Plant Rhizospheres Harbour Specific Fungal Groups and Form a Stable Co-Occurrence Pattern in the Saline-Alkali Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Information
2.2. Sample Collection and Physiochemical Analysis
2.3. DNA Extraction, Sequencing, and Bioinformatics Analysis
2.4. Construction of Co-Occurrence Network
2.5. Statistical Analyses
3. Results
3.1. Soil Properties
3.2. Soil Fungal Composition
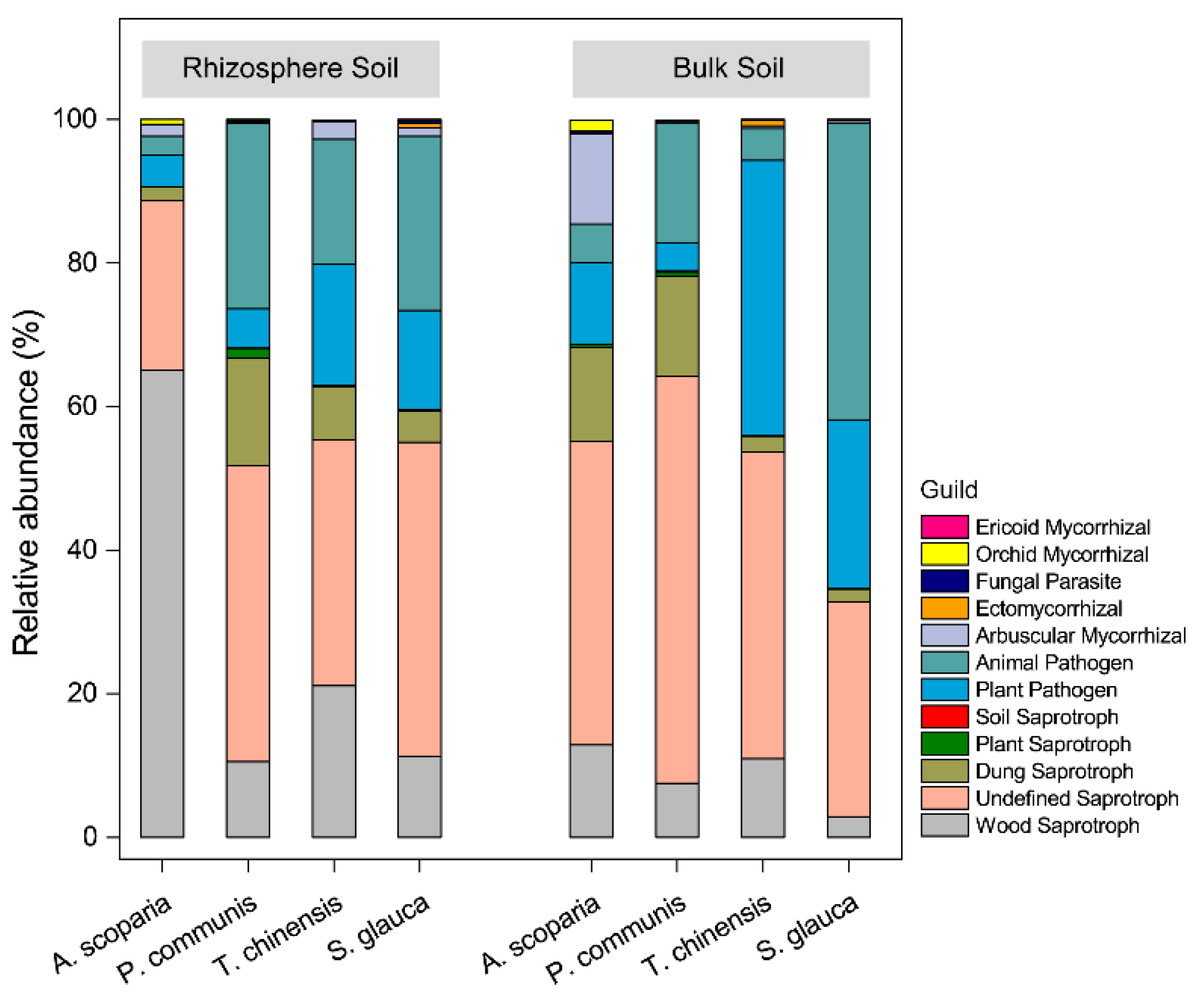
3.3. Functional Guilds
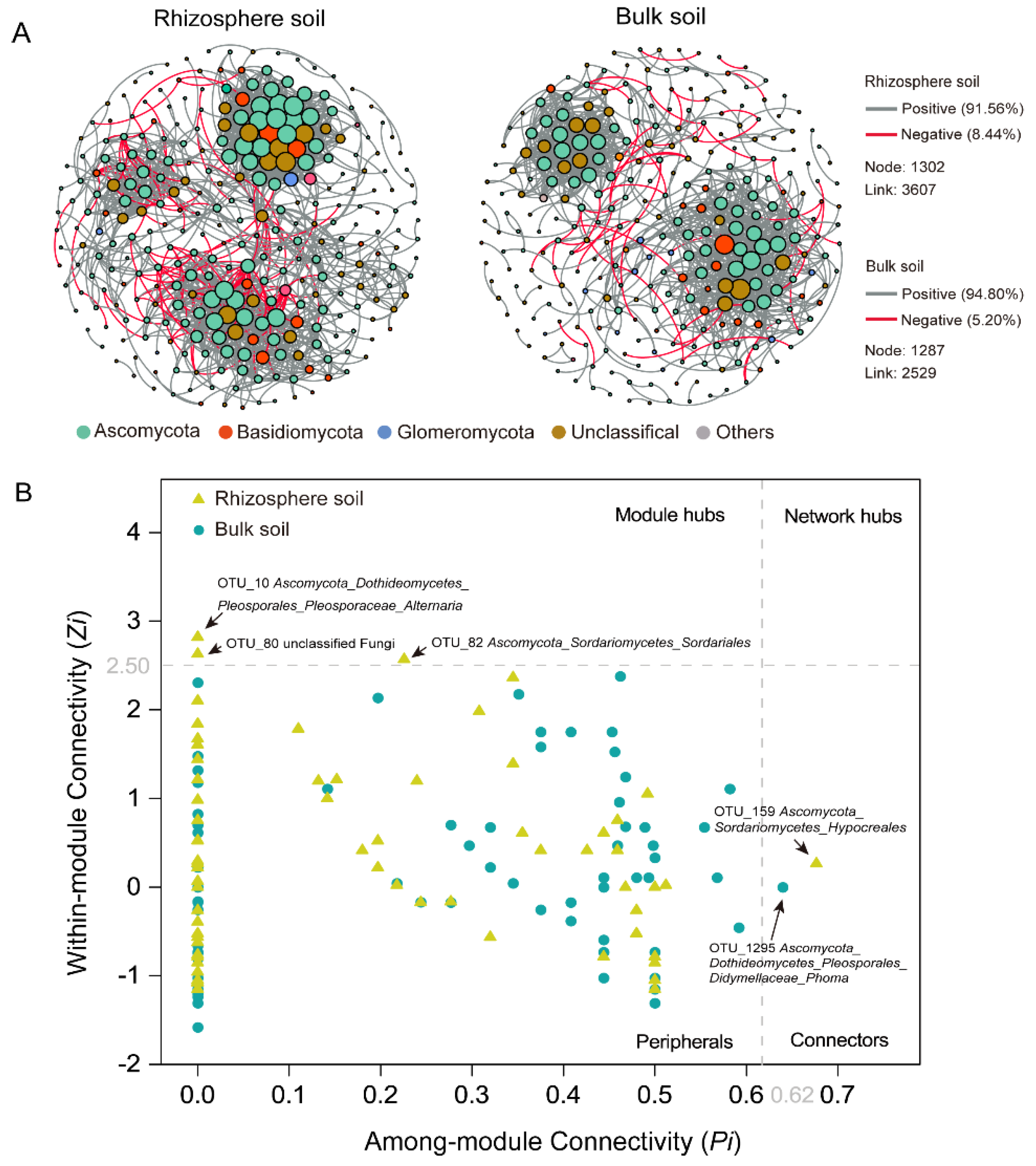
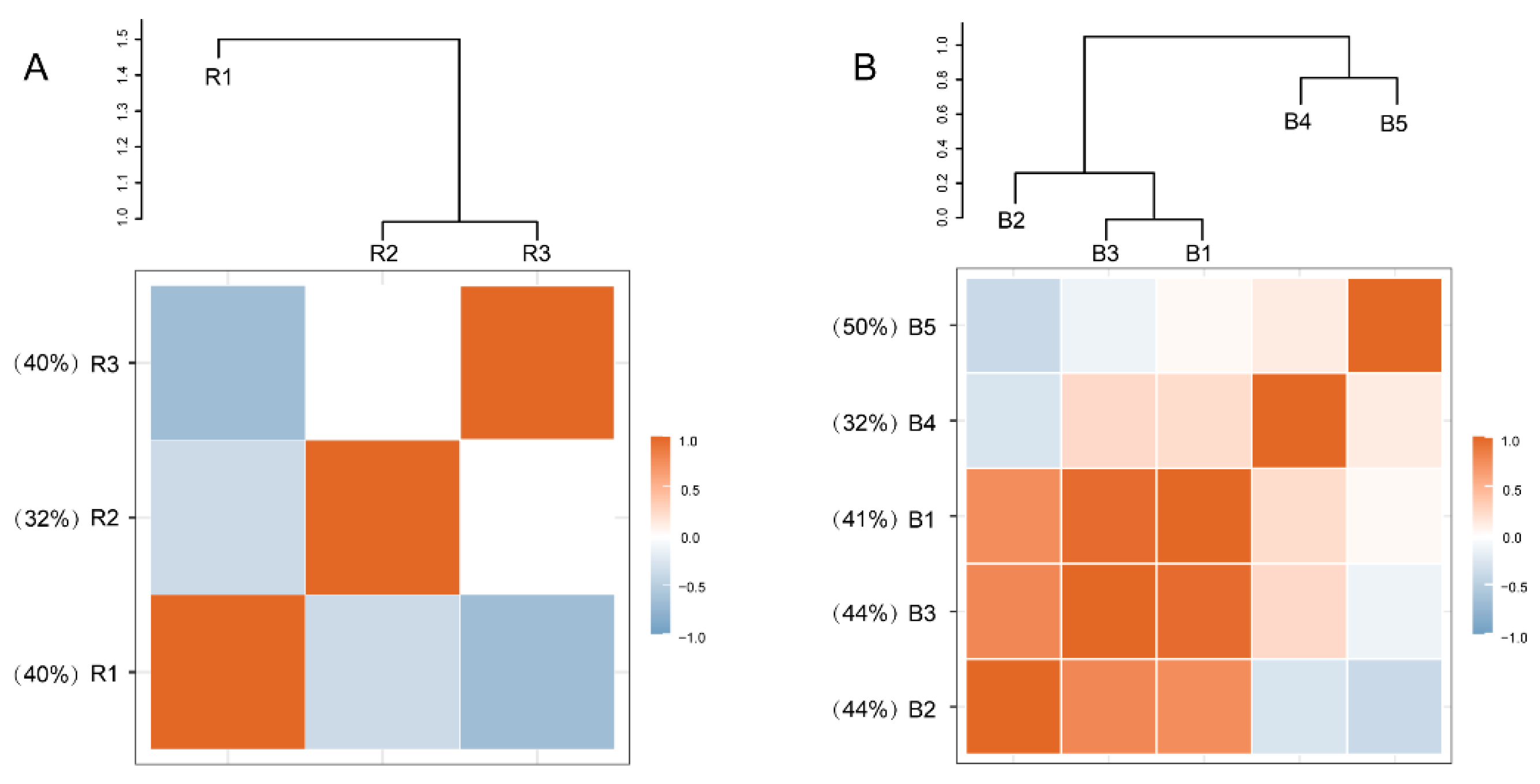
3.4. Fungal Co-Occurrence Network Analyses
4. Discussion
4.1. Differences in Fungal Composition between the Rhizosphere and Bulk Soil of Salt-Tolerant Plants Were Not Always Detected
4.2. Rhizosphere Harboured a More Complex Fungal Co-Occurrence Network
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cho, K.H.; Beon, M.S.; Jeong, J.C. Dynamics of soil salinity and vegetation in a reclaimed area in Saemangeum, Republic of Korea. Geoderma 2018, 321, 42–51. [Google Scholar] [CrossRef]
- Yu, P.J.; Liu, S.W.; Yang, H.T.; Fan, G.H.; Zhou, D.W. Short-term land use conversions influence the profile distribution of soil salinity and sodicity in northeastern China. Ecol. Indic. 2018, 88, 79–87. [Google Scholar] [CrossRef]
- Yadav, S.; Irfan, M.; Ahmad, A.; Hayat, S. Causes of salinity and plant manifestations to salt stress: A review. J. Environ. Biol. 2011, 32, 667–685. [Google Scholar] [PubMed]
- Pitman, M.G.; Läuchli, A. Global impact of salinity and agricultural ecosystems. In Salinity: Environment Plants Molecules; Läuchli, A., Lüttge, U., Eds.; Springer: Dordrecht, The Netherlands, 2002; pp. 3–20. [Google Scholar]
- Sahab, S.; Suhani, I.; Srivastava, V.; Chauhan, P.S.; Singh, R.P.; Prasad, V. Potential risk assessment of soil salinity to agroecosystem sustainability: Current status and management strategies. Sci. Total Environ. 2021, 764, 144164. [Google Scholar] [CrossRef]
- Vargas, R.; Pankova, E.I.; Balyuk, S.A.; Krasilnikov, P.V.; Khasankhanova, G.M. Handbook for Saline Soil Management; FAO: Rome, Italy, 2018. [Google Scholar]
- Jesus, J.M.; Danko, A.S.; Fiúza, A.; Borges, M.T. Phytoremediation of salt-affected soils: A review of processes, applicability, and the impact of climate change. Environ. Sci. Pollut. Res. 2015, 22, 6511. [Google Scholar] [CrossRef]
- Liu, H.Q.; Lu, X.B.; Li, Z.H.; Tian, C.Y.; Song, J. The role of root-associated microbes in growth stimulation of plants under saline conditions. Land Degrad. Dev. 2021, 32, 3471–3486. [Google Scholar] [CrossRef]
- Li, N.; Shao, T.Y.; Zhou, Y.J.; Cao, Y.C.; Hu, H.Y.; Sun, Q.K.; Long, X.H.; Yue, Y.; Gao, X.M.; Rengel, Z. Effects of planting Melia azedarach L. on soil properties and microbial community in saline-alkali soil. Land Degrad. Dev. 2021, 32, 2951–2961. [Google Scholar] [CrossRef]
- Martin, F.M.; Uroz, S.; Barker, D.G. Ancestral alliances: Plant mutualistic symbioses with fungi and bacteria. Science 2017, 356, eaad4501. [Google Scholar] [CrossRef]
- Rodriguez, P.A.; Rothballer, M.; Chowdhury, S.P.; Nussbaumer, T.; Falter-Braun, P. Systems biology of plant-microbiome interactions. Mol. Plant 2019, 12, 804–821. [Google Scholar] [CrossRef]
- Feng, Q.J.; Cao, S.L.; Liao, S.J.; Wassie, M.; Sun, X.Y.; Chen, L.; Xie, Y. Fusarium equiseti-inoculation altered rhizosphere soil microbial community, potentially driving perennial ryegrass growth and salt tolerance. Sc. Total Environ. 2023, 871, 162153. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Guo, D.F. Response of soil fungi community structure to salt vegetation succession in the Yellow River Delta. Curr. Microbiol. 2016, 73, 595–601. [Google Scholar] [CrossRef]
- Carneiro, B.; Cardoso, P.; Figueira, E.; Lopes, I.; Venâncio, C. Forward-looking on new microbial consortia: Combination of rot fungi and rhizobacteria on plant growth-promoting abilities. Appl. Soil Ecol. 2023, 182, 104689. [Google Scholar] [CrossRef]
- Li, X.N.; Han, S.J.; Wang, G.Y.; Liu, X.Y.; Erick, A.; Xie, Y.; Fu, J.M. The fungus Aspergillus aculeatus enhances salt-stress tolerance, metabolite accumulation, and improves forage quality in perennial ryegrass. Front. Microbiol. 2017, 8, 1664. [Google Scholar] [CrossRef] [PubMed]
- Guan, B.; Zhang, H.X.; Wang, X.H.; Yang, S.S.; Chen, M.; Hou, A.X.; Grace, A.C.; Han, G.X. Salt is a main factor shaping community composition of arbuscular mycorrhizal fungi along a vegetation successional series in the Yellow River Delta. Catena 2020, 185, 104318. [Google Scholar] [CrossRef]
- Turner, T.R.; Ramakrishnan, K.; Walshaw, J.; Heavens, D.; Alston, M.; Swarbreck, D.; Osbourn, A.; Grant, A.; Poole, P.S. Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 2013, 7, 2248–2258. [Google Scholar] [CrossRef]
- Williams, A.; de Vries, F.T. Plant root exudation under drought: Implications for ecosystem functioning. New Phytol. 2020, 225, 1899–1905. [Google Scholar] [CrossRef]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef]
- Fan, K.K.; Weisenhorn, P.; Gilbert, J.A.; Chu, H.Y. Wheat rhizosphere harbors a less complex and more stable microbial co-occurrence pattern than bulk soil. Soil Biol. Biochem. 2018, 125, 251–260. [Google Scholar] [CrossRef]
- Chen, J.; Xu, H.; He, D.; Li, Y.D.; Luo, T.S.; Yang, H.G.; Lin, M.X. Historical logging alters soil fungal community composition and network in a tropical rainforest. Forest Ecol. Manag. 2019, 433, 228–239. [Google Scholar] [CrossRef]
- Lima-Mendez, G.; Faust, K.; Henry, N.; Decelle, J.; Colin, S.; Carcillo, F.; Chaffron, S.; Ignacio-Espinosa, J.C.; Roux, S.; Vincent, F.; et al. Determinants of community structure in the global plankton interactome. Science 2015, 348, 1262073. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.H.; Yu, Z.B.; Yu, L.; Wang, D.W.; Guan, Y.A.; Liu, X.H.; Gu, C.H.; Cui, B.S. In-situ organic phosphorus mineralization in sediments in coastal wetlands with different flooding periods in the Yellow River Delta, China. Sci. Total Environ. 2019, 682, 417–425. [Google Scholar] [CrossRef]
- Zhao, Q.Q.; Bai, J.H.; Zhang, G.L.; Jia, J.; Wang, W.; Wang, X. Effects of water and salinity regulation measures on soil carbon sequestration in coastal wetlands of the Yellow River Delta. Geoderma 2018, 319, 219–229. [Google Scholar] [CrossRef]
- Liu, S.; Hou, X.; Yang, M.; Chen, F.Y.; Coxixo, A.; Wu, X.; Zhang, Y.Q. Factors driving the relationships between vegetation and soil properties in the Yellow River Delta, China. Catena 2018, 165, 279–285. [Google Scholar] [CrossRef]
- Fan, X.; Pedroli, B.; Liu, G.; Liu, Q.; Liu, H.; Shu, L. Soil salinity development in the yellow river delta in relation to groundwater dynamics. Land Degrad. Dev. 2012, 23, 175–189. [Google Scholar] [CrossRef]
- Riley, D.; Barber, S.A. Effect of ammonium and nitrate fertilization on phosphorus uptake as related to root-induced pH changes at the root-soil interface. Soil Sci. Soc. Am. J. 1971, 35, 301–306. [Google Scholar] [CrossRef]
- Liu, F.D.; Mo, X.; Kong, W.J.; Song, Y. Soil bacterial diversity, structure, and function of Suaeda salsa in rhizosphere and non-rhizosphere soils in various habitats in the Yellow River Delta, China. Sci. Total Environ. 2020, 740, 140144. [Google Scholar] [CrossRef]
- Bao, S. Soil and Agricultural Chemistry Analysis; Agriculture Publication: Beijing, China, 2000; pp. 25–103. [Google Scholar]
- Ramette, A.; Tiedje, J.M. Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc. Natl. Acad. Sci. USA 2007, 104, 2761–2766. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Deng, Y.; Jiang, Y.H.; Yang, Y.F.; He, Z.L.; Luo, F.; Zhou, J.Z. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef]
- Olesen, J.M.; BascompSte, J.; Dupont, Y.L.; Jordano, P. The modularity of pollination networks. Proc. Natl. Acad. Sci. USA 2007, 104, 19891–19896. [Google Scholar] [CrossRef] [PubMed]
- Tylianakis, J.M.; Morris, R.J. Ecological networks across environmental gradients. Annu. Rev. Ecol. Evol. Syst. 2017, 48, 25–48. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Xiong, Y.W.; Li, X.W.; Wang, T.T.; Gong, Y.; Zhang, C.M.; Xing, K.; Qin, S. Root exudates-driven rhizosphere recruitment of the plant growth-promoting rhizobacterium Bacillus flexus KLBMP 4941 and its growth-promoting effect on the coastal halophyte Limonium sinense under salt stress. Ecotox. Environ. Saf. 2020, 194, 110374. [Google Scholar] [CrossRef]
- Hinsinger, P.; Bengough, A.G.; Vetterlein, D.; Young, I.M. Rhizosphere: Biophysics, biogeochemistry and ecological relevance. Plant Soil 2009, 321, 117–152. [Google Scholar] [CrossRef]
- Koranda, M.; Schnecker, J.; Kaiser, C.; Fuchslueger, L.; Kitzler, B.; Stange, C.F.; Sessitsch, A.; Zechmeister-Boltenstern, S.; Richtera, A. Microbial processes and community composition in the rhizosphere of European beech–the influence of plant C exudates. Soil Biol. Biochem. 2011, 43, 551–558. [Google Scholar] [CrossRef]
- Chaudhary, D.R.; Gautam, R.K.; Yousuf, B.; Mishra, A.; Jha, B. Nutrients, microbial community structure and functional gene abundance of rhizosphere and bulk soils of halophytes. Appl. Soil Ecol. 2015, 91, 16–26. [Google Scholar] [CrossRef]
- Mohammadi, M.H.; Khataar, M.; Shekari, F. Effect of soil salinity on the wheat and bean root respiration rate at low matric suctions. Paddy Water Environ. 2017, 15, 639–648. [Google Scholar] [CrossRef]
- Huang, Y.; Fan, G.H.; Zhou, D.W.; Pang, J.Y. Phenotypic plasticity of four Chenopodiaceae species with contrasting saline–sodic tolerance in response to increased salinity-sodicity. Ecol. Evol. 2019, 9, 1545–1553. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, C.J.; de Vries, F.T.; van Dongen, B.E.; Bardgett, R.D. Root traits explain rhizosphere fungal community composition among temperate grassland plant species. New Phytol. 2021, 229, 1492–1507. [Google Scholar] [CrossRef]
- Baldrian, P.; Kolařík, M.; Štursová, M.; Kopecký, J.; Valášková, V.; Větrovský, T.; Žifčáková, L.; Šnajdr, J.; Rídl, J.; Vlček, Č.; et al. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 2012, 6, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Polyakova, O.; Billor, N. Impact of deciduous tree species on litterfall quality, decomposition rates and nutrient circulation in pine stands. Forest Ecol. Manag. 2007, 253, 11–18. [Google Scholar] [CrossRef]
- Aati, H.Y.; Perveen, S.; Orfali, R.; Al-Taweel, A.M.; Aati, S.; Wanner, J.; Khan, A.; Mehmood, R. Chemical composition and antimicrobial activity of the essential oils of Artemisia absinthium, Artemisia scoparia, and Artemisia sieberi grown in Saudi Arabia. Arab. J. Chem. 2020, 13, 8209–8217. [Google Scholar] [CrossRef]
- Van Geel, M.; Yu, K.; Ceulemans, T.; Peeters, G.; van Acker, K.; Geerts, W.; Ramos, M.A.; Serafim, C.; Kastendeuch, P.; Najjar, G.; et al. Variation in ectomycorrhizal fungal communities associated with Silver linden (Tilia tomentosa) within and across urban areas. FEMS Microbiol. Ecol. 2018, 94, fiy207. [Google Scholar] [CrossRef]
- Abdel Latef, A.A.H.; Miransari, M. The role of arbuscular mycorrhizal fungi in alleviation of salt stress. In Use of Microbes for the Alleviation of Soil Stresses: Volume 2: Alleviation of Soil Stress by PGPR and Mycorrhizal Fungi; Springer: New York, NY, USA, 2014; pp. 23–38. [Google Scholar] [CrossRef]
- Dickie, I.A.; Martínez-García, L.B.; Koele, N.; Grelet, G.A.; Tylianakis, J.M.; Peltzer, D.A.; Richardson, S.J. Mycorrhizas and mycorrhizal fungal communities throughout ecosystem development. Plant Soil 2013, 367, 11–39. [Google Scholar] [CrossRef]
- Hajiboland, R.; Aliasgharzadeh, N.; Laiegh, S.F.; Poschenrieder, C. Colonization with arbuscular mycorrhizal fungi improves salinity tolerance of tomato (Solanum lycopersicum L.) plants. Plant Soil 2010, 331, 313–327. [Google Scholar] [CrossRef]
- Luo, Z.B.; Li, K.; Gai, Y.; Göbel, C.; Wildhagen, H.; Jiang, X.N.; Feußner, I.; Rennenberg, H.; Polle, A. The ectomycorrhizal fungus (Paxillus involutus) modulates leaf physiology of poplar towards improved salt tolerance. Environ. Exp. Bot. 2011, 72, 304–311. [Google Scholar] [CrossRef]
- Cao, D.; Shi, F.C.; Koike, T.; Lu, Z.H.; Sun, J.K. Halophyte plant communities affecting enzyme activity and microbes in saline soils of the Yellow River Delta in China. CLEAN—Soil Air Water 2014, 42, 1433–1440. [Google Scholar] [CrossRef]
- Decker, J.P. Salt secretion by Tamarix pentandra Pall. Forest Sci. 1961, 7, 214–217. [Google Scholar] [CrossRef]
- Koyro, H.W.; Geissler, N.; Hussin, S.; Debez, A.; Huchzermeyer, B. Strategies of halophytes to survive in a salty environment. In Abiotic Stress and Plant Responses; Khan, N.A., Singh, S., Eds.; I.K. International Publishing: New Delhi, India, 2008; pp. 83–104. [Google Scholar]
- Li, Y.M.; Yang, Y.; Wu, T.; Zhang, H.; Wei, G.H.; Li, Z.F. Rhizosphere bacterial and fungal spatial distribution and network pattern of Astragalus mongholicus in representative planting sites differ the bulk soil. Appl. Soil Ecol. 2021, 168, 104114. [Google Scholar] [CrossRef]
- Yan, Y.; Kuramae, E.E.; de Hollander, M.; Klinkhamer, P.G.L.; van Veen, J.A. Functional traits dominate the diversity-related selection of bacterial communities in the rhizosphere. ISME J. 2017, 11, 56–66. [Google Scholar] [CrossRef]
- Mendes, L.W.; Kuramae, E.E.; Navarrete, A.A.; van Veen, J.A.; Tsai, S.M. Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 2014, 8, 1577–1587. [Google Scholar] [CrossRef] [PubMed]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.G.; Zhang, J.; Liu, Y.; Shi, P.; Wei, G.H. Co-occurrence patterns of soybean rhizosphere microbiome at a continental scale. Soil Biol. Biochem. 2018, 118, 178–186. [Google Scholar] [CrossRef]
- Wang, S.; Wang, X.B.; Han, X.G.; Deng, Y. Higher precipitation strengthens the microbial interactions in semi-arid grassland soils. Glob. Ecol. Biogeogr. 2018, 27, 570–580. [Google Scholar] [CrossRef]
- Coyte, K.Z.; Schluter, J.; Foster, K.R. The ecology of the microbiome: Networks, competition, and stability. Science 2015, 350, 663–666. [Google Scholar] [CrossRef]
- Kang, S.; Niu, J.M.; Zhang, Q.; Zhang, X.F.; Han, G.D.; Zhao, M.L. Niche differentiation is the underlying mechanism maintaining the relationship between community diversity and stability under grazing pressure. Glob. Ecol. Conserv. 2020, 24, e01246. [Google Scholar] [CrossRef]
- Zhou, J.Z.; Deng, Y.; Luo, F.; He, Z.L.; Yang, Y.F. Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. MBio 2011, 2, e00122-11. [Google Scholar] [CrossRef]
- Zhang, W.W.; Lu, Z.T.; Yang, K.; Zhu, J.J. Impacts of conversion from secondary forests to larch plantations on the structure and function of microbial communities. Appl. Soil Ecol. 2017, 111, 73–83. [Google Scholar] [CrossRef]
- Schoch, C.L.; Crous, P.W.; Groenewald, J.Z.; Boehm, E.W.A.; Burgess, T.I.; de Gruyter, J.; de Hoog, G.S.; Dixon, L.J.; Grube, M.; Gueidan, C.; et al. A class-wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 2009, 64, 1–15. [Google Scholar] [CrossRef] [PubMed]



| Site Properties | Site 1 | Site 2 | Site 3 | Site 4 | |
|---|---|---|---|---|---|
| Dominant plant | A. scoparia | P. communis | T. chinensis | S. glauca | |
| EC (μs cm−1) | BS | 228.1 ± 45.2 c | 3052.4 ± 1434.2 bc * | 6362.2 ± 3143.2 b * | 10,995.4 ± 976.9 a |
| RS | 259.8 ± 33.5 b | 1105.1 ± 314.6 b | 1467.7 ± 805.8 b | 11,708.2 ± 1623.1 a | |
| pH | BS | 8.64 ± 0.032 a * | 8.16 ± 0.144 b * | 8.79 ± 0.430 a | 7.94 ± 0.077 b |
| RS | 8.53 ± 0.08 ab | 8.44 ± 0.088 ab | 8.81 ± 0.338 a | 8.11 ± 0.233 b | |
| SWC (%) | BS | 7.10 ± 0.702 b | 19.24 ± 1.58 a | 16.39 ± 1.31 a | 17.96 ± 1.57 a |
| RS | 8.41 ± 4.59 b | 17.45 ± 4.05 a | 18.12 ± 3.32 a | 19.96 ± 2.04 a | |
| SOC (g kg−1) | BS | 4.04 ± 0.367 b | 6.29 ± 1.62 a | 1.62 ± 0.339 c | 5.77 ± 0.527 ab |
| RS | 4.47 ± 1.10 a | 6.69 ± 1.94 a | 1.85 ± 0.130 b | 6.34 ± 0.886 a | |
| TN (g kg−1) | BS | 0.491 ± 0.049 b | 0.618 ±0.122 ab | 0.279 ± 0.047 c | 0.637 ± 0.052 a |
| RS | 0.534 ± 0.082 a | 0.626 ± 0.116 a | 0.320 ± 0.027 b | 0.599 ± 0.075 a | |
| C/N | BS | 8.34 ± 0.832 a | 10.05 ± 1.18 a | 6.00 ± 1.66 b | 9.06 ± 0.360 a ** |
| RS | 8.36 ± 1.61 a | 10.49 ± 1.32 a | 5.84 ± 0.709 b | 10.57 ± 0.602 a | |
| AN (mg kg−1) | BS | 66.95 ± 14.91 a | 92.34 ± 23.08 a | 26.48 ± 14.68 b | 61.33 ± 16.81 ab |
| RS | 49.12 ± 14.70 b | 95.90 ± 21.47 a | 32.93 ± 22.02 b | 66.54 ± 11.86 ab | |
| AP (mg kg−1) | BS | 13.66 ± 0.692 c * | 37.60 ± 4.77 a | 13.52 ± 0.558 c | 23.22 ± 3.82 b |
| RS | 17.98 ± 3.06 b | 37.30 ± 8.74 a | 13.26 ± 0.454 b | 19.88 ± 3.17 b | |
| AK (mg kg−1) | BS | 125.3 ± 20.32 b * | 239.1 ± 27.79 a | 163.9 ± 42.06 b | 227.2 ± 16.36 a |
| RS | 233.3 ± 60.54 a | 208.3 ± 48.37 a | 171.4 ± 8.71 a | 203.0 ± 19.23 a | |
| Index | Rhizosphere Soil | Bulk Soil | ||
|---|---|---|---|---|
| Empirical | Randomized | Empirical | Randomized | |
| Average degree (avgK) | 6.219 | − | 5.946 | − |
| Average clustering coefficient (avgCC) | 0.412 | 0.104 ± 0.016 | 0.381 | 0.127 ± 0.015 |
| Average path distance (GD) | 3.923 | 2.868 ± 0.048 | 4.617 | 2.855 ± 0.042 |
| Centralization of degree (CD) | 0.134 | 0.134 | 0.156 | 0.156 |
| Centralization of betweenness (CB) | 0.207 | 0.083 ± 0.011 | 0.223 | 0.112 ± 0.014 |
| Centralization of eigenvector centrality (CE) | 0.269 | 0.193 ± 0.016 | 0.221 | 0.213 ± 0.012 |
| Harmonic geodesic distance (HD) | 3.047 | 2.535 ± 0.026 | 3.222 | 2.509 ± 0.025 |
| Modularity (M) | 0.555 | 0.326 ± 0.010 | 0.454 | 0.324 ± 0.008 |
| Bulk Soil | Rhizosphere Soil | |||||||
|---|---|---|---|---|---|---|---|---|
| Module | B1 | B2 | B3 | B4 | B5 | R1 | R2 | R3 |
| SWC | −0.732 *** | −0.938 *** | −0.914 *** | −0.246 | 0.132 | 0.421 | 0.430 | −0.661 ** |
| pH | 0.253 | 0.490 * | 0.368 | 0.604 ** | −0.240 | −0.218 | −0.214 | −0.035 |
| EC | −0.756 *** | −0.601 ** | −0.463 * | 0.099 | 0.612 ** | 0.019 | 0.800 *** | −0.047 |
| SOC | 0.129 | −0.250 | −0.161 | −0.840 *** | 0.101 | 0.660 ** | −0.126 | −0.055 |
| TN | 0.191 | −0.187 | −0.079 | −0.791 *** | 0.219 | 0.572 ** | −0.201 | 0.061 |
| C/N | 0.110 | −0.142 | −0.109 | −0.750 *** | −0.063 | 0.601 ** | −0.049 | −0.055 |
| AN | 0.368 | −0.024 | −0.018 | −0.730 *** | −0.049 | 0.730 *** | −0.350 | −0.283 |
| AP | −0.142 | −0.600 ** | −0.614 ** | −0.783 *** | −0.151 | 0.799 *** | −0.570 ** | −0.396 |
| AK | −0.470 * | −0.702 *** | −0.628 ** | −0.507 * | 0.230 | 0.050 | −0.224 | 0.242 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Li, J.; Hou, R.; Zhang, Y.; Gong, H.; Sun, Y.; Ouyang, Z.; Sun, Z. Plant Rhizospheres Harbour Specific Fungal Groups and Form a Stable Co-Occurrence Pattern in the Saline-Alkali Soil. Agronomy 2023, 13, 1036. https://doi.org/10.3390/agronomy13041036
Liu Z, Li J, Hou R, Zhang Y, Gong H, Sun Y, Ouyang Z, Sun Z. Plant Rhizospheres Harbour Specific Fungal Groups and Form a Stable Co-Occurrence Pattern in the Saline-Alkali Soil. Agronomy. 2023; 13(4):1036. https://doi.org/10.3390/agronomy13041036
Chicago/Turabian StyleLiu, Zhen, Jing Li, Ruixing Hou, Yitao Zhang, Huarui Gong, Yanfei Sun, Zhu Ouyang, and Zhigang Sun. 2023. "Plant Rhizospheres Harbour Specific Fungal Groups and Form a Stable Co-Occurrence Pattern in the Saline-Alkali Soil" Agronomy 13, no. 4: 1036. https://doi.org/10.3390/agronomy13041036
APA StyleLiu, Z., Li, J., Hou, R., Zhang, Y., Gong, H., Sun, Y., Ouyang, Z., & Sun, Z. (2023). Plant Rhizospheres Harbour Specific Fungal Groups and Form a Stable Co-Occurrence Pattern in the Saline-Alkali Soil. Agronomy, 13(4), 1036. https://doi.org/10.3390/agronomy13041036








