Recognition of Tomato Leaf Diseases Based on DIMPCNET
Abstract
1. Introduction
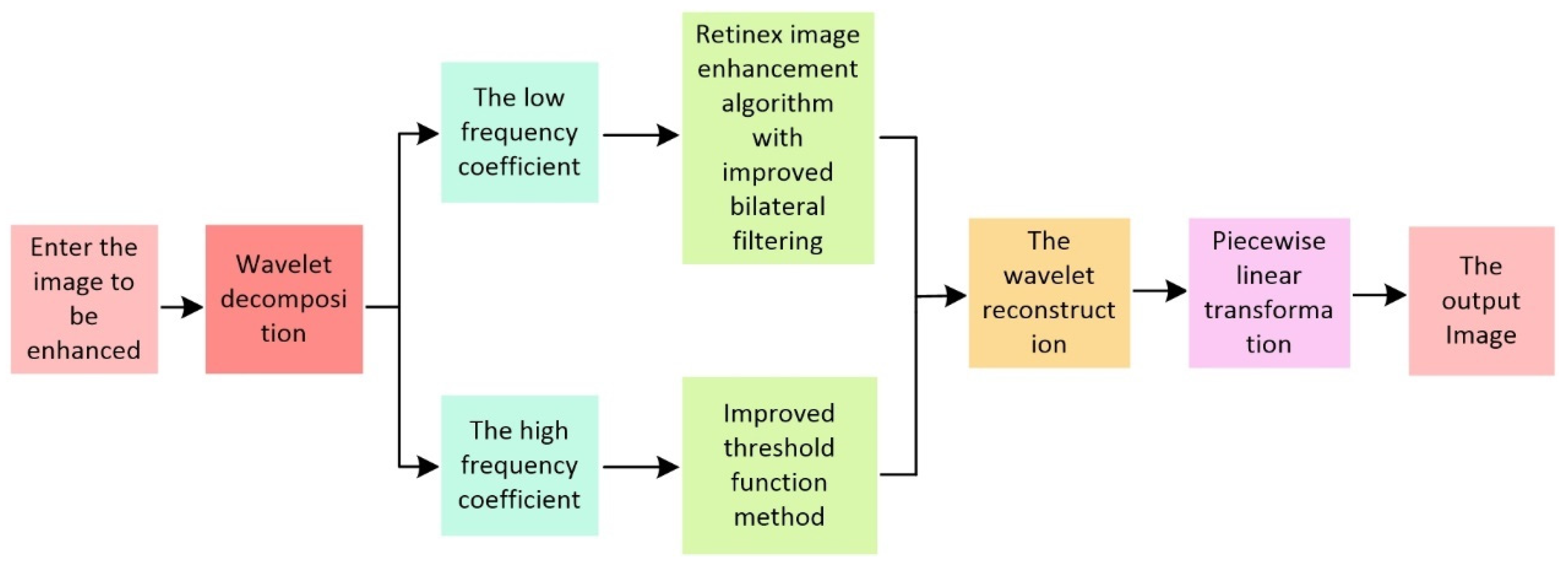
- In order to optimize the performance of disease features, an improved IBFTF image enhancement algorithm is proposed, in which wavelet transform is used to combine image enhancement and de-noising. Firstly, the noise image is decomposed by wavelet, obtaining the low frequency (LF) and high frequency (HF) coefficients. The HF coefficients were denoised using an enhanced threshold function method, while the LF coefficients were denoised using an improved bilateral filtering Retinex image enhancing algorithm. Then, the rebuilt image is obtained by inverse wavelet transform. Finally, to boost the comparison of the rebuilt image, linear segmentation processing is utilized. In comparison to existing algorithms, this approach can remove the halo effect, image blur, detail loss, and grayscale phenomenon from the image and effectively remove noise;
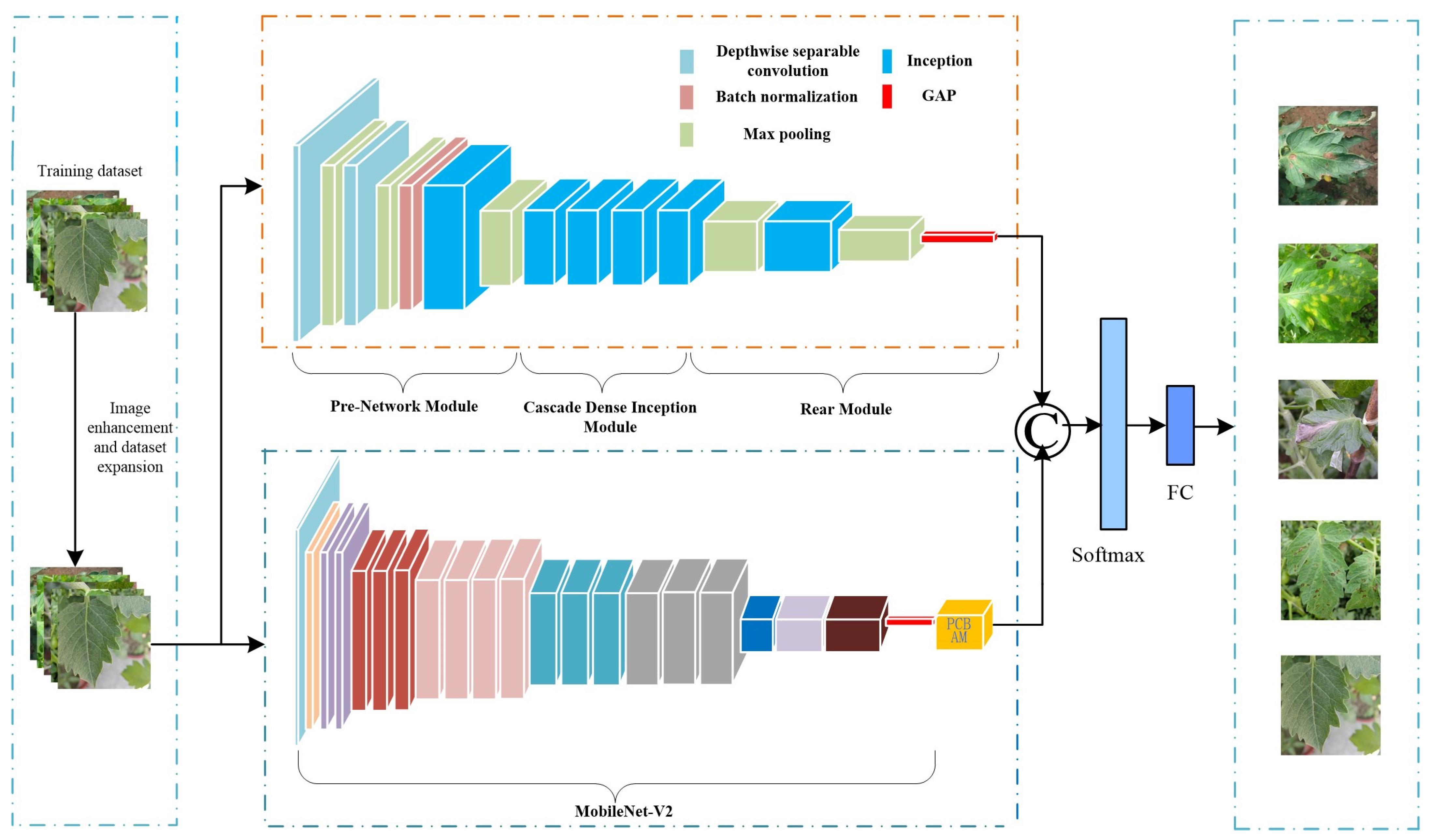
- Aiming at the problems of complex background of natural scene images, small differences in disease classification, and difficult model recognition, we propose a method of tomato leaf disease recognition based on DIMPCNET. The method is described below.
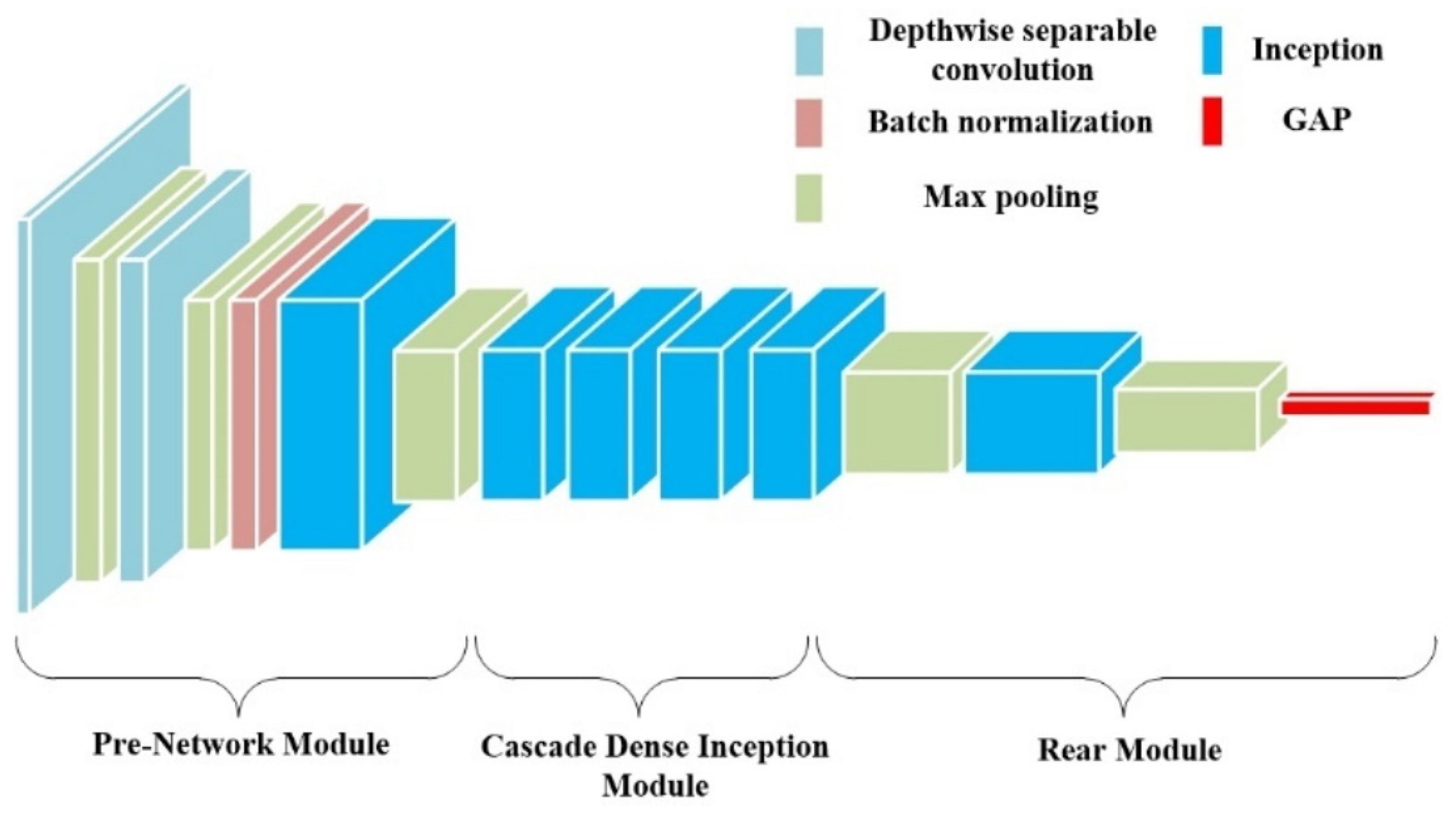
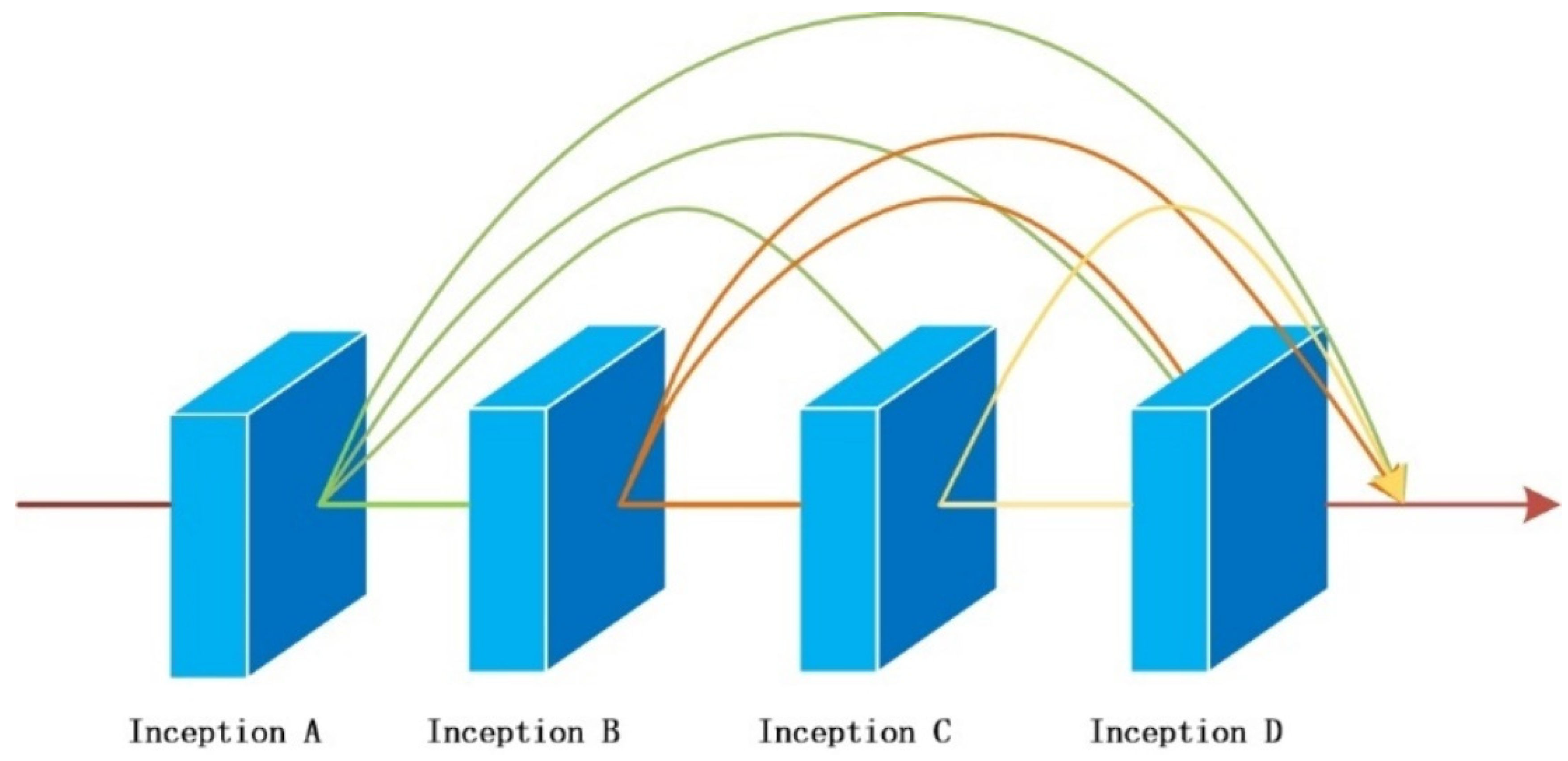
- A new depth Convolutional neural network module DI is proposed, which uses depth separable convolution to construct the first two layers of convolution, reducing the number of parameters and avoiding overfitting of the model. We introduce the initial structure into the model to enhance the extraction performance of multi-scale pathological features and adopt a dense connection strategy for the four initial structures to improve the response of the network to features and alleviate gradient disappearance. We have made progress in extracting the depth characteristics of the disease area, reducing the similarity difference between different disease leaves, solving the problem of indistinguishable caused by highly similar disease leaves, and enhancing the overall performance of the model;
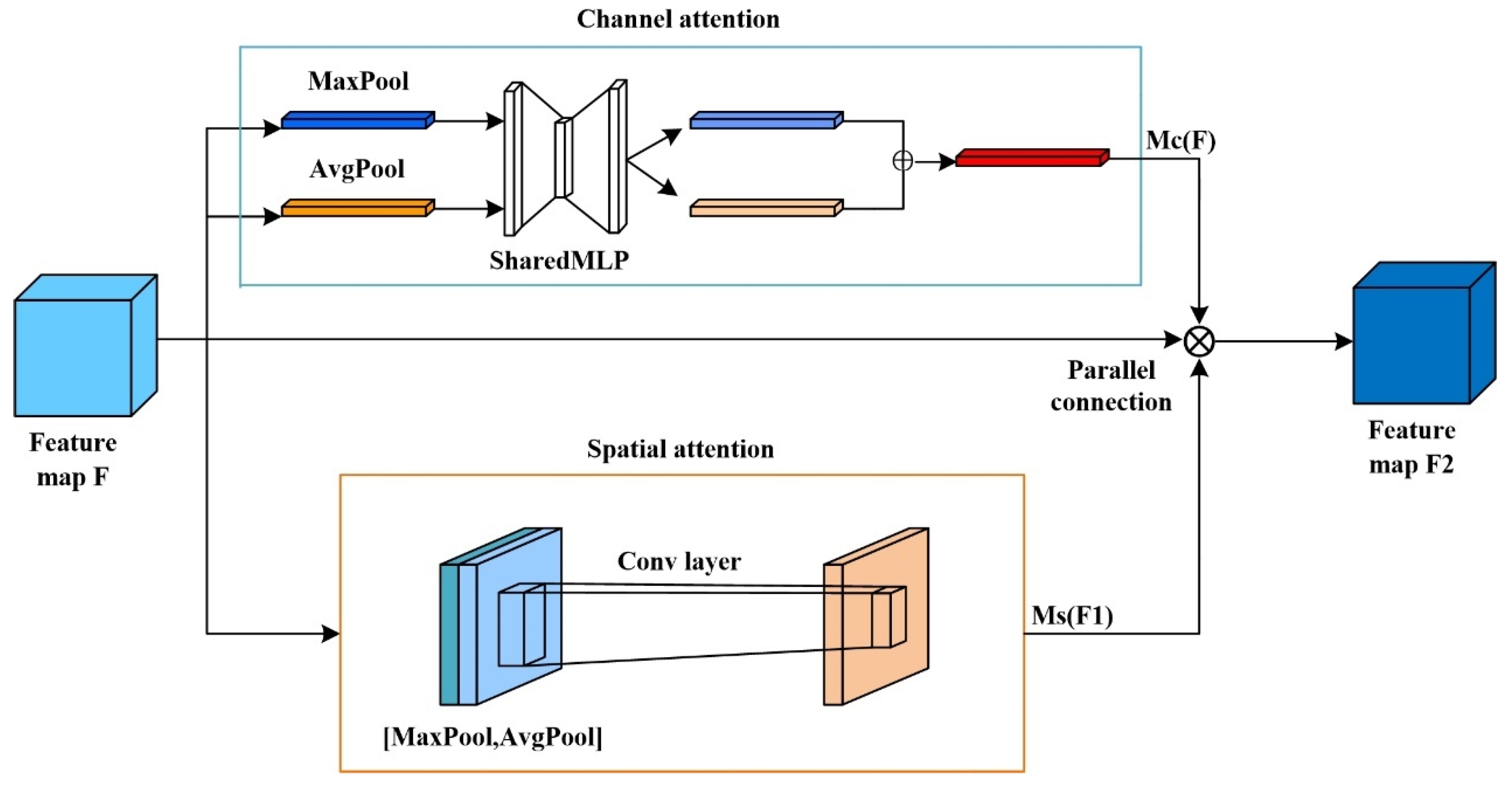
- A new hybrid attention module, PCBAM, is proposed. It connects channel attention and spatial attention in parallel, which solves the interference problem caused by serial connection, effectively improves the feature extraction ability of the model, weakens unnecessary features, and reduces the influence of complex background on the discrimination results;
- The method proposed in this paper achieved recognition accuracy of 94.44% and an F1 value of 0.9475 for the identification of five types of tomato leaf diseases. It is effective to identify tomato leaf diseases with complex backgrounds and high similarity between classes. This allows agricultural experts and scholars to better apply this technology to the prevention and control of tomato diseases so as to effectively alleviate food production problems to a certain extent.
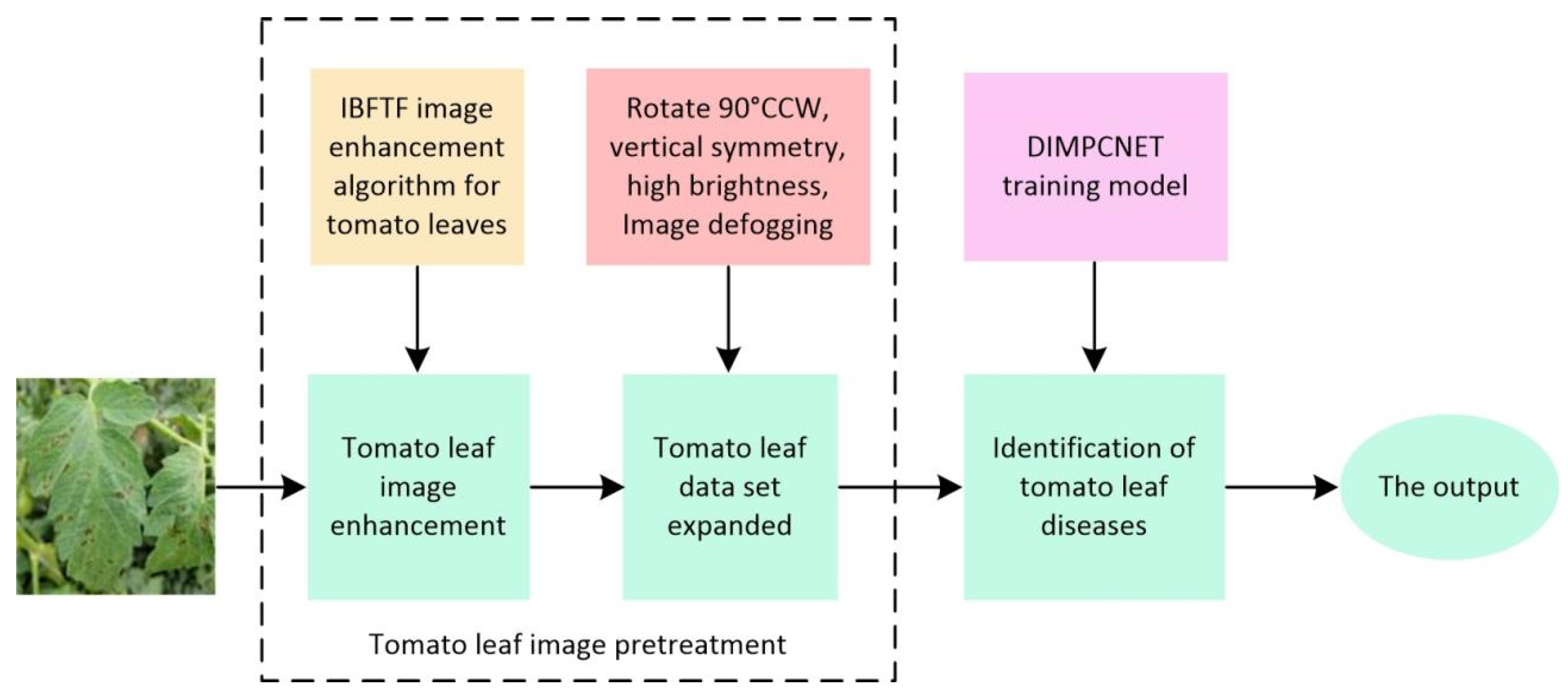
2. Materials and Methods
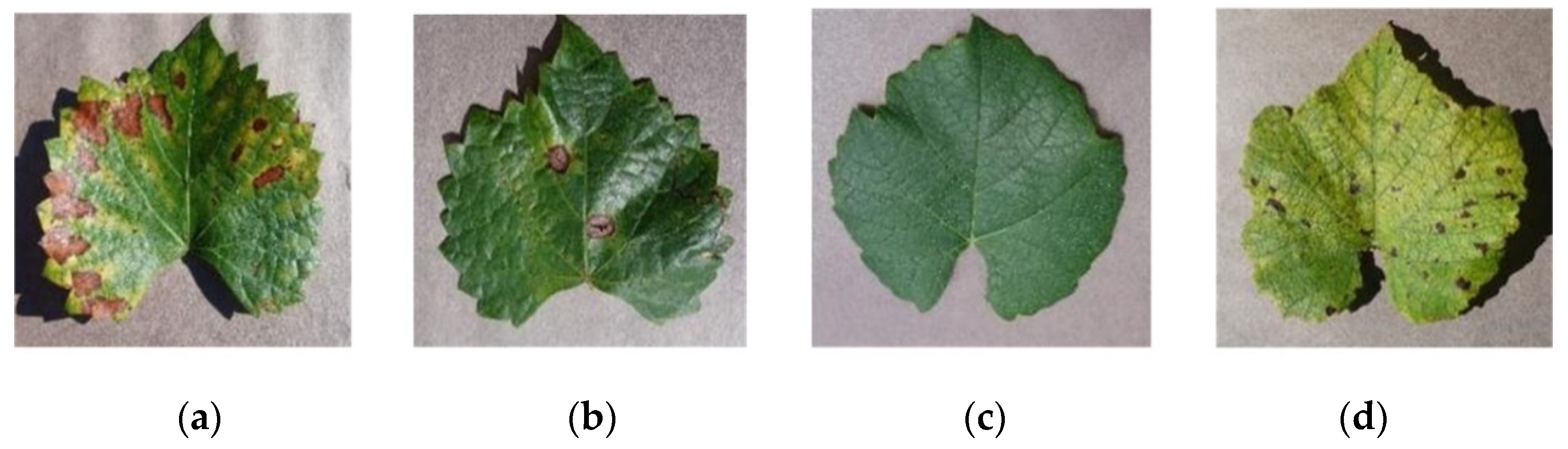
2.1. Data Acquisition
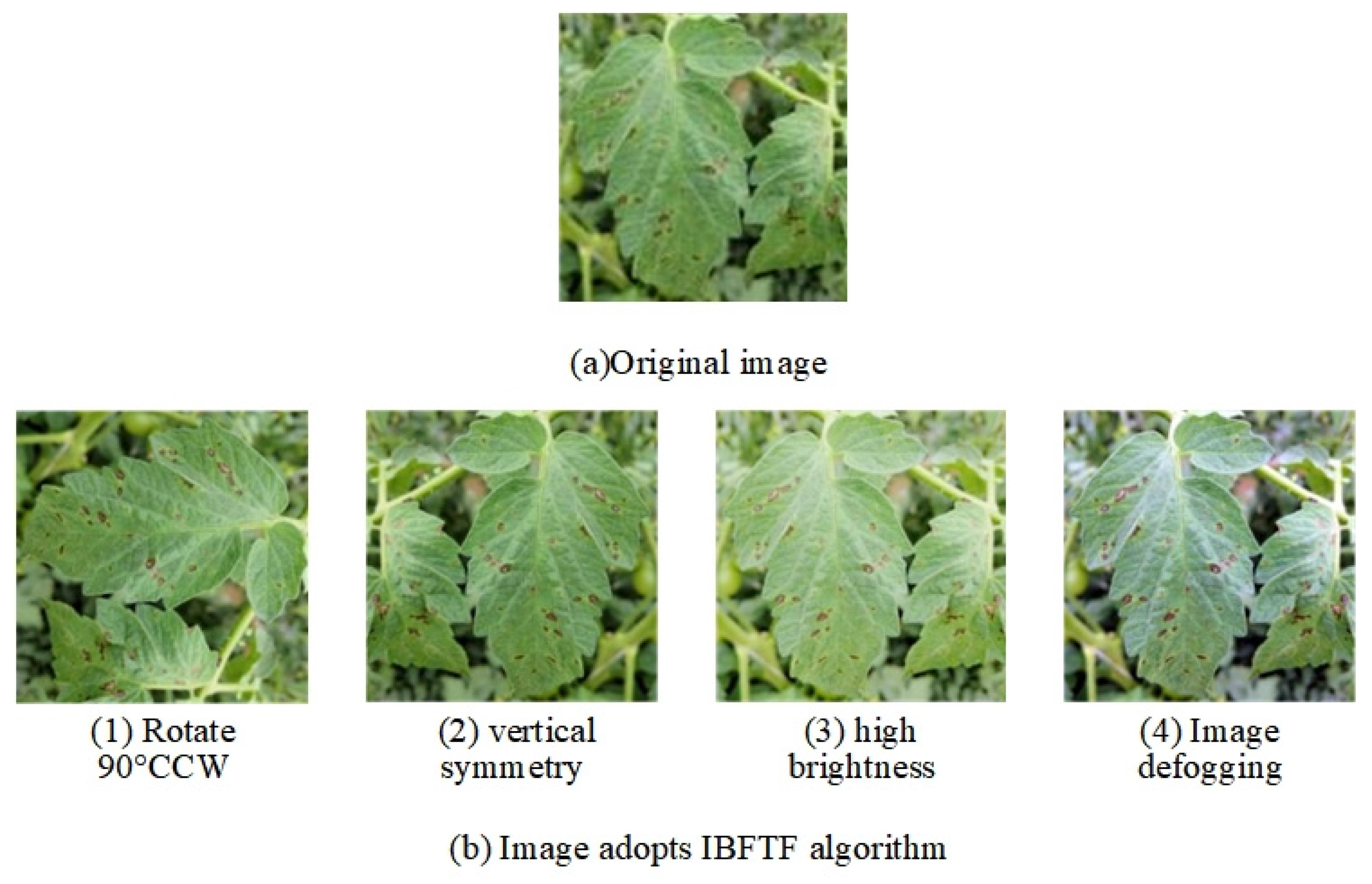
2.2. Image Augmentation
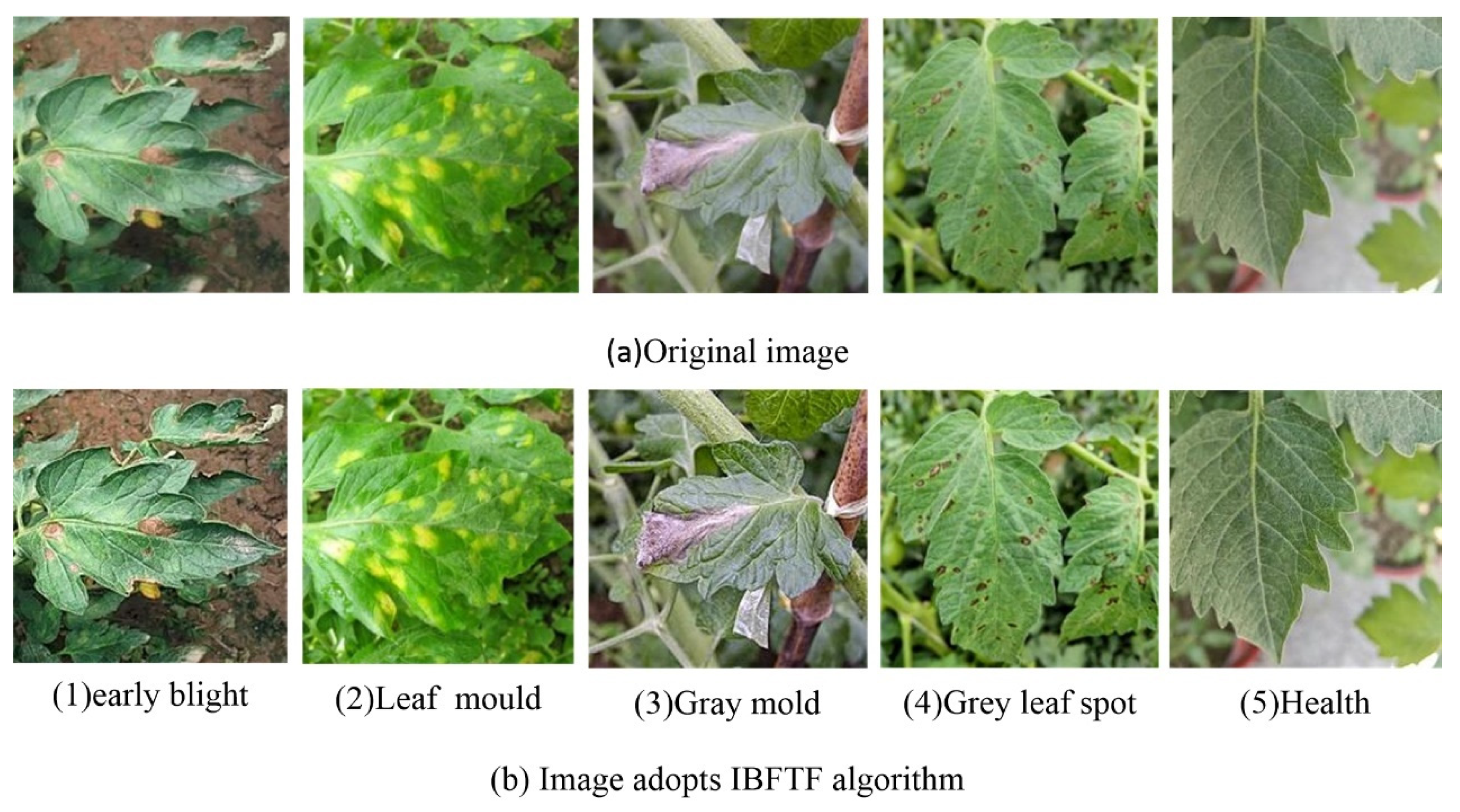
2.3. Tomato Leaf Image Enhancement Based on IBFTF
- The noise image is decomposed using wavelets, and both the low and HF coefficients are calculated;
- The LF coefficients are handled by the enhanced bilateral filtering Retinex image-enhancing method;
- The method of improved threshold function is designed to handle the HF coefficient;
- The reconstructed image is obtained by wavelet reconstruction of both LF and HF coefficients;
- The reconstructed image is processed by a piecewise linear transformation, and the enhanced image is obtained.
| Algorithm 1: IBFTF image enhancement |
| Input: Image |
| 1. Decompose the noisy image into LF and HF coefficients |
| 2. The image is launched |
| 3. uses enhanced bilateral filtering processing |
| 4. , sets the filtering window parameter , and the original bilateral filtering window size is |
| 5. The enhanced threshold function is used to estimate the HF wavelet coefficient in three parts |
| 6. Process using three-segment piecewise linear transformation |
| 7. and are reconstructed using 2D discrete wavelet |
| 8. Output reconstructed image |
2.3.1. Wavelet Decomposition and Reconstruction
2.3.2. Improved Retinex Algorithm for Bilateral Filtering
2.3.3. Wavelet HF Coefficient Processing
2.3.4. Contrast Enhancement
3. Tomato Leaf Disease Recognition Based on DIMPCNET Model
3.1. DI Module
3.1.1. Depth-Separable Convolutional Layer
3.1.2. Cascading Dense Inception Modules
3.2. The MPC Module
4. Experimental Outcomes and Analysis
4.1. Environment and Setup for Experiments
4.2. Evaluation Index
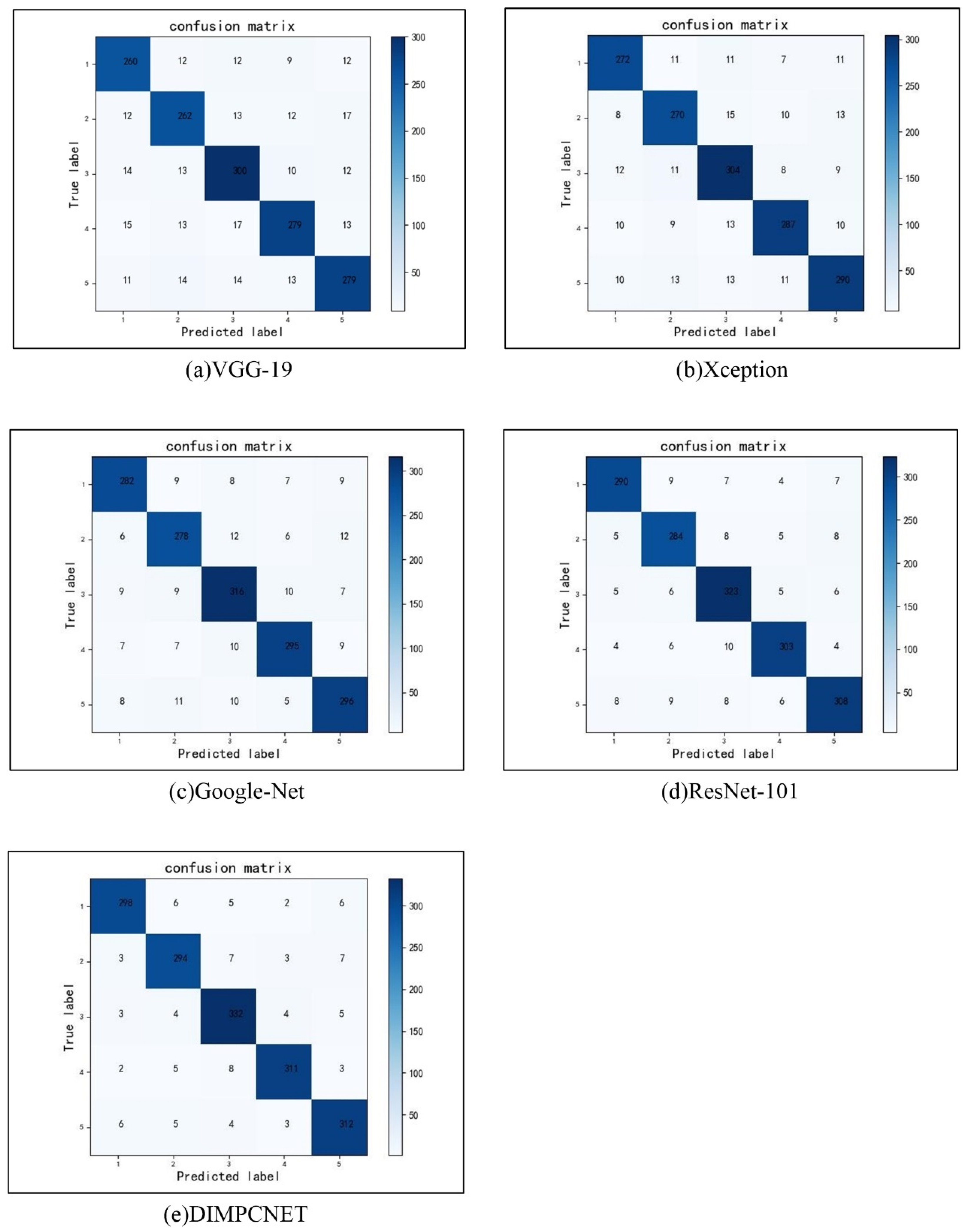
4.3. Comparison with Traditional Convolutional Neural Networks
4.4. Comparison with the Latest Convolutional Neural Networks
4.5. Ablation Experiment
4.6. Attention Module Experiment
4.7. Effectiveness of DIMPCNET on Other Leaf Disease Datasets
5. Conclusions and Prospect
- This study proposes the utilization of DIMPCNET for the identification of tomato leaf diseases. DIMPCNET is capable of efficiently extracting multi-dimensional and multi-scale features from diseased leaves, effectively addressing the challenges posed by complex background environments and the similarity of disease spots. Notably, DIMPCNET employs a unique two-branch classification structure, where each branch utilizes basic convolutional neural networks as feature extractors. The branches can be symmetric or asymmetric. Following feature extraction, a bilinear pooling method is employed to fuse the features. The DIMPCNET method demonstrates high accuracy in image classification tasks, outperforming traditional convolutional neural networks;
- To combat the challenge of low precision in identifying pests and diseases with fine-grained multi-classification, a novel approach has been developed. The existing CBAM module, which utilizes serial connections, has been enhanced by introducing a parallel PCBAM attention module. This new module aims to address the interference problem caused by serial connections and improve the accuracy of recognition. By leveraging the parallel structure, the proposed solution aims to minimize interference and enhance the precision of pest and disease identification;
- In order to assess the effectiveness and robustness of the PCBAM module, a comprehensive analysis was conducted to compare its recognition performance with other attention mechanisms in the DIMNET model. The recognition abilities of channel attention, spatial attention, CBAM, RCBAM, and PCBAM were thoroughly evaluated and compared. The results unequivocally demonstrate that the PCBAM module outperforms other attention mechanisms in accurately classifying diseases with fine-grained details. Furthermore, it exhibits remarkable generalization capabilities across different convolutional neural network models. These findings underscore the efficacy of the PCBAM module in precisely identifying and categorizing diseases, thus making it a valuable asset in the field of agricultural disease management;
- Compared with the current popular classification method of tomato leaf disease, our recognition accuracy was 94.44%, and the F1 value was 0.9475. This method achieves better performance. It has obvious advantages in that it can effectively remove image noise and reduce the negative impact of complex background environments and disease similarity on disease recognition so as to effectively prevent and control tomato diseases and improve tomato yield;
- At present, we only consider one disease on a single leaf, but if there are multiple diseases on a single leaf, how to identify them can be seen as a future direction. Next, we will further simplify the training process and consider improving the efficiency of the model. In addition, considering the shortcomings of relying solely on image modality to complete the tomato leaf disease recognition process, we plan to consider using a richer multi-modal dataset to achieve tomato leaf disease recognition.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kong, J.; Wang, H.; Wang, X.; Jin, X.; Fang, X.; Lin, S. Multi-stream hybrid architecture based on cross-level fusion strategy for fine-grained crop species recognition in precision agriculture. Comput. Electron. Agric. 2021, 185, 106134. [Google Scholar] [CrossRef]
- Shao, W.; Liu, L.; Jiang, J.; Yan, Y. Low-light-level image enhancement based on fusion and Retinex. J. Mod. Opt. 2020, 67, 1190–1196. [Google Scholar] [CrossRef]
- Qin, Y.; Luo, F.; Li, M. A Medical Image Enhancement Method Based on Improved Multi-Scale Retinex Algorithm. J. Med. Imaging Health Inform. 2020, 10, 152–157. [Google Scholar] [CrossRef]
- Liu, J.; Wang, S.; Wang, X.; Ju, M.; Zhang, D. A Review of Remote Sensing Image Dehazing. Sensors 2021, 21, 3926. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, P.; Quan, L.; Yi, C.; Lu, C. Underwater image enhancement based on deep learning and image formation model. arXiv 2021, arXiv:2101.00991. [Google Scholar]
- Wu, W.; Weng, J.; Zhang, P.; Wang, X.; Yang, W.; Jiang, J. URetinex-Net: Retinex-Based Deep Unfolding Network for Low-Light Image Enhancement. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 19–20 June 2022; pp. 5901–5910. [Google Scholar]
- Kaur, K.; Jindal, N.; Singh, K. Fractional derivative based Unsharp masking approach for enhancement of digital images. Multimed. Tools Appl. 2020, 80, 3645–3679. [Google Scholar] [CrossRef]
- Hou, Y.; Li, Q.; Zhang, C.; Lu, G.; Ye, Z.; Chen, Y.; Wang, L.; Cao, D. The State-of-the-Art Review on Applications of Intrusive Sensing, Image Processing Techniques, and Machine Learning Methods in Pavement Monitoring and Analysis. Engineering 2020, 7, 845–856. [Google Scholar] [CrossRef]
- Oktavianto, B.; Purboyo, T.W. A study of histogram equalization techniques for image enhancement. Int. J. Appl. Eng. Res. 2018, 13, 1165–1170. [Google Scholar]
- Li, P.; Huang, Y.; Yao, K. Multi-algorithm Fusion of RGB and HSV Color Spaces for Image Enhancement. In Proceedings of the 2018 37th Chinese Control Conference (CCC), Wuhan, China, 25–27 July 2018. [Google Scholar] [CrossRef]
- Land, E.H.; McCann, J.J. Lightness and Retinex Theory. J. Opt. Soc. Am. 1971, 61, 1–11. [Google Scholar] [CrossRef]
- Jobson, D.; Rahman, Z.; Woodell, G. Properties and performance of a center/surround retinex. IEEE Trans. Image Process. 1997, 6, 451–462. [Google Scholar] [CrossRef]
- Hu, W.W.; Wang, R.G.; Fang, S.; Hu, Q. Retinex image enhancement algorithm based on bilateral filtering. Graph. J. 2010, 31, 104–109. [Google Scholar]
- Zhang, X.; Zhao, L. Image enhancement algorithm based on improved Retinex. J. Nanjing Univ. Sci. Technol. 2016, 40, 24–28. [Google Scholar]
- Tan, L.; Chen, Y.; Zhang, W. Multi-focus Image Fusion Method based on Wavelet Transform. J. Phys. Conf. Ser. 2019, 1284, 012068. [Google Scholar] [CrossRef]
- Li, X.; Cong, Z.; Zhang, Y. Rail Track Edge Detection Methods Based on Improved Hough Transform. In Proceedings of the IEEE International Conference on Power Electronics, Computer Applications (ICPECA), Shenyang, China, 22–24 January 2021; pp. 12–16. [Google Scholar]
- Donoho, D. De-noising by soft-thresholding. IEEE Trans. Inf. Theory 1995, 41, 613–627. [Google Scholar] [CrossRef]
- Jianhui, X.; Li, T. Image Denoising Method Based on Improved Wavelet Threshold Transform. In Proceedings of the 2019 IEEE Symposium Series on Computational Intelligence (SSCI), Xiamen, China, 6–9 December 2019; pp. 1064–1067. [Google Scholar] [CrossRef]
- Lei, S.; Lu, M.; Lin, J.; Zhou, X.; Yang, X. Remote sensing image denoising based on improved semi-soft threshold. Signal Image Video Process. 2020, 15, 73–81. [Google Scholar] [CrossRef]
- Panchal, P.; Raman, V.C.; Mantri, S. Plant diseases detection and classification using machine learning models. In Proceedings of the 2019 4th International Conference on Computational Systems and Information Technology for Sustainable Solution (CSITSS), Bengaluru, India, 20–21 December 2019; IEEE: Piscataway, NJ, USA, 2019; Volume 4, pp. 1–6. [Google Scholar]
- Ramesh, S.; Hebbar, R.; Niveditha, M.; Pooja, R.; Shashank, N.; Vinod, P.V. Plant disease detection using machine learning. In Proceedings of the 2018 International Conference on Design Innovations for 3Cs Compute Communicate Control (ICDI3C), Bengaluru, India, 24–26 April 2018; pp. 41–45. [Google Scholar]
- Rashmi, N.; Shetty, C. A Machine Learning Technique for Identification of Plant Diseases in Leaves. In Proceedings of the 6th International Conference on Inventive Computation Technologies (ICICT), Lalitpur, Nepal, 26–28 April 2023; pp. 481–484. [Google Scholar]
- Reza, Z.N.; Nuzhat, F.; Mahsa, N.A.; Ali, M.H. Detecting Jute Plant Disease Using Image Processing and Machine Learning. In Proceedings of the 3rd International Conference on Electrical Engineering and Information Communication Technology (ICEEICT), Dhaka, Bangladesh, 22–24 September 2016; pp. 1–6. [Google Scholar]
- Akhtar, A.; Khanum, A.; Khan, S.A.; Shaukat, A. Automated Plant Disease Analysis (APDA): Performance Comparison of Machine Learning Techniques. In Proceedings of the 2013 11th International Conference on Frontiers of Information Technology, Islamabad, Pakistan, 16–18 December 2013; pp. 60–65. [Google Scholar] [CrossRef]
- Wang, J.; Ning, F.; Lu, S. Study on apple leaf disease identification method based on support vector machine. Shandong Agric. Sci. 2015, 47, 122–125. [Google Scholar]
- Qin, F.; Liu, D.X.; Sun, B.D. Recognition of four different alfalfa leaf diseases based on image processing technology. China Agric. Univ. 2016, 21, 65–75. [Google Scholar]
- Xie, C.; He, Y. Spectrum and Image Texture Features Analysis for Early Blight Disease Detection on Eggplant Leaves. Sensors 2016, 16, 676. [Google Scholar] [CrossRef]
- Chai, A.L.; Li, B.J.; Shi, Y.X. Recognition of tomato foliage disease based on computer vision technology. Acta Hortic. Sinica 2010, 37, 1423–1430. [Google Scholar]
- Zhang, Y.L.; Yuan, H.; Zhang, Q.Q. Apple leaf disease recognition method based on color feature and difference histogram. Jiangsu Agric. Sci. 2017, 45, 171–174. [Google Scholar]
- Xia, Y.Q.; Bing, W.; Jun, Z. Identification of wheat leaf disease based on random forest method. J. Graph. 2018, 39, 57. [Google Scholar]
- Wu, Y. Identification of Maize Leaf Diseases based on Convolutional Neural Network. J. Phys. Conf. Ser. 2021, 1748, 032004. [Google Scholar] [CrossRef]
- Ding, R.; Zhou, P. Identification of typical crop leaf diseases based on convolutional neural network. Packag. J. 2018, 10, 74–80. [Google Scholar]
- Alom, M.Z.; Taha, T.M.; Yakopcic, C.; Westberg, S.; Sidike, P.; Nasrin, M.S.; Asari, V.K. The history began from alexnet: A comprehensive survey on deep learning approaches. arXiv 2018, arXiv:1803.01164. [Google Scholar]
- Guo, X.Q.; Fan, T.J.; Shu, X. Tomato leaf disease image recognition based on improved Multi-scale AlexNet. Trans. Chin. Soc. Agric. Eng. 2019, 35, 162–169. [Google Scholar]
- Ni, L.; Zou, W. Recognition of animal species based on improved Xception by SE module. Navig. Control 2020, 19, 106–111. [Google Scholar]
- Zhang, S.; Huang, W.; Zhang, C. Three-channel convolutional neural networks for vegetable leaf disease recognition. Cogn. Syst. Res. 2019, 53, 31–41. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, S.; Zhang, C.; Wang, X.; Shi, Y. Cucumber Leaf Disease Identification with Global Pooling Dilated Con-volutional Neural Network. Comput. Electron. Agric. 2019, 162, 422–430. [Google Scholar] [CrossRef]
- Gulzar, Y. Fruit Image Classification Model Based on MobileNetV2 with Deep Transfer Learning Technique. Sustainability 2023, 15, 1906. [Google Scholar] [CrossRef]
- Mamat, N.; Othman, M.F.; Abdulghafor, R.; Alwan, A.A.; Gulzar, Y. Enhancing Image Annotation Technique of Fruit Clas-sification Using a Deep Learning Approach. Sustainability 2023, 15, 901. [Google Scholar] [CrossRef]
- Aggarwal, S.; Gupta, S.; Gupta, D.; Gulzar, Y.; Juneja, S.; Alwan, A.A.; Nauman, A. An Artificial Intelligence-Based Stacked Ensemble Approach for Prediction of Protein Subcellular Localization in Confocal Microscopy Images. Sustainability 2023, 15, 1695. [Google Scholar] [CrossRef]
- Gulzar, Y.; Hamid, Y.; Soomro, A.B.; Alwan, A.A.; Journaux, L. A Convolution Neural Network-Based Seed Classification System. Symmetry 2020, 12, 2018. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, G.; Chen, A.; Yi, J.; Zhang, W.; Hu, Y. Identification of tomato leaf diseases based on combination of ABCK-BWTR and B-ARNet. Comput. Electron. Agric. 2020, 178, 105730. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, S.; Zhou, G.; Hu, Y.; Li, L. Identification of tomato leaf diseases based on multi-channel automatic orien-tation recurrent attention network. Comput. Electron. Agr. 2023, 205, 107605. [Google Scholar] [CrossRef]
- Sun, J.; Tan, W.; Mao, H.; Wu, X.; Chen, Y.; Wang, L. Recognition of multiple plant leaf diseases based on improved convo-lutional neural network. Trans. Chin. Soc. Agric. Eng. 2017, 33, 209–215. [Google Scholar]
- Gibran, M.; Wibowo, A. Convolutional Neural Network Optimization for Disease Classification Tomato Plants Through Leaf Image. In Proceedings of the 5th International Conference on Informatics and Computational Sciences (ICICoS), Semarang, Indonesia, 24–25 November 2021; pp. 116–121. [Google Scholar]
- Cai, C.; Wang, Q.; Cai, W.; Yang, Y.; Hu, Y.; Li, L.; Wang, Y.; Zhou, G. Identification of grape leaf diseases based on VN-BWT and Siamese DWOAM-DRNet. Eng. Appl. Artif. Intell. 2023, 123, 106341. [Google Scholar] [CrossRef]
- Kong, J.; Wang, H.; Yang, C.; Jin, X.; Zuo, M.; Zhang, X. A Spatial Feature-Enhanced Attention Neural Network with High-Order Pooling Repre-sentation for Application in Pest and Disease Recognition. Agriculture 2022, 12, 500. [Google Scholar] [CrossRef]
- Mohideen, S.K.; Perumal, S.A.; Sathik, M.M. Image De-Noising Using Discrete Wavelet Transform. Int. J. Comput. Sci. Netw. Secur. 2008, 8, 213–216. [Google Scholar]
- Fang, S.; Yang, J.R.; Cao, Y.; Wu, P.F.; Rao, R.Z. Local Multi-Scale Retinex Algorithm for Image Guided Filtering. J. Image Graph. 2012, 17, 748–755. [Google Scholar]
- Ouhami, M.; Es-Saady, Y.; Hajji, M.E.; Hafiane, A.; Canals, R.; Yassa, M.E. Deep transfer learning models for tomato disease detection. In Proceedings of the 9th International Conference, ICISP 2020, Marrakesh, Morocco, 4–6 June 2020; pp. 65–73. [Google Scholar]
- McNeely-White, D.; Beveridge, J.R.; Draper, B.A. Inception and ResNet features are (almost) equivalent. Cogn. Syst. Res. 2019, 59, 312–318. [Google Scholar] [CrossRef]
- Li, Z.; Yang, Y.; Li, Y.; Guo, R.; Yang, J.; Yue, J. A solanaceae disease recognition model based on SE-Inception. Comput. Electron. Agric. 2020, 178, 105792. [Google Scholar] [CrossRef]
- Jangapally, T. Image Classification Using Network Inception-Architecture & Applications. JIDPTS 2021, 4, 6–9. [Google Scholar]
- Zhang, Y.; Hou, Y.; OuYang, K.; Zhou, S. Multi-scale signed recurrence plot based time series classification using inception architectural networks. Pattern Recognit. 2021, 123, 108385. [Google Scholar] [CrossRef]
- Li, S.; Ma, H. A Siamese inception architecture network for person re-identification. Mach. Vis. Appl. 2017, 28, 725–736. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Szegedy, C.; Wei, L.; Yangqing, J.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Fu, J.; Zheng, H.; Mei, T. Look Closer to See Better: Recurrent Attention Convolutional Neural Network for Fine-Grained Image Recognition. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 4476–4484. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018. [Google Scholar] [CrossRef]
- Prado, J.; Yandun, F.; Torriti, M.T.; Cheein, F.A. Overcoming the Loss of Performance in Unmanned Ground Vehicles Due to the Terrain Variability. IEEE Access 2018, 6, 17391–17406. [Google Scholar] [CrossRef]

| Disease Type | Before Data Enhancement | After Data Enhancement | Proportion |
|---|---|---|---|
| early blight | 312 | 1560 | 19.04% |
| leaf mold | 314 | 1570 | 19.16% |
| gray mold | 356 | 1780 | 21.73% |
| grey leaf | 323 | 1615 | 19.71% |
| health | 333 | 1665 | 20.32% |
| Experimental Environment Configuration | Parameter Values Enhancement |
|---|---|
| CPU | Intel(R) Xeon(R) Gold 6271C CPU@2.60 GHz |
| GPU | NVIDIA Tesla V100 GPU32 G |
| RAM | 32 G |
| Magnetic disk | 100 G |
| Deep learning framework | PyTorch |
| Operating System | Windows 10(64-bits) |
| Others | Python 3.7.1CUDA10.1 |
| Parameter Name | Parameter Values |
|---|---|
| Image Size | 224 × 224 |
| Batch size | 60 |
| Learning rate | 0.001 |
| Magnetic disk | 100 G |
| Epoch | 200 |
| Module | Input | Precision | Recall | F1 | (ms) | Accuracy (%) |
|---|---|---|---|---|---|---|
| VGG-19 | 224 | 0.8333 | 0.8525 | 0.8427 | 33.56 | 84.25 |
| Xception | 224 | 0.8718 | 0.8718 | 0.8718 | 32.92 | 86.87 |
| GoogleNet | 224 | 0.9038 | 0.8952 | 0.8995 | 40.26 | 89.56 |
| ResNet-101 | 224 | 0.9295 | 0.9148 | 0.9220 | 39.53 | 92.06 |
| DIMPCNET | 224 | 0.9551 | 0.9400 | 0.9475 | 31.68 | 94.44 |
| Module | Input | Precision | Recall | F1 | (ms) | Accuracy (%) |
|---|---|---|---|---|---|---|
| VGG-19 | 224 | 0.8298 | 0.8508 | 0.8401 | 33.39 | 84.06 |
| Xception | 224 | 0.8679 | 0.8648 | 0.8663 | 34.79 | 86.12 |
| GoogleNet | 224 | 0.9011 | 0.8883 | 0.8947 | 38.46 | 89.78 |
| ResNet-101 | 224 | 0.9279 | 0.9109 | 0.9193 | 37.87 | 92.71 |
| DIMPCNET | 224 | 0.9467 | 0.9337 | 0.9402 | 32.21 | 93.82 |
| Module | Input | Precision | Recall | F1 | (ms) | Accuracy (%) |
|---|---|---|---|---|---|---|
| Multi-ScaleAlexNet | 224 | 0.9163 | 0.9159 | 0.9134 | 35.34 | 91.96 |
| B-ARNet | 224 | 0.8572 | 0.8565 | 0.8568 | 33.85 | 85.49 |
| DWOAM-DRNet | 224 | 224 | 0.9281 | 0.9264 | 0.9272 | 36.62 |
| M−AORANet | 224 | 0.9369 | 0.9360 | 0.9364 | 38.64 | 93.59 |
| DIMPCNET | 224 | 0.9551 | 0.9400 | 0.9475 | 31.68 | 94.44 |
| Module | Accuracy | F1 Score | Param (M) | ||||
|---|---|---|---|---|---|---|---|
| Early Blight | Leaf Mold | Gray Mold | Grey Leaf Spot | Health | |||
| VGG + VGG | 88.45% | 0.8957 | 0.8745 | 0.8871 | 0.9007 | 0.8949 | 54.02 |
| VGG + DI | 91.44% | 0.9256 | 0.9157 | 0.9161 | 0.9195 | 0.9039 | 56.02 |
| VGG + MPC | 92.68% | 0.9321 | 0.9335 | 0.9266 | 0.9312 | 0.9205 | 46.02 |
| DI + MPC | 94.29% | 0.9469 | 0.9301 | 0.9384 | 0.9458 | 0.9326 | 48.02 |
| Network | Top1 Accuracy/% | Forward Propagation Time/ms | Model Size/ MB | Number of Parameters | Accuracy↑ (↑, Percentage Increase) |
|---|---|---|---|---|---|
| DIMNET | 90.48% | 7.18 | 56.1 | ||
| DIMNET+channel attention | 90.97% | 7.18 | 86.8 | 0.49% | |
| DIMNET+spatial attention | 91.36% | 7.15 | 86.1 | 0.88% | |
| DIMNET+CBAM | 93.21% | 7.33 | 86.9 | 2.73% | |
| DIMNET+RCBAM | 92.38% | 7.33 | 86.9 | 1.90% | |
| DIMNET+SE | 94.58% | 7.33 | 92.6 | 4.10% | |
| DIMNET+PCBAM | 95.70% | 7.15 | 86.9 | 5.22% |
| Grape | Input | Precision | Recall | F1 | TA (ms) | Accuracy |
|---|---|---|---|---|---|---|
| VGG-19 | 224 | 0.8639 | 0.8747 | 0.8693 | 31.97 | 86.91 |
| Xception | 224 | 0.8925 | 0.8714 | 0.8818 | 31.97 | 88.16 |
| GoogleNet | 224 | 0.9216 | 0.9309 | 0.9262 | 31.42 | 92.27 |
| ResNet-101 | 224 | 0.9595 | 0.9513 | 0.9554 | 33.27 | 95.86 |
| DIMPCNET | 224 | 0.9756 | 0.9715 | 0.9735 | 31.42 | 97.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, D.; Li, W.; Zhao, H.; Zhou, G.; Cai, C. Recognition of Tomato Leaf Diseases Based on DIMPCNET. Agronomy 2023, 13, 1812. https://doi.org/10.3390/agronomy13071812
Peng D, Li W, Zhao H, Zhou G, Cai C. Recognition of Tomato Leaf Diseases Based on DIMPCNET. Agronomy. 2023; 13(7):1812. https://doi.org/10.3390/agronomy13071812
Chicago/Turabian StylePeng, Ding, Wenjiao Li, Hongmin Zhao, Guoxiong Zhou, and Chuang Cai. 2023. "Recognition of Tomato Leaf Diseases Based on DIMPCNET" Agronomy 13, no. 7: 1812. https://doi.org/10.3390/agronomy13071812
APA StylePeng, D., Li, W., Zhao, H., Zhou, G., & Cai, C. (2023). Recognition of Tomato Leaf Diseases Based on DIMPCNET. Agronomy, 13(7), 1812. https://doi.org/10.3390/agronomy13071812







