A Comparative Transcriptome Analysis of Avocado Embryogenic Lines Susceptible or Resistant to Rosellinia necatrix Exudate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Preparation of Crude Filtrate from Rosellinia necatrix
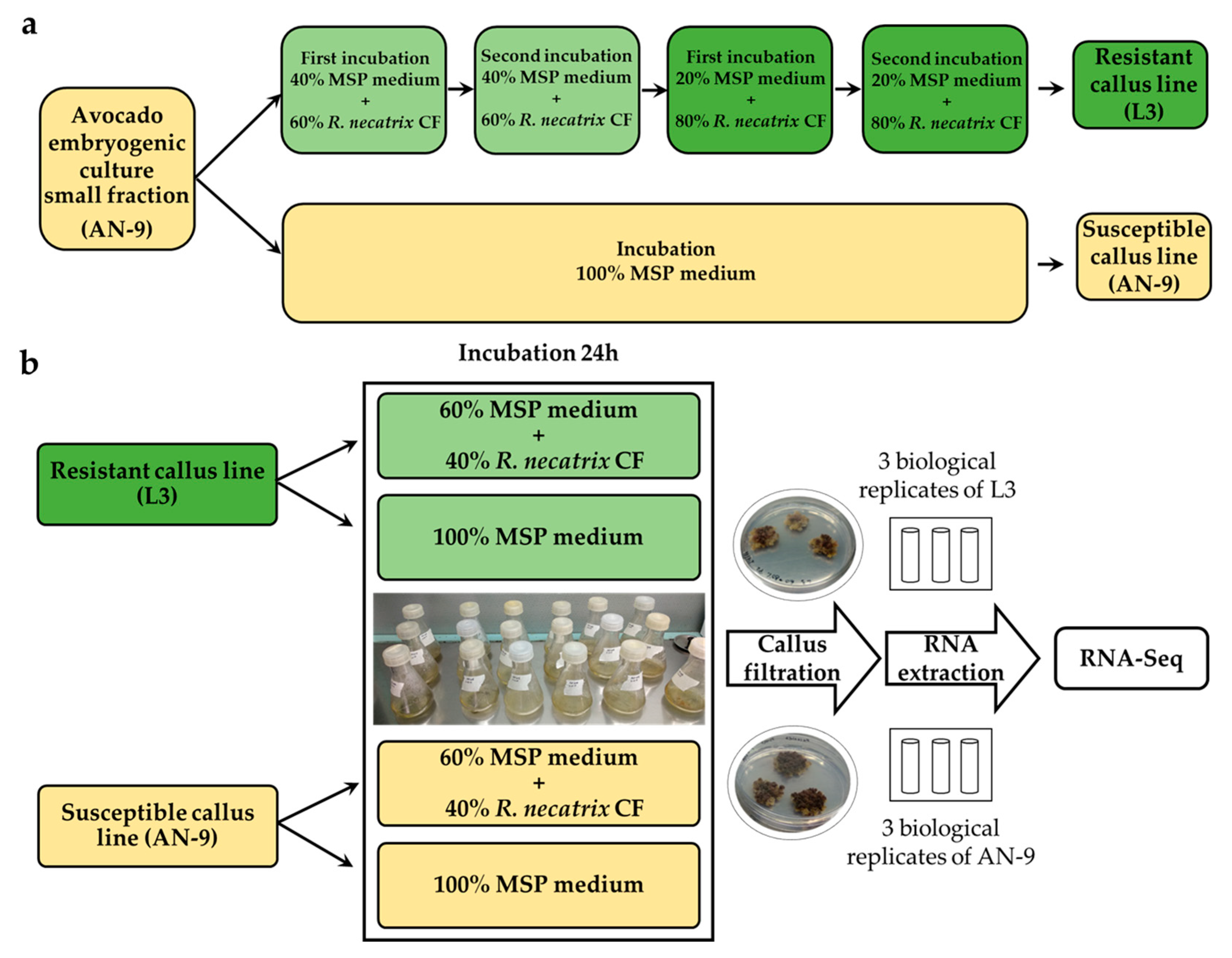
2.3. Obtainment of a Callus Line Resistant to Fungal Filtrate
2.4. Experimental Design
2.5. RNA Extraction
2.6. Transcriptome Analysis
2.7. Gene Predictions and Annotation
2.8. Quantitative Real Time-PCR
3. Results
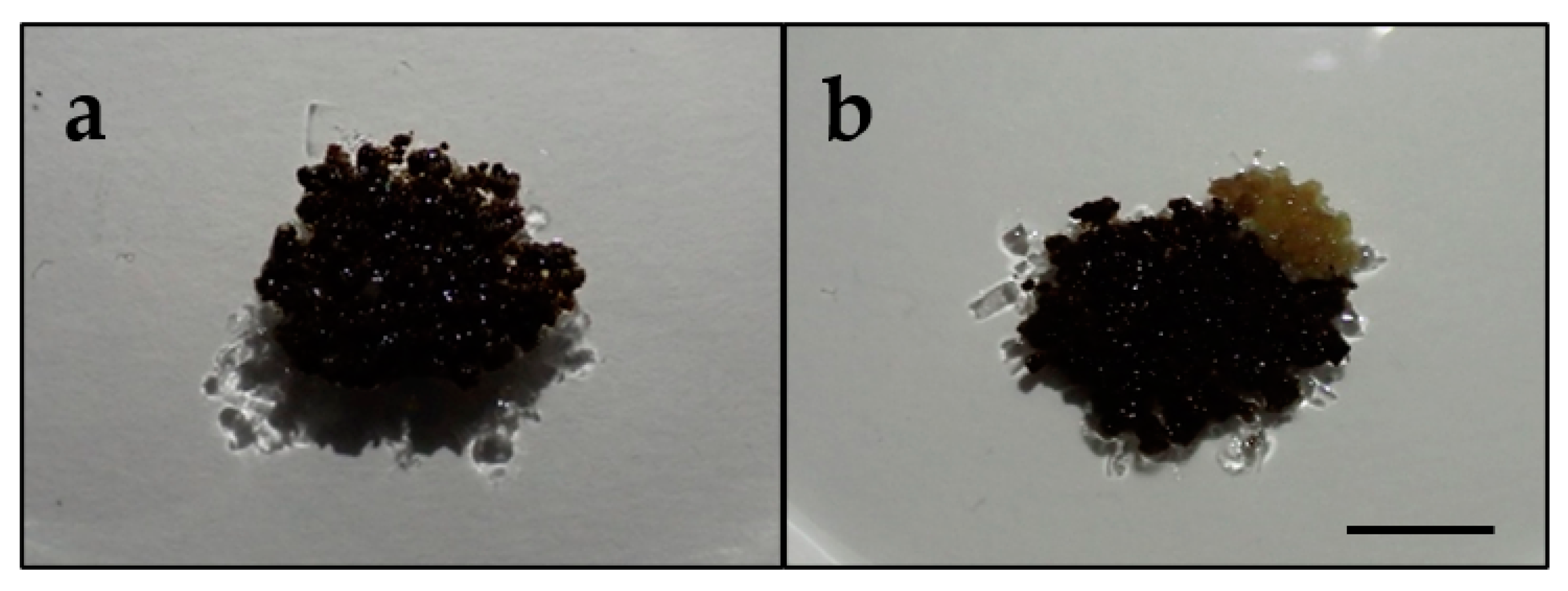
3.1. Obtaining a Callus Line Resistant to Rosellinia necatrix Filtrate
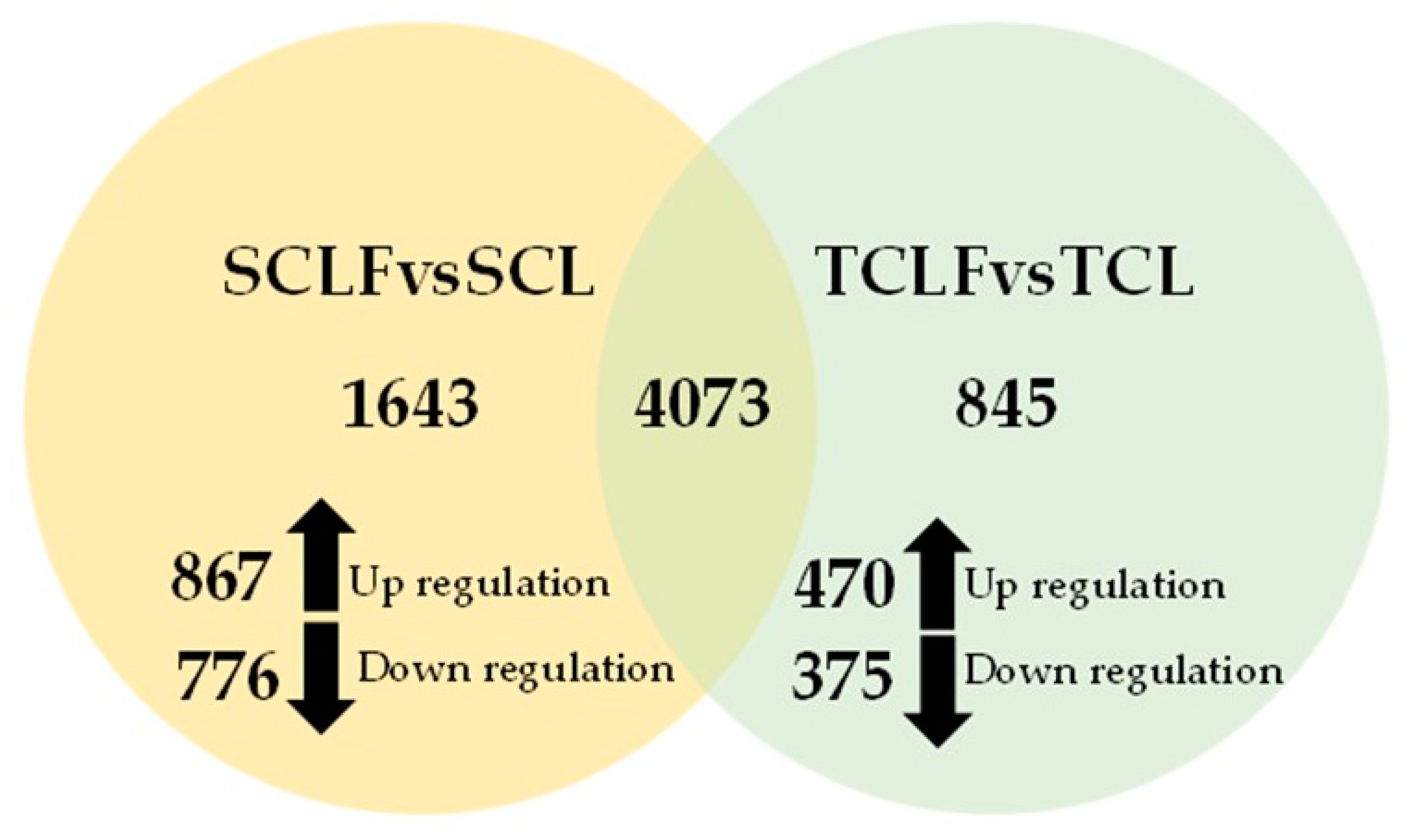
3.2. Comparative Transcriptome Analysis of Resistant and Susceptible Callus Lines Exposed to Fungal Filtrate
3.3. RNA-Seq Analysis Validation
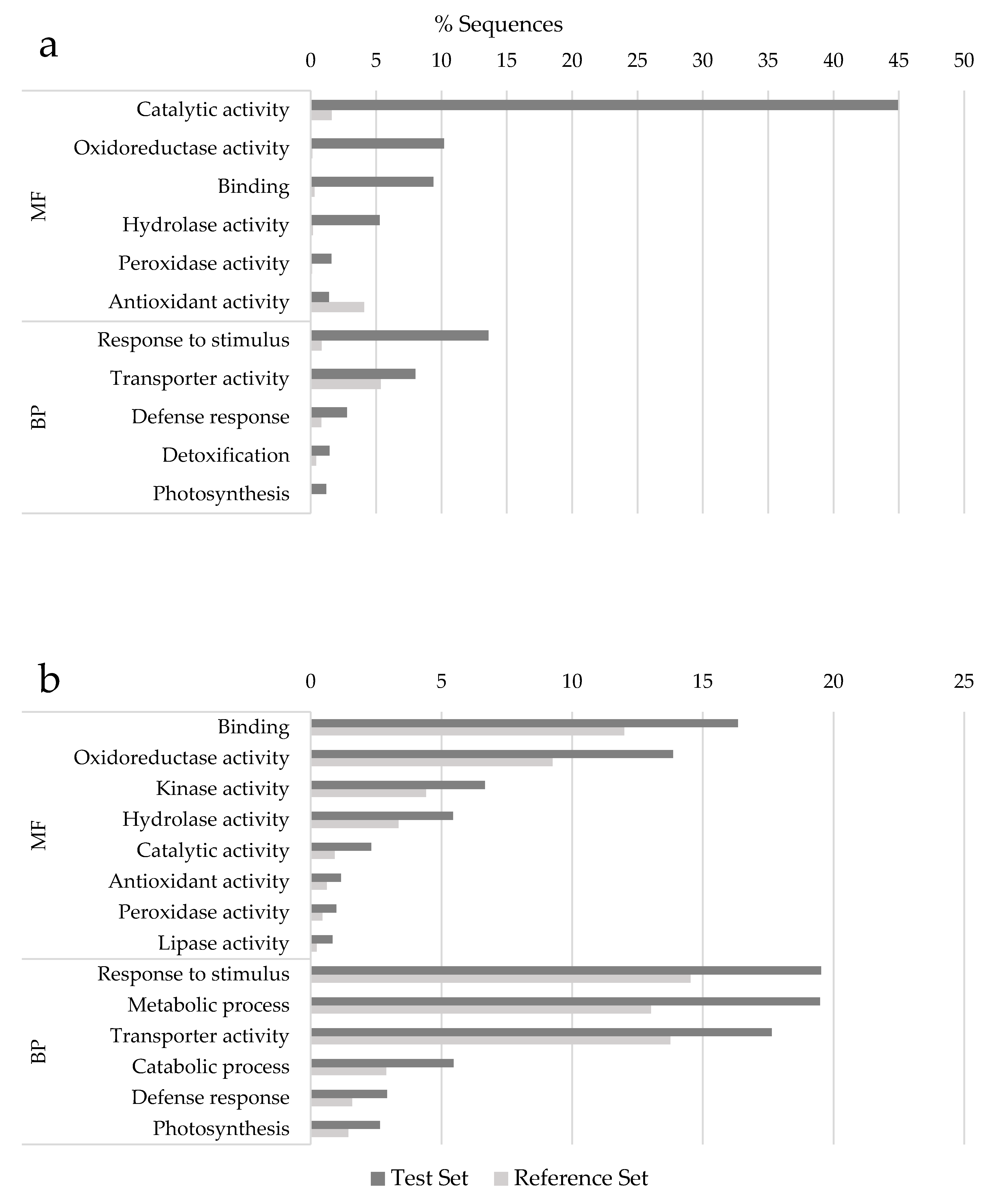
3.4. Functional Annotation and Pathway Analysis of Differentially Expressed Genes (DEGs)
3.5. Potential Genes Related to Avocado Embryogenic Callus Defense against Fungal Exudates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- FAOSTAT. Data. 2023. Available online: https://www.fao.org/faostat/es/#data (accessed on 21 February 2023).
- López-Herrera, C.J.; Zea-Bonilla, T. Effects of benomyl, carbendazim, fluazinam and thiophanate methyl on white root rot of avocado. Crop Prot. 2007, 26, 1186–1192. [Google Scholar] [CrossRef]
- Khan, A.H. Biology and pathogenicity of Rosellinia necatrix (Hart.) Berl. Biologia 1955, 5, 199–425. [Google Scholar]
- Pliego, C.; López-Herrera, C.; Ramos, C.; Cazorla, F.M. Developing tools to unravel the biological secrets of Rosellinia necatrix, an emergent threat to woody crops. Mol. Plant Pathol. 2012, 13, 226–239. [Google Scholar] [CrossRef]
- Pérez-Jiménez, R.M. A review of the biology and pathogenicity of Rosellinia necatrix–the cause of white root rot disease of fruit trees and other plants. J. Phytopathol. 2006, 154, 257–266. [Google Scholar] [CrossRef]
- Arjona-Girona, I.; López-Herrera, C.J. First report of Rosellinia necatrix causing white root rot in mango trees in Spain. Plant Dis. 2018, 102, 2639. [Google Scholar] [CrossRef]
- Edwards, R.L.; Maitland, D.J.; Scowen, I.J.; De Sousa, A.J.T.; Whalley, A.J.S. Metabolites of the higher fungi. Part 32. A phytotoxic bicyclo[4.1.0]hept-3-en-2-one from the fungus Rosellinia necatrix Prill. J. Chem. Soc. Perkin Trans. 2001, 1, 537–542. [Google Scholar] [CrossRef]
- Kimura, Y.; Nakajima, H.; Hamasaki, T. Structure of rosellichalasin, a new metabolite produced by Rosellinia necatrix. Agric. Biol. Chem. 1989, 53, 1699–1701. [Google Scholar] [CrossRef]
- Kshirsagar, A.; Reid, A.J.; McColl, S.M.; Saunders, V.A.; Whalley, A.J.S.; Evans, E.H. The effect of fungal metabolites on leaves as detected by chlorophyll fluorescence. New Phytol. 2001, 151, 451–457. [Google Scholar] [CrossRef]
- Kanematsu, S.; Hayashi, T.; Kudo, A. Isolation of Rosellinia necatrix mutants with impaired cytochalasin E production and its pathogenicity. Jpn. J. Phytopathol. 1997, 63, 425–431. [Google Scholar] [CrossRef] [Green Version]
- Zumaquero, A.; Kanematsu, S.; Nakayashiki, H.; Matas, A.; Martínez-Ferri, E.; Barceló-Muñóz, A.; Pliego-Alfaro, F.; López-Herrera, C.; Cazorla, F.M.; Pliego, C. Transcriptome analysis of the fungal pathogen Rosellinia necatrix during infection of a susceptible avocado rootstock identifies potential mechanisms of pathogenesis. BMC Genom. 2019, 20, 1016. [Google Scholar] [CrossRef] [Green Version]
- Ten Hoopen, G.M.; Krauss, U. Biology and control of Rosellinia bunodes, Rosellinia necatrix and Rosellinia pepo: A review. Crop Prot. 2006, 25, 89–107. [Google Scholar] [CrossRef]
- Zumaquero, A.; Martínez-Ferri, E.; Matas, A.J.; Reeksting, B.; Olivier, N.A.; Pliego-Alfaro, F.; Barceló, A.; van den Berg, N.; Pliego, C. Rosellinia necatrix infection induces differential gene expression between tolerant and susceptible avocado rootstocks. PLoS ONE 2019, 14, e0212359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engelbrecht, J.; van den Berg, N. Expression of defence-related genes against Phytophthora cinnamomi in five avocado rootstocks. S. Afr. J. Sci. 2013, 109, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Reeksting, B.J.; Coetzer, N.; Mahomed, W.; Engelbrecht, J.; van den Berg, N. De novo sequencing, assembly, and analysis of the root transcriptome of Persea americana (Mill.) in response to Phytophthora cinnamomi and flooding. PLoS ONE 2014, 9, e86399. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, N.; Mahomed, W.; Olivier, N.A.; Swart, V.; Crampton, B.G. Transcriptome analysis of an incompatible Persea americana-Phytophthora cinnamomi interaction reveals the involvement of SA- and JA-pathways in a successful defense response. PLoS ONE 2018, 13, e0205705. [Google Scholar] [CrossRef]
- Daniel, S.; Barz, W. Elicitor-induced metabolic changes in cell cultures of chickpea (Cicer arietinum L.) cultivars resistant and susceptible to Ascochyta rabiei. Planta 1990, 182, 279–286. [Google Scholar] [CrossRef]
- Ramirez-Suero, M.; Bénard-Gellon, M.; Chong, J.; Laloue, H.; Stempien, E.; Abou-Mansour, E.; Fontaine, F.; Larignon, P.; Mazet-Kieffer, F.; Farine, S.; et al. Extracellular compounds produced by fungi associated with Botryosphaeria dieback induce differential defence gene expression patterns and necrosis in Vitis vinifera cv. Chardonnay cells. Protoplasma 2014, 251, 1417–1426. [Google Scholar] [CrossRef] [Green Version]
- Bénard-Gellon, M.; Farine, S.; Goddard, M.L.; Schmitt, M.; Stempien, E.; Pensec, F.; Laloue, H.; Mazet-Kieffer, F.; Fontaine, F.; Larignon, P.; et al. Toxicity of extracellular proteins from Diplodia seriata and Neofusicoccum parvum involved in grapevine Botryosphaeria dieback. Protoplasma 2015, 252, 679–687. [Google Scholar] [CrossRef]
- Lima, M.R.M.; Ferreres, F.; Dias, A.C.P. Response of Vitis vinifera cell cultures to Phaeomoniella chlamydospora: Changes in phenolic production, oxidative state and expression of defence-related genes. Eur. J. Plant Pathol. 2012, 132, 133–146. [Google Scholar] [CrossRef]
- Larkin, P.J.; Scowcroft, W.R. Somaclonal variation—A novel source of variability from cell cultures for plant improvement. Theor. Appl. Genet. 1981, 60, 197–214. [Google Scholar] [CrossRef]
- Svabova, L.; Lebeda, A. In vitro selection for improved plant resistance to toxin-producing pathogens. J. Phytopathol. 2005, 153, 52–64. [Google Scholar] [CrossRef]
- Rai, M.K.; Kalia, R.K.; Singh, R.; Gangola, M.P.; Dhawan, A.K. Developing stress tolerant plants through in vitro selection—An overview of the recent progress. Environ. Exp. Bot. 2011, 71, 89–98. [Google Scholar] [CrossRef]
- Krishna, H.; Alizadeh, M.; Singh, D.; Singh, U.; Chauhan, N.; Eftekhari, M.; Sadh, R.K. Somaclonal variations and their applications in horticultural crops improvement. 3 Biotech 2016, 6, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anil, V.S.; Bennur, S.; Lobo, S. Somaclonal variations for crop improvement: Selection for disease resistant variants in vitro. Plant Sci. Today 2018, 5, 44–54. [Google Scholar] [CrossRef]
- Lestari, E.G. In vitro selection and somaclonal variation for biotic and abiotic stress tolerance. Biodiversitas 2006, 7, 297–301. [Google Scholar] [CrossRef]
- Jain, S.M. Tissue culture-derived variation in crop improvement. Euphytica 2001, 118, 153–166. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Pliego-Alfaro, F.; Murashige, T. Somatic embryogenesis in avocado (Persea americana Mill.) in vitro. Plant Cell Tissue Organ Cult. 1988, 112, 61–66. [Google Scholar] [CrossRef]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, 115. [Google Scholar] [CrossRef] [Green Version]
- Kõressaar, T.; Lepamets, M.; Kaplinski, L.; Raime, K.; Andreson, R.; Remm, M. Primer3_masker: Integrating masking of template sequence with primer design software. Bioinformatics 2018, 34, 1937–1938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Abbas, H.K.; Boyette, C.D.; Hoagland, R.E. Phytotoxicity of Fusarium, other fungal isolates, and of the phytotoxins fumonisin, fusaric acid and moniliformin to jimsonweed. Phytoprotection 1995, 76, 17–25. [Google Scholar] [CrossRef] [Green Version]
- Edwards, R.L.; Maitland, D.J.; Whalley, A.J.S. Metabolites of the higher fungi. Part 24. Cytochalasin N, O, P, Q, and R. New cytochalasins from the fungus Hypoxylon terricola Mill. J. Chem. Soc. Perkin Trans. 1989, 1, 57–65. [Google Scholar] [CrossRef]
- Whalley, A.J.S. The xylariaceous way of life. Mycol. Res. 1996, 100, 897–922. [Google Scholar] [CrossRef]
- Knogge, W. Molecular basis of specificity in host/fungus interactions. Eur. J. Plant Pathol. 1996, 102, 807–816. [Google Scholar] [CrossRef]
- Salehi, M.; Moieni, A.; Safaie, N.; Farhadi, S. Whole fungal elicitors boost paclitaxel biosynthesis induction in Corylus avellana cell culture. PLoS ONE 2020, 15, e0236191. [Google Scholar] [CrossRef]
- Young, D.H.; Michelotti, E.L.; Swindell, C.S.; Krauss, N.E. Antifungal properties of Taxol and various analogues. Experientia 1992, 48, 882–885. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.A.; Singh, S.K.; Kumar, B.S.; Doble, M. Synthesis, anti-fungal activity evaluation and QSAR studies on podophyllotoxin derivatives. Open Chem. 2007, 5, 880–897. [Google Scholar] [CrossRef] [Green Version]
- Barz, W.; Daniel, S.; Hinderer, W.; Jaques, U.; Kessmann, H.; Köster, J.K.; Tiemann, K. Elicitation and metabolism of phytoalexins in plant cell cultures. In Plant Cell Biotechnology; NATO ASI Series; Pais, M.S.S., Mavituna, F., Novais, J.M., Eds.; Springer: Berlin/Heidelberg, Germany, 1988; Volume 18. [Google Scholar] [CrossRef]
- Naveed, Z.A.; Ali, G.S. Comparative transcriptome analysis between a resistant and a susceptible wild tomato accession in response to Phytophthora parasitica. Int. J. Mol. Sci. 2018, 19, 3735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, X.; Gao, Z.; Zhao, L.; Zhang, S.; Wu, J.; Yang, Q.; Liu, S.; Chen, X. Comparative transcriptome analysis of resistant and susceptible wheat in response to Rhizoctonia cerealis. BMC Plant Biol. 2022, 22, 235. [Google Scholar] [CrossRef]
- Ramírez-Tejero, J.A.; Jiménez-Ruiz, J.; Serrano, A.; Belaj, A.; León, L.; de la Rosa, R.; Mercado -Blanco, J.; Luque, F. Verticillium wilt resistant and susceptible olive cultivars express a very different basal set of genes in roots. BMC Genom. 2021, 22, 229. [Google Scholar] [CrossRef]
- Yang, Q.; Yang, L.; Wang, Y.; Chen, Y.; Hu, K.; Yang, W.; Zuo, S.; Xu, J.; Kang, Z.; Xiao, X.; et al. A High-Quality genome of Rhizoctonia solani, a devastating fungal pathogen with a wide host range. Mol. Plant-Microbe Interact. 2022, 35, 954–958. [Google Scholar] [CrossRef] [PubMed]
- Khandagale, K.; Roylawar, P.; Kulkarni, O.; Khambalkar, P.; Ade, A.; Kulkarni, A.; Singh, M.; Gawande, S. Comparative Transcriptome Analysis of Onion in Response to Infection by Alternaria porri (Ellis) Cifferi. Front. Plant Sci. 2022, 13, 857306. [Google Scholar] [CrossRef] [PubMed]
- Chittem, K.; Yajima, W.R.; Goswami, R.S.; del Río-Mendoza, L.E. Transcriptome analysis of the plant pathogen Sclerotinia sclerotiorum interaction with resistant and susceptible canola (Brassica napus) lines. PLoS ONE 2020, 15, e0229844. [Google Scholar] [CrossRef] [Green Version]
- Lagrimini, L.M.; Vaughn, J.; Erb, W.A.; Miller, S.A. Peroxidase overproduction in tomato: Wound-induced polyphenol deposition and disease resistance. HortScience 1993, 28, 218–221. [Google Scholar] [CrossRef] [Green Version]
- Kawasaki, T.; Koita, H.; Nakatsubo, T.; Hasegawa, K.; Wakabayashi, K.; Takahashi, H.; Umemura, K.; Umezawa, T.; Shimamoto, K. Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 230–235. [Google Scholar] [CrossRef] [Green Version]
- van den Berg, N.; Christie, J.B.; Aveling, T.A.S.; Engelbrecht, J. Callose and β-1, 3-glucanase inhibit Phytophthora cinnamomi in a resistant avocado rootstock. Plant Pathol. 2018, 67, 1150–1160. [Google Scholar] [CrossRef]
- Jayasankar, S.; Litz, R.E. Characterization of embryogenic mango cultures selected for resistance to Colletotrichum gloeosporoides culture filtrate and phytotoxin. Theor. Appl. Genet. 1998, 96, 823–831. [Google Scholar] [CrossRef]
- Singh, R.; Sindhu, A.; Singal, H.R.; Singh, R. Biochemical basis of resistance in chickpea (Cicer arietinum L.) against Fusarium wilt. Acta Phytopathol. Entomol. Hung. 2003, 38, 13–19. [Google Scholar] [CrossRef]
- Jayasankar, S.; Li, Z.; Gray, D.J. In vitro selection of Vitis vinífera ‘Chardonnay’ with Elsinoe ampelina culture filtrate is accompanied by fungal resistance and enhanced secretion of chitinase. Planta 2000, 211, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Jayasankar, S.; Li, Z.; Gray, D.J. Constitutive expression of Vitis vinifera thaumatin-like protein after in vitro selection and its role in anthracnose resistance. Funct. Plant Biol. 2003, 30, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Ferri, E.; Moreno-Ortega, G.; van den Berg, N.; Pliego, C. Mild water stress-induced priming enhance tolerance to Rosellinia necatrix in susceptible avocado rootstocks. BMC Plant Biol. 2019, 19, 458. [Google Scholar] [CrossRef] [Green Version]
- Müller, M.; Munné-Bosch, S. Ethylene response factors: A key regulatory hub in hormone and stress signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jashni, M.K.; Mehrabi, R.; Collemare, J.; Mesarich, C.H.; de Wit, P.J.G.M. The battle in the apoplast: Further insights into the roles of proteases and their inhibitors in plant–pathogen interactions. Front. Plant Sci. 2015, 6, 584. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Gene ID | Description | Resistant Callus Line (L3) | Susceptible Callus Line (AN-9) | ||
|---|---|---|---|---|---|
| RNA-Seq FC | qRT-PCR FC | RNA-Seq FC | qRT-PCR FC | ||
| Pag64949 | Proteinase inhibitor | 48.50 | 38.54 ± 2.00 a | 22.29 | 14.60 ± 1.43 b |
| Pag115909 | Tumor related protein | 6.79 | 11.59 ± 0.40 a | 5.20 | 6.61 ± 0.58 b |
| MSTRG.18398 | Pathogenesis-related protein P2 | ND | −1.64 ± 0.19 | ND | −1.99 ± 0.20 |
| Pag191108 | Lignin-forming anionic peroxidase | 78.36 | 64.88 ± 3.93 a | 26.95 | 21.30 ± 5.46 b |
| Pag291312 | Heat shock transcription factor 30 | 19.75 | 25.15 ± 5.55 b | 86.46 | 62.00 ± 11.23 a |
| Pag275152 | Translocon-associated protein subunit beta | −2.88 | −1.76 ± 0.11 | −4.79 | −2.52 ± 0.38 |
| Gene ID | Description (Blast NCBI) | TCLFvsTCL FC | SCLFvsSCL FC |
|---|---|---|---|
| Cell wall | |||
| Pag55749 | Cinnamoyl-CoA reductase 1 | 94.23 | 79.56 |
| Pag191108 | Lignin-forming anionic peroxidase | 78.36 | 26.95 |
| Pag66455 | Glucan 1,3-beta-glucosidase A-like protein | 43.23 | 29.77 |
| Pag116557 | Endonuclease-8-like protein | 45.04 | 8.59 |
| Pag44234 | Basic endochitinase-like-protein | 2.33 | ND |
| Detoxification and disease-resistance proteins | |||
| Pag255058 | Disease resistance protein RPM1-like | 18.05 | ND |
| Pag319424 | Pleiotropic drug resistance protein 3-like protein isoform X1 | 10.08 | 5.72 |
| Pag279361 | Detoxification 49 protein | 7.65 | 4.96 |
| Pag352089 | Putative disease resistance protein | 4.76 | ND |
| Pag297678 | Pleiotropic drug resistance protein 2-like protein | 2.06 | ND |
| Pathogenesis-related proteins | |||
| Pag289080 | Pathogenesis-related protein PR-4-like protein | 18.65 | 15.79 |
| Pag154162 | Thaumatin-like protein | 9.44 | 6.37 |
| Pag168264 | Pathogenesis-related protein STH-21 | 7.97 | 4.64 |
| Pag154170 | Thaumatin-like protein 1 | 7.67 | 5.11 |
| Pag103568 | Pathogenesis-related genes transcriptional activator PTI5-like | 2.31 | ND |
| Protease and protease inhibitor activity | |||
| Pag64949 | Proteinase inhibitor | 48.50 | 22.29 |
| Pag341255 | Subtilisin-like protein protease | 41.13 | 7.70 |
| Pag231368 | Cysteine proteinase RD21A-like protein | 7.66 | 4.94 |
| Pag26660 | Serine carboxypeptidase-like protein | 6.11 | ND |
| Pag136044 | Subtilisin-like protein protease SBT5.3 | 5.72 | ND |
| Pag214427 | Subtilisin-like protease SBT5.6 | 2.60 | ND |
| Pag297773 | Serine carboxypeptidase 24 | 2.00 | ND |
| Transcription factor MYB, WRKY, MYC and NAC | |||
| Pag51890 | Transcription factor MYB1R1-like protein isoform X2 | 27.15 | ND |
| Pag121997 | RWR3 transcription factor MYB108-like protein 1 | 19.25 | ND |
| Pag345989 | Putative WRKY transcription factor 75 | 17.15 | 6.68 |
| Pag306446 | WRKY64 | 12.48 | 5.64 |
| Pag307121 | Putative WRKY transcription factor 72 Isoform X1 | 7.43 | 4.58 |
| Pag28154 | WRKY transcription factor 55 | 3.22 | ND |
| Pag223376 | NAC domain-containing protein 96-like protein | 3.22 | ND |
| Pag389501 | SANT/Myb domain-containing protein | 2.64 | ND |
| Pag46518 | WRKY67 | 2.56 | ND |
| Pag240398 | MYB108-like protein | 2.56 | ND |
| Pag190993 | MYB family transcription factor APL | 2.46 | ND |
| Pag201669 | NAC domain-containing protein | 2.32 | ND |
| Pag276502 | SANT/Myb domain-containing protein | 2.31 | ND |
| Pag262640 | NAC domain-containing protein | 2.16 | ND |
| Pag296108 | Transcription factor MYC2-like protein | 2.01 | ND |
| Hormonal regulation | |||
| Pag26291 | Ethylene-responsive transcription factor (ERF) 020 | 60.92 | 45.99 |
| Pag169786 | Abscisic acid-insensitive 5-like protein 6 | 37.65 | ND |
| Pag204839 | Ethylene-responsive transcription factor ERF109-like protein | 35.06 | ND |
| Pag204844 | AP2/ERF domain-containing protein | 33.32 | 23.44 |
| Pag174105 | Abscisic acid 8’hydroxylase 2 | 22.62 | 11.51 |
| Pag212858 | Gibberellin 20 oxidase | 12.74 | 3.58 |
| Pag120094 | Auxin-responsive protein SAUR71-like protein | 8.02 | 3.97 |
| Pag197469 | Ethylene-responsive transcription factor 1B-like protein | 7.05 | 2.18 |
| Pag157588 | Auxin-induced protein | 5.51 | ND |
| Pag70205 | Auxin transporte-like protein 5 | 4.22 | ND |
| Pag101088 | AP2/ERF domain-containing protein | 3.03 | ND |
| Pag75125 | Ethylene-responsive transcription factor RAP2-11 like protein | 2.80 | ND |
| Pag46880 | Gibberellic acid methyltransferase 2 isoform X2 | 2.54 | ND |
| Pag227350 | Auxin-responsive protein SAUR71 | 2.26 | ND |
| Pag75115 | Ethylene-responsive transcription factor 1A | 2.09 | ND |
| Pag362340 | Auxin-induced protein AUX22 | 2.04 | ND |
| Redox homeostasis | |||
| Pag378827 | Cytochrome P450 704B1 | 35.03 | 8.60 |
| Pag254946 | Peroxidase 68 | 34.50 | ND |
| Pag236665 | Beta-amyrin 28-oxidase-like protein | 27.32 | ND |
| Pag268581 | Protein DMR6-Like oxygenase 2-like protein | 26.89 | 16.06 |
| Pag364610 | Protein DMR6-Like oxygenase 2-like protein | 25.61 | 18.95 |
| Pag264980 | Cytochrome P450 71A1 | 23.62 | 16.60 |
| Pag323404 | P450 domain-containing protein | 17.10 | ND |
| Pag260376 | Cytochrome P450 94C1-like protein | 16.83 | 14.05 |
| Pag255004 | Cationic peroxidase 1-like protein | 14.79 | 8.31 |
| Pag88265 | Cytochrome P450 78a5-Like protein | 13.07 | 9.38 |
| Pag171020 | Allene oxidase synthase 1 | 11.22 | 7.42 |
| Pag281952 | Cytocrome B6-f complex iron-sulfur subunit 2. | 10.23 | ND |
| Pag171015 | Allene oxidase synthase 1 | 9.80 | 7.62 |
| Pag34770 | Protein DMR6-Like oxygenase 2 | 8.60 | 4.90 |
| Pag339287 | Plant peroxidase | 7.21 | ND |
| Pag236679 | Cytochrome P450 | 6.53 | ND |
| Pag19781 | Cytochrome P450 | 5.24 | 3.29 |
| Pag357969 | NADPH-cytochrome P450 reductase-like | 3.86 | ND |
| Pag369777 | Cytochrome P450 87A3 | 2.83 | ND |
| Pag377126 | Cytochrome P450 cyp72A219-like protein | 2.09 | ND |
| Gene ID | Annotation | TCLFvsTCL FC | Gene ID | Annotation | SCLFvsSCL FC |
|---|---|---|---|---|---|
| Pag246776 | Hypothetical protein | 188.08 | Pag344126 | Protein P21-like protein | 644.07 |
| Pag92571 | Heme peroxidase | 177.29 | Pag234412 | Hypothetical protein CKAN_00293900 | 287.37 |
| Pag162432 | Photosystem II reaction center W protein | 145.81 | Pag242850 | Nicotianamine synthase | 260.95 |
| Pag21497 | Protein chlororespiratory reduction 7 | 140.12 | Pag92571 | Heme peroxidase | 242.25 |
| Pag147146 | Putative transcription factor bHLH041 | 131.20 | Pag108159 | Glyco-hydro 9 domain-containing protein | 182.57 |
| Pag321196 | Phosphoribosylformylglycinamidine cyclo-ligase, chloroplastic | 130.83 | Pag81308 | Small heat shock protein 22K | 166.87 |
| Pag275645 | Vacuolar amino acid transporter protein | 118.48 | Pag102350 | Sugar/inositol transport protein | 155.56 |
| Pag308947 | Protein P21-like | 113.78 | Pag44452 | ABC transporter G family member | 153.64 |
| Pag127237 | Hypothetical protein CKAN_02343900 | 111.18 | Pag246776 | Hypothetical protein | 143.79 |
| Pag55749 | Cinnamoyl-CoA reductase | 94.23 | Pag102899 | F-box protein FBW2 | 135.27 |
| Pag369420 | Omega-hydroxypalmitate O-feruloyl transferase | 88.77 | Pag308947 | Protein P21-like | 117.13 |
| Pag102899 | F-box protein FBW2 | 87.72 | Pag275645 | Vacuolar amino acid transporter protein | 112.13 |
| Pag380545 | Monocopper oxidase-like protein SKU5 | 82.92 | Pag129297 | 17.3 kDa class II heat shock protein | 103.24 |
| Pag147154 | Putative transcription factor bHLH041 | 79.00 | Pag21497 | Protein chlororespiratory reduction 7 | 102.12 |
| Pag191108 | Lignin-forming anionic peroxidase | 78.36 | Pag104432 | Transcription initiation factor IIF subunit alpha | 100.13 |
| Pag112733 | Cysteine-rich receptor-like protein kinase 2 | 76.61 | Pag147146 | Putative transcription factor bHLH041 | 90.97 |
| Pag385754 | TOX high mobility group box family member 4-A, Putative isoform | 74.68 | Pag288015 | Alpha carbonic anhydrase 1. chloroplastic | 88.85 |
| Pag226385 | Ethylene-responsive transcription factor ERF096 | 63.20 | Pag365527 | Heat shock factor protein HSF30 | 88.83 |
| Pag26291 | Ethylene-responsive transcription factor ERF020 | 60.92 | Pag47864 | 22.0 kDa class IV heat shock protein | 87.44 |
| Pag48634 | Lysine histidine transporter-like 5 | 60.09 | Pag291312 | Heat shock factor protein HSF30 | 86.46 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreno-Pérez, A.; Zumaquero, A.; Martínez-Ferri, E.; López-Herrera, C.; Pliego-Alfaro, F.; Palomo-Ríos, E.; Pliego, C. A Comparative Transcriptome Analysis of Avocado Embryogenic Lines Susceptible or Resistant to Rosellinia necatrix Exudate. Agronomy 2023, 13, 1354. https://doi.org/10.3390/agronomy13051354
Moreno-Pérez A, Zumaquero A, Martínez-Ferri E, López-Herrera C, Pliego-Alfaro F, Palomo-Ríos E, Pliego C. A Comparative Transcriptome Analysis of Avocado Embryogenic Lines Susceptible or Resistant to Rosellinia necatrix Exudate. Agronomy. 2023; 13(5):1354. https://doi.org/10.3390/agronomy13051354
Chicago/Turabian StyleMoreno-Pérez, Ana, Adela Zumaquero, Elsa Martínez-Ferri, Carlos López-Herrera, Fernando Pliego-Alfaro, Elena Palomo-Ríos, and Clara Pliego. 2023. "A Comparative Transcriptome Analysis of Avocado Embryogenic Lines Susceptible or Resistant to Rosellinia necatrix Exudate" Agronomy 13, no. 5: 1354. https://doi.org/10.3390/agronomy13051354
APA StyleMoreno-Pérez, A., Zumaquero, A., Martínez-Ferri, E., López-Herrera, C., Pliego-Alfaro, F., Palomo-Ríos, E., & Pliego, C. (2023). A Comparative Transcriptome Analysis of Avocado Embryogenic Lines Susceptible or Resistant to Rosellinia necatrix Exudate. Agronomy, 13(5), 1354. https://doi.org/10.3390/agronomy13051354






