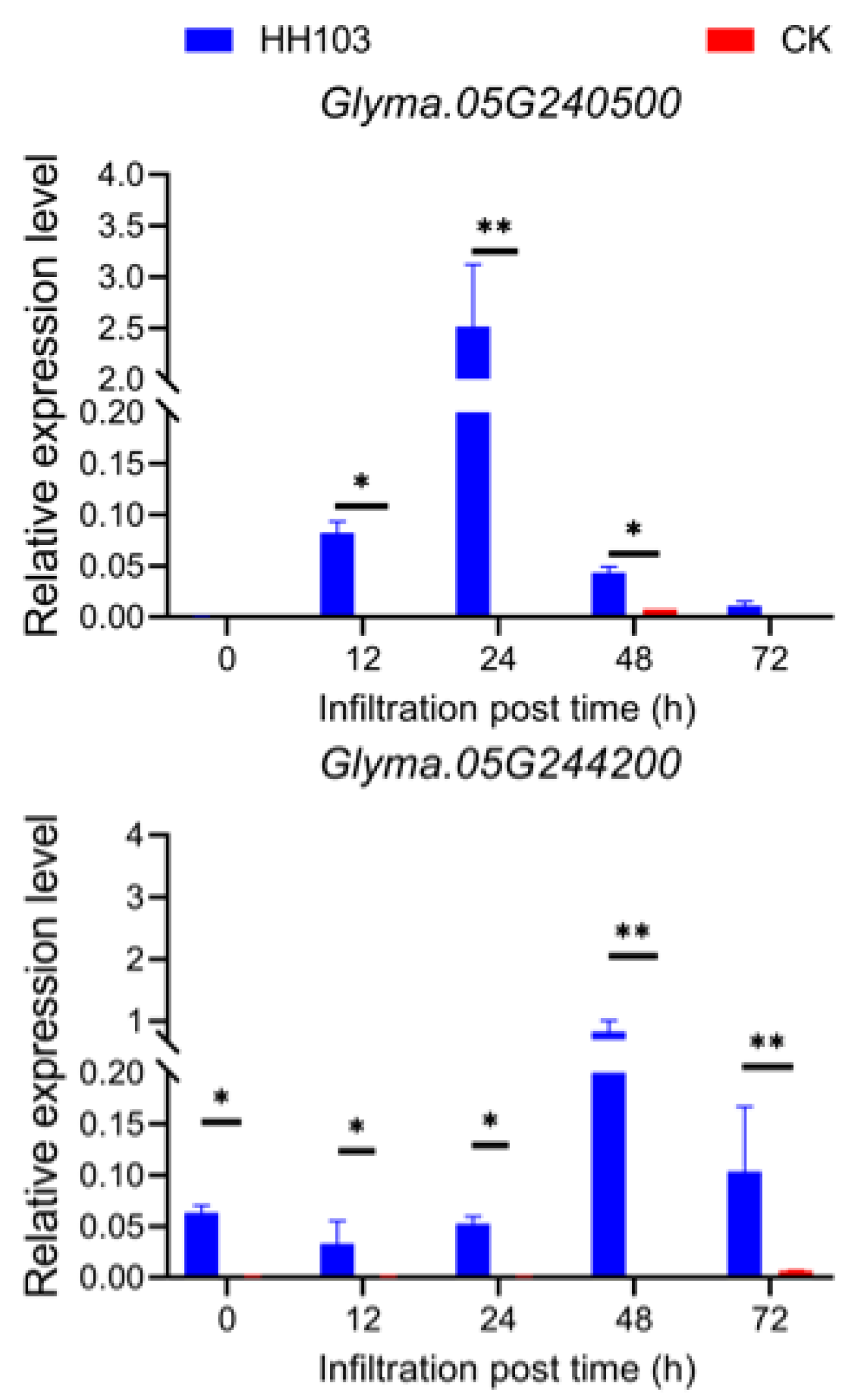
3.4. Candidate Gene Prediction and qRT-PCR Validation
3.5. Glyma.05g240500 Promotes Nodulation in Transgenic Root Hairy Plants and Targeted Plant Nucleus and Cytosol
3.6. Haplotype Analysis of Glyma.05g240500 in CSSL Population
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dong, L.; Fang, C.; Cheng, Q.; Su, T.; Kou, K.; Kong, L.; Zhang, C.; Li, H.; Hou, Z.; Zhang, Y.; et al. Genetic basis and adaptation trajectory of soybean from its temperate origin to tropics. Nat. Commun. 2021, 12, 5445. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R. Soybean: Market Driven Research Needs. In Genetics and Genomics of Soybean; Springer: New York, NY, USA, 2008; pp. 3–15. [Google Scholar]
- Cassman, K.G.; Dobermann, A. Nitrogen and the future of agriculture: 20 years on. Ambio 2022, 51, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Good, A.G.; Beatty, P.H. Fertilizing Nature: A Tragedy of Excess in the Commons. PLoS Biol. 2011, 9, e1001124. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zheng, X.; Wei, X.; Kai, Z.; Xu, Y. Excessive application of chemical fertilizer and organophosphorus pesticides induced total phosphorus loss from planting causing surface water eutrophication. Sci. Rep. 2021, 11, 23015. [Google Scholar] [CrossRef]
- Waqas, M.; Hawkesford, M.J.; Geilfus, C.-M. Feeding the world sustainably: Efficient nitrogen use. Trends Plant Sci. 2023, 28, 505–508. [Google Scholar] [CrossRef]
- Sachs, J.L.; Quides, K.W.; Wendlandt, C.E. Legumes versus rhizobia: A model for ongoing conflict in symbiosis. New Phytol. 2018, 219, 1199–1206. [Google Scholar] [CrossRef]
- Yang, J.; Lan, L.; Jin, Y.; Yu, N.; Wang, D.; Wang, E. Mechanisms underlying legume–rhizobium symbioses. J. Inter. Plant Biol. 2022, 64, 244–267. [Google Scholar] [CrossRef]
- Pérez-Pizá, M.C.; Cejas, E.; Zilli, C.; Prevosto, L.; Mancinelli, B.; Santa-Cruz, D.; Yannarelli, G.; Balestrasse, K. Enhancement of soybean nodulation by seed treatment with non–thermal plasmas. Sci. Rep. 2020, 10, 4917. [Google Scholar] [CrossRef]
- Vogel, J.T.; Liu, W.; Olhoft, P.; Crafts-Brandner, S.J.; Pennycooke, J.C.; Christiansen, N. Soybean Yield Formation Physiology—A Foundation for Precision Breeding Based Improvement. Front. Plant Sci. 2021, 12, 719706. [Google Scholar] [CrossRef]
- Ravelombola, W.; Qin, J.; Shi, A.; Song, Q.; Yuan, J.; Wang, F.; Chen, P.; Yan, L.; Feng, Y.; Zhao, T.; et al. Genome-wide association study and genomic selection for yield and related traits in soybean. PLoS ONE 2021, 16, e0255761. [Google Scholar] [CrossRef]
- Liang, H.-Z.; Li, W.-D.; Wang, H.; Fang, X.-J. Genetic effects on seed traits in soybean. Acta Genet. Sin. 2005, 32, 1199–1204. [Google Scholar] [PubMed]
- Xia, Z.; Zhai, H.; Lü, S.; Wu, H.; Zhang, Y. Recent Achievement in Gene Cloning and Functional Genomics in Soybean. Sci. World J. 2013, 2013, 281367. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Pan, X.; Wang, F.; Liu, C.; Wang, X.; Li, Y.; Zhang, Q. Novel QTL and Meta-QTL Mapping for Major Quality Traits in Soybean. Front. Plant Sci. 2021, 12, 774270. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Jin, H.; Zhu, Y.-H.; Siddique, K.H.M. Nodule Formation and Nitrogen Use Efficiency Are Important for Soybean to Adapt to Water and P Deficit Conditions. Agriculture 2022, 12, 1326. [Google Scholar]
- Tominaga, A.; Gondo, T.; Akashi, R.; Zheng, S.; Arima, S.; Suzuki, A. Quantitative trait locus analysis of symbiotic nitrogen fixation activity in the model legume Lotus japonicus. J. Plant Res. 2012, 125, 395–406. [Google Scholar] [CrossRef]
- Huo, X.; Li, X.; Du, H.; Kong, Y.; Tian, R.; Li, W.; Zhang, C. Genetic loci and candidate genes of symbiotic nitrogen fixation–related characteristics revealed by a genome-wide association study in soybean. Mol. Breed. 2019, 39, 127. [Google Scholar] [CrossRef]
- Kamfwa, K.; Cichy, K.A.; Kelly, J.D. Identification of quantitative trait loci for symbiotic nitrogen fixation in common bean. Theor. Appl. Genet. 2019, 132, 1375–1387. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, M.; Chen, Q.; Xin, D.; Sun, X. Mapping quantitative trait loci related to nodule number in soybean (Glycine max (L.) Merr.) in response to the Sinorhizobium (Ensifer) fredii HH103 NopT type III effector. J. Plant Interact. 2021, 16, 126–135. [Google Scholar] [CrossRef]
- Wang, J.; Wang, J.; Ma, C.; Zhou, Z.; Yang, D.; Zheng, J.; Wang, Q.; Li, H.; Zhou, H.; Sun, Z.; et al. QTL Mapping and Data Mining to Identify Genes Associated With the Sinorhizobium fredii HH103 T3SS Effector NopD in Soybean. Front. Plant Sci. 2020, 11, 453. [Google Scholar] [CrossRef]
- Ni, H.; Peng, Y.; Wang, J.; Wang, J.; Yuan, Y.; Fu, T.; Zhu, Z.; Zhang, J.; Pan, X.; Cui, Z.; et al. Mapping of Quantitative Trait Loci Underlying Nodule Traits in Soybean (Glycine max (L.) Merr.) and Identification of Genes Whose Expression Is Affected by the Sinorhizobium fredii HH103 Effector Proteins NopL and NopT. Agronomy 2022, 12, 946. [Google Scholar] [CrossRef]
- Xin, D.; Qi, Z.; Jiang, H.; Hu, Z.; Zhu, R.; Hu, J.; Han, H.; Hu, G.; Liu, C.; Chen, Q. QTL Location and Epistatic Effect Analysis of 100-Seed Weight Using Wild Soybean (Glycine soja Sieb. & Zucc.) Chromosome Segment Substitution Lines. PLoS ONE 2016, 11, e0149380. [Google Scholar]
- Wang, J.; Wang, J.; Liu, C.; Ma, C.; Li, C.; Zhang, Y.; Qi, Z.; Zhu, R.; Shi, Y.; Zou, J.; et al. Identification of Soybean Genes Whose Expression is Affected by the Ensifer fredii HH103 Effector Protein NopP. Int. J. Mol. Sci. 2018, 19, 3438. [Google Scholar] [CrossRef]
- Gremaud, M.F.; Harper, J.E. Selection and initial characterization of partially nitrate tolerant nodulation mutants of soybean. Plant Physiol. 1989, 89, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Wang, J.; Ma, C.; Ma, S.; Zheng, H.; Tian, H.; Wang, X.; Wang, Y.; Jiang, H.; Wang, J.; Zhang, Z.; et al. Genetic variation in GmCRP contributes to nodulation in soybean (Glycine max Merr.). Crop. J. 2023, 11, 332–344. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.; Chen, L.; Fu, Y.; Li, C.; Qi, Z.; Zou, J.; Zhu, R.; Li, S.; Wei, W.; et al. Mining for genes encoding proteins associated with NopL of Sinorhizobium fredii HH103 using quantitative trait loci in soybean (Glycine max Merr.) recombinant inbred lines. Plant Soil. 2018, 431, 245–255. [Google Scholar] [CrossRef]
- Kereszt, A.; Li, D.; Indrasumunar, A.; Nguyen, C.; Nontachaiyapoom, S.; Kinkema, M.; Kinkema, M. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2007, 2, 948–952. [Google Scholar] [CrossRef]
- Li, F.; Ni, H.; Yan, W.; Xie, Y.; Liu, X.; Tan, X.; Zhang, L.; Zhang, S. Overexpression of an aquaporin protein from Aspergillus glaucus confers salt tolerance in transgenic soybean. Transgenic. Res. 2021, 30, 727–737. [Google Scholar] [CrossRef]
- Zhao, Y.; Cui, Y.; Huang, S.; Yu, J.; Wang, X.; Xin, D.; Li, X.; Liu, Y.; Dai, Y.; Qi, Z.; et al. Genome-Wide Analysis of the Glucose-6-Phosphate Dehydrogenase Family in Soybean and Functional Identification of GmG6PDH2 Involvement in Salt Stress. Front. Plant Sci. 2020, 11, 214. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Song, S.; Wang, J.; Yang, X.; Zhang, X.; Xin, X.; Liu, C.; Zou, J.; Cheng, X.; Zhang, N.; Hu, Y.; et al. GsRSS3L, a Candidate Gene Underlying Soybean Resistance to Seedcoat Mottling Derived from Wild Soybean (Glycine soja Sieb. and Zucc). Int. J. Mol. Sci. 2022, 23, 7577. [Google Scholar] [CrossRef]
- Wang, J.; Feng, H.; Jia, X.; Ma, S.; Ma, C.; Wang, Y.; Pan, S.; Chen, Q.; Xin, D.; Liu, C. Identifications of QTLs and Candidate Genes Associated with Pseudomonas syringae Responses in Cultivated Soybean (Glycine max) and Wild Soybean (Glycine soja). Int. J. Mol. Sci. 2023, 24, 4618. [Google Scholar] [CrossRef] [PubMed]
- Cunicelli, M.; Olukolu, B.A.; Sams, C.; Schneider, L.; West, D.; Pantalone, V. Mapping and identification of QTL in 5601T × U99-310255 RIL population using SNP genotyping: Soybean seed quality traits. Mol. Biol. Rep. 2022, 49, 6623–6632. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-H.; Reif, J.C.; Hong, H.-L.; Li, H.-H.; Liu, Z.-X.; Ma, Y.-S.; Li, J.; Tian, Y.; Li, Y.-F.; Li, W.-B.; et al. Genome-wide association mapping of QTL underlying seed oil and protein contents of a diverse panel of soybean accessions. Plant Sci. 2018, 266, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Huang, Z.; Ma, R.; Chen, J.; Zhang, Z.; Yrjälä, K. Genome-wide identification and analysis of the heat shock transcription factor family in moso bamboo (Phyllostachys edulis). Sci. Rep. 2021, 11, 16492. [Google Scholar] [CrossRef]
- Li, P.-S.; Yu, T.-F.; He, G.-H.; Chen, M.; Zhou, Y.-B.; Chai, S.-C.; Xu, Z.-S.; Ma, Y.-Z. Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genom. 2014, 15, 1009. [Google Scholar] [CrossRef]
- Yang, X.; Zhu, W.; Zhang, H.; Liu, N.; Tian, S. Heat shock factors in tomatoes: Genome-wide identification, phylogenetic analysis and expression profiling under development and heat stress. PeerJ 2016, 4, e1961. [Google Scholar] [CrossRef]
- Nover, L.; Bharti, K.; Döring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 2001, 6, 177–189. [Google Scholar] [CrossRef]
- Xiang, J.; Ran, J.; Zou, J.; Zhou, X.; Liu, A.; Zhang, X.; Peng, Y.; Tang, N.; Luo, G.; Chen, X. Heat shock factor OsHsfB2b negatively regulates drought and salt tolerance in rice. Plant Cell Rep. 2013, 32, 1795–1806. [Google Scholar] [CrossRef]
- Kim, T.; Samraj, S.; Jiménez, J.; Gómez, C.; Liu, T.; Begcy, K. Genome-wide identification of heat shock factors and heat shock proteins in response to UV and high intensity light stress in lettuce. BMC Plant Biol. 2021, 21, 185. [Google Scholar] [CrossRef]
- Gomez-Pastor, R.; Burchfiel, E.T.; Thiele, D.J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Bio. 2018, 19, 4–19. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Liu, W.; Yu, L.; Guo, Z.; Chen, Z.; Jiang, S.; Xu, H.; Fang, H.; Wang, Y.; Zhang, Z.; et al. HEAT SHOCK FACTOR A8a Modulates Flavonoid Synthesis and Drought Tolerance. Plant Physiol. 2020, 184, 1273–1290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Agyenim-Boateng, K.; Zhang, S.; Gu, Y.; Qi, J.; Azam, M.; Ma, C.; Li, Y.; Feng, Y.; Liu, Y.; et al. QTL Mapping for Seed Tocopherol Content in Soybean. Agronomy 2023, 13, 1188. [Google Scholar] [CrossRef]
- Qi, Z.; Huang, L.; Zhu, R.; Xin, D.; Liu, C.; Han, X.; Jiang, H.; Hong, W.; Hu, G.; Zheng, H.; et al. A High-Density Genetic Map for Soybean Based on Specific Length Amplified Fragment Sequencing. PLoS ONE 2014, 9, e104871. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Dong, L.; Fang, C.; Liu, S.; Kong, L.; Cheng, Q.; Chen, L.; Su, T.; Nan, H.; Zhang, D.; et al. Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 2020, 52, 428–436. [Google Scholar] [CrossRef]
- Lam, H.-M.; Xu, X.; Liu, X.; Chen, W.; Yang, G.; Wong, F.; Li, M.; He, W.; Qin, N.; Wang, B.; et al. Resequencing of 31 Wild and Cultivated Soybean Genomes Identifies Patterns of Genetic Diversity and Selection. Nat. Genet. 2010, 42, 1053–1059. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Sun, Y.; Shan, Z.; He, J.; Wang, N.; Gai, J.; Li, Y. Natural variation in the promoter of GsERD15B affects salt tolerance in soybean. Plant. Biotechnol. J. 2020, 19, 1155–1169. [Google Scholar] [CrossRef]
- Chen, Q.J.; Zhou, H.M.; Chen, J.; Wang, X.C. Using a modified TA cloning method to create entry clones. Anal. Biochem. 2006, 358, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Kurose, T.; Hino, T.; Tanaka, K.; Kawamukai, M.; Niwa, Y.; Kimura, T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007, 104, 34–41. [Google Scholar] [CrossRef]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Schmidhuber, S.; Ludwig, W.; Schleifer, K.H. Construction of a DNA probe for the specific identification of Streptococcus oralis. J. Clin. Microbiol. 1988, 26, 1042–1044. [Google Scholar] [CrossRef] [PubMed]




| CSSLs (n = 195) | Parents (Average) | |||||
|---|---|---|---|---|---|---|
| Traits | Average | Standard Deviation | Coefficient of Variation | SN14 | Zyd00006 | |
| HH103 | Nodule number | 48.38974 | 32.93076 | 0.68053 | 22.6 ± 55.3 | 12.6 ± 30.3 * |
| Nodule dry weight (g) | 0.06418 | 0.03929 | 0.61216 | 0.02566 ± 0.000788 | 0.00974 ± 0.000012508 * | |
| QTL | Chr/LG | Start Position | End Position | LOD | PVE (%) | ADD |
|---|---|---|---|---|---|---|
| qNN-1 | Chr02/D1b | 43,852,453 | 43,880,682 | 3.5025 | 6.5895 | 32.6421 |
| qNDW-1 | Chr05/A1 | 41,425,769 | 41,907,158 | 3.2988 | 8.8072 | −0.0184 |
| qNDW-2 | Chr20/I | 47,302,953 | 47,897,802 | 2.6613 | 7.3791 | 0.0185 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, H.; Tian, S.; Zhang, G.; Huo, J.; Tian, H.; Peng, Y.; Yu, K.; Chen, Q.; Wang, J.; Xin, D.; et al. QTL Mapping and Functional Identification of Candidate Genes Regulated by Sinorhizobium fredii HH103 and Associated with Nodulation Traits in Soybean. Agronomy 2023, 13, 2037. https://doi.org/10.3390/agronomy13082037
Ni H, Tian S, Zhang G, Huo J, Tian H, Peng Y, Yu K, Chen Q, Wang J, Xin D, et al. QTL Mapping and Functional Identification of Candidate Genes Regulated by Sinorhizobium fredii HH103 and Associated with Nodulation Traits in Soybean. Agronomy. 2023; 13(8):2037. https://doi.org/10.3390/agronomy13082037
Chicago/Turabian StyleNi, Hejia, Siyi Tian, Guoqing Zhang, Jingyi Huo, Huilin Tian, Yang Peng, Kaixin Yu, Qingshan Chen, Jinhui Wang, Dawei Xin, and et al. 2023. "QTL Mapping and Functional Identification of Candidate Genes Regulated by Sinorhizobium fredii HH103 and Associated with Nodulation Traits in Soybean" Agronomy 13, no. 8: 2037. https://doi.org/10.3390/agronomy13082037
APA StyleNi, H., Tian, S., Zhang, G., Huo, J., Tian, H., Peng, Y., Yu, K., Chen, Q., Wang, J., Xin, D., & Liu, C. (2023). QTL Mapping and Functional Identification of Candidate Genes Regulated by Sinorhizobium fredii HH103 and Associated with Nodulation Traits in Soybean. Agronomy, 13(8), 2037. https://doi.org/10.3390/agronomy13082037








