The dsRNA Delivery, Targeting and Application in Pest Control
Abstract
1. Introduction
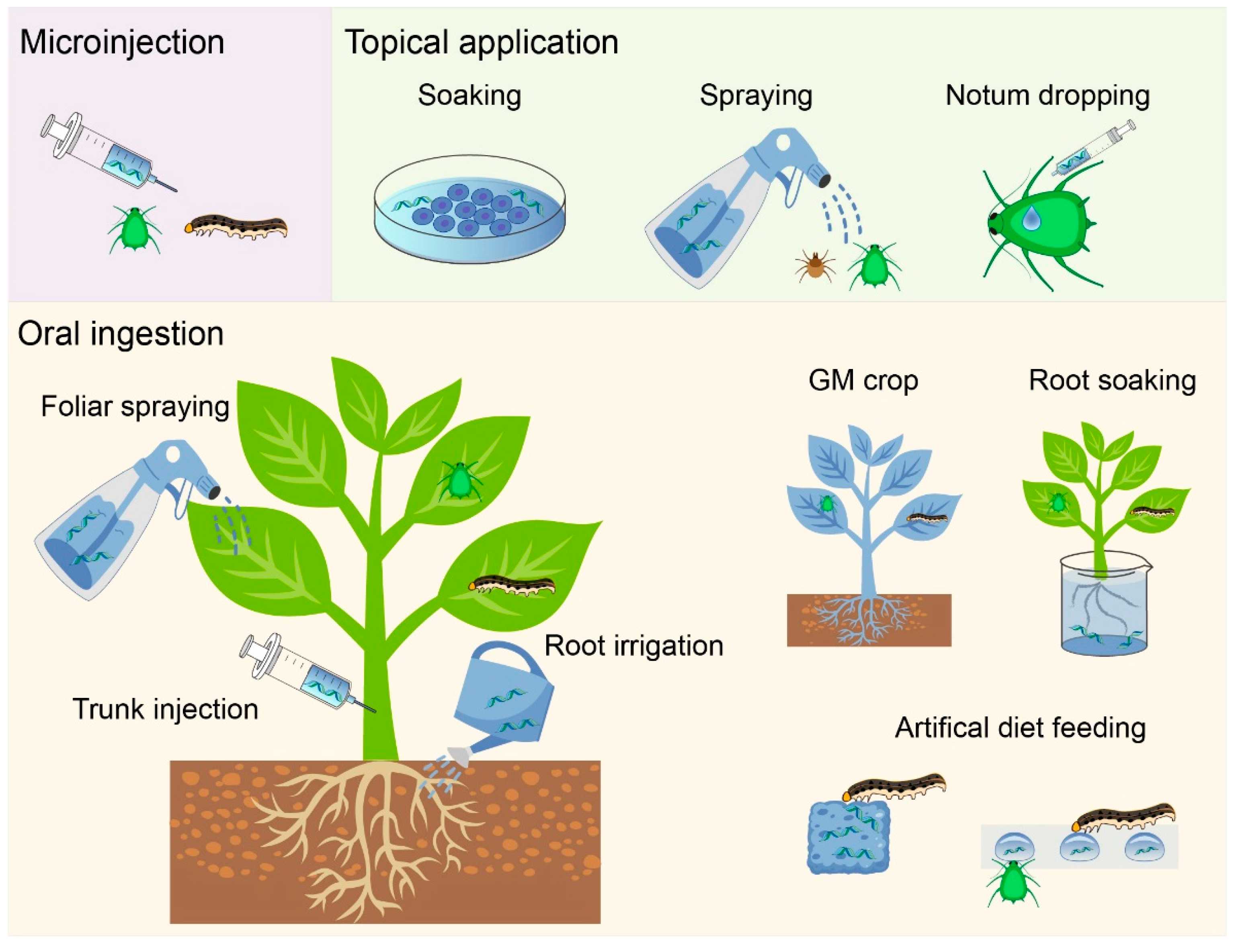
2. Double-Stranded RNA Delivery Methods
2.1. Microinjection
2.2. Ingestion
2.3. Soaking/Topical Application
2.4. Nanoparticle-Mediated dsRNA Delivery
3. Target Genes for RNAi in Pest Control
3.1. Targeting Energy Metabolism
3.2. Targeting Essential Cellular Components
3.3. Targeting Hormone Homeostasis
3.4. Targeting Chitin Metabolism
3.5. Targeting the Digestive System
3.6. Targeting Immunity, Detoxification, and Pesticide Resistance Molecules
3.7. Other Target Genes
4. RNAi-Based Products for Pest Management
5. Conclusions and Future Perspectives
- (1)
- Selecting efficient RNAi target genes and delivery methods according to different pests;
- (2)
- Enhancing the persistence and stability of dsRNA in the environment and its subsequent effects;
- (3)
- Facilitating plant uptake and systemic movement of dsRNA spray;
- (4)
- Solving the shortage of genetic transformation technology in some crops and expanding the range of transplastomic plants expressing dsRNA;
- (5)
- Developing more RNAi products against a wide range of insect species;
- (6)
- Searching for synergistic effects, for example, dsRNAs targeting multiple genes, and combing of RNAi with other pest control methods;
- (7)
- Establishing a consensus regulatory framework for the commercialization of RNAi pesticides;
- (8)
- Reducing the costs for large-scale application of RNAi biopesticides.
Supplementary Materials
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhu, K.Y.; Palli, S.R. Mechanisms, Applications, and Challenges of Insect RNA Interference. Annu. Rev. Entomol. 2020, 65, 293–311. [Google Scholar] [CrossRef] [PubMed]
- Preall, J.B.; Sontheimer, E.J. RNAi: RISC gets loaded. Cell 2005, 123, 543–545. [Google Scholar] [CrossRef] [PubMed]
- Nandety, R.S.; Kuo, Y.W.; Nouri, S.; Falk, B.W. Emerging strategies for RNA interference (RNAi) applications in insects. Bioengineered 2015, 6, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Peng, Y.; Pu, J.; Fu, W.; Wang, J.; Han, Z. Variation in RNAi efficacy among insect species is attributable to dsRNA degradation in vivo. Insect Biochem. Mol. Biol. 2016, 77, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Singh, I.K.; Singh, S.; Mogilicherla, K.; Shukla, J.N.; Palli, S.R. Comparative analysis of double-stranded RNA degradation and processing in insects. Sci. Rep. 2017, 7, 17059. [Google Scholar] [CrossRef] [PubMed]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef]
- Joga, M.R.; Zotti, M.J.; Smagghe, G.; Christiaens, O. RNAi Efficiency, Systemic Properties, and Novel Delivery Methods for Pest Insect Control: What We Know So Far. Front. Physiol. 2016, 7, 553. [Google Scholar] [CrossRef]
- Yu, N.; Christiaens, O.; Liu, J.; Niu, J.; Cappelle, K.; Caccia, S.; Huvenne, H.; Smagghe, G. Delivery of dsRNA for RNAi in insects: An overview and future directions. Insect Sci. 2013, 20, 4–14. [Google Scholar] [CrossRef]
- Yan, S.; Ren, B.-Y.; Shen, J. Nanoparticle-mediated double-stranded RNA delivery system: A promising approach for sustainable pest management. Insect Sci. 2021, 28, 21–34. [Google Scholar] [CrossRef]
- Garbatti Factor, B.; de Moura Manoel Bento, F.; Figueira, A. Methods for Delivery of dsRNAs for Agricultural Pest Control: The Case of Lepidopteran Pests. Methods Mol. Biol. 2022, 2360, 317–345. [Google Scholar]
- Whyard, S.; Singh, A.D.; Wong, S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 2009, 39, 824–832. [Google Scholar] [CrossRef]
- Kennerdell, J.R.; Carthew, R.W. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 1998, 95, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, D.H.; Taylor, C.E.; Wu, K.; Siegfried, B.D. Delivery of gene-specific dsRNA by microinjection and feeding induces RNAi response in Sri Lanka weevil, Myllocerus undecimpustulatus undatus Marshall. Pest Manag. Sci. 2020, 76, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Fan, Y.; Zhang, J.; Cooper, A.M.; Silver, K.; Li, D.; Li, T.; Ma, E.; Zhu, K.Y.; Zhang, J. Contributions of dsRNases to differential RNAi efficiencies between the injection and oral delivery of dsRNA in Locusta migratoria. Pest Manag. Sci. 2019, 75, 1707–1717. [Google Scholar] [CrossRef]
- Prentice, K.; Smagghe, G.; Gheysen, G.; Christiaens, O. Nuclease activity decreases the RNAi response in the sweetpotato weevil Cylas puncticollis. Insect Biochem. Mol. Biol. 2019, 110, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Gurusamy, D.; Howell, J.L.; Chereddy, S.; Koo, J.; Palli, S.R. Transport of orally delivered dsRNA in southern green stink bug, Nezara viridula. Arch. Insect Biochem. Physiol. 2020, 104, e21692. [Google Scholar] [CrossRef]
- Hu, J.; Xia, Y. Increased virulence in the locust-specific fungal pathogen Metarhizium acridum expressing dsRNAs targeting the host F(1) F(0)-ATPase subunit genes. Pest Manag. Sci. 2019, 75, 180–186. [Google Scholar] [CrossRef]
- Ma, Z.Z.; Zhou, H.; Wei, Y.L.; Yan, S.; Shen, J. A novel plasmid-Escherichia coli system produces large batch dsRNAs for insect gene silencing. Pest Manag. Sci. 2020, 76, 2505–2512. [Google Scholar] [CrossRef]
- Wu, M.; Zhang, Q.; Dong, Y.; Wang, Z.; Zhan, W.; Ke, Z.; Li, S.; He, L.; Ruf, S.; Bock, R.; et al. Transplastomic tomatoes expressing double-stranded RNA against a conserved gene are efficiently protected from multiple spider mites. New Phytol. 2023, 237, 1363–1373. [Google Scholar] [CrossRef]
- Mao, Y.B.; Cai, W.J.; Wang, J.W.; Hong, G.J.; Tao, X.Y.; Wang, L.J.; Huang, Y.P.; Chen, X.Y. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 2007, 25, 1307–1313. [Google Scholar] [CrossRef]
- Zhang, J.; Khan, S.A.; Hasse, C.; Ruf, S.; Heckel, D.G.; Bock, R. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science 2015, 347, 991–994. [Google Scholar] [CrossRef]
- Jin, S.; Singh, N.D.; Li, L.; Zhang, X.; Daniell, H. Engineered chloroplast dsRNA silences cytochrome p450 monooxygenase, V-ATPase and chitin synthase genes in the insect gut and disrupts Helicoverpa zea larval development and pupation. Plant Biotechnol. J. 2015, 13, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Khan, S.A.; Heckel, D.G.; Bock, R. Next-Generation Insect-Resistant Plants: RNAi-Mediated Crop Protection. Trends Biotechnol. 2017, 35, 871–882. [Google Scholar] [CrossRef]
- Fletcher, S.J.; Reeves, P.T.; Hoang, B.T.; Mitter, N. A Perspective on RNAi-Based Biopesticides. Front. Plant Sci. 2020, 11, 51. [Google Scholar] [CrossRef] [PubMed]
- Cagliari, D.; Dias, N.P.; Galdeano, D.M.; Dos Santos, E.; Smagghe, G.; Zotti, M.J. Management of Pest Insects and Plant Diseases by Non-Transformative RNAi. Front. Plant Sci. 2019, 10, 1319. [Google Scholar] [CrossRef] [PubMed]
- Castellanos, N.L.; Smagghe, G.; Sharma, R.; Oliveira, E.E.; Christiaens, O. Liposome encapsulation and EDTA formulation of dsRNA targeting essential genes increase oral RNAi-caused mortality in the Neotropical stink bug Euschistus heros. Pest Manag. Sci. 2019, 75, 537–548. [Google Scholar] [CrossRef]
- Clemens, J.C.; Worby, C.A.; Simonson-Leff, N.; Muda, M.; Maehama, T.; Hemmings, B.A.; Dixon, J.E. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA 2000, 97, 6499–6503. [Google Scholar] [CrossRef]
- Sivakumar, S.; Rajagopal, R.; Venkatesh, G.R.; Srivastava, A.; Bhatnagar, R.K. Knockdown of aminopeptidase-N from Helicoverpa armigera larvae and in transfected Sf21 cells by RNA interference reveals its functional interaction with Bacillus thuringiensis insecticidal protein Cry1Ac. J. Biol. Chem. 2007, 282, 7312–7319. [Google Scholar] [CrossRef]
- Zheng, Y.; Hu, Y.; Yan, S.; Zhou, H.; Song, D.; Yin, M.; Shen, J. A polymer/detergent formulation improves dsRNA penetration through the body wall and RNAi-induced mortality in the soybean aphid Aphis glycines. Pest Manag. Sci. 2019, 75, 1993–1999. [Google Scholar] [CrossRef]
- Pridgeon, J.W.; Zhao, L.; Becnel, J.J.; Strickman, D.A.; Clark, G.G.; Linthicum, K.J. Topically applied AaeIAP1 double-stranded RNA kills female adults of Aedes aegypti. J. Med. Entomol. 2008, 45, 414–420. [Google Scholar] [CrossRef]
- San Miguel, K.; Scott, J.G. The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest Manag. Sci. 2016, 72, 801–809. [Google Scholar] [CrossRef]
- Dias, N.; Cagliari, D.; Kremer, F.S.; Rickes, L.N.; Nava, D.E.; Smagghe, G.; Zotti, M. The South American Fruit Fly: An Important Pest Insect With RNAi-Sensitive Larval Stages. Front. Physiol. 2019, 10, 794. [Google Scholar] [CrossRef]
- Chao, Z.; Zhongzheng, M.; Yunhui, Z.; Yan, S.; Shen, J. RNA interference cannot be operated in lepidopteran insect? A nanocarrier breaks bottlenecks at all developmental stages of Spodoptera frugiperda. J. Insect Physiol. 2022. preprint. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, J.; Zhu, K.Y. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Mol. Biol. 2010, 19, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Liu, S.; Wan, F.; Jian, Y.; Guo, H.; Chen, J.; Ning, Y.; Ding, W. Graphene oxide-acaricide nanocomposites advance acaricidal activity of acaricides against Tetranychus cinnabarinus by directly inhibiting the transcription of a cuticle protein gene. Environ. Sci. Nano 2021, 8, 3122–3137. [Google Scholar] [CrossRef]
- Yan, S.; Qian, J.; Cai, C.; Ma, Z.; Li, J.; Yin, M.; Ren, B.; Shen, J. Spray method application of transdermal dsRNA delivery system for efficient gene silencing and pest control on soybean aphid Aphis glycines. J. Pest Sci. 2020, 93, 449–459. [Google Scholar] [CrossRef]
- He, B.; Chu, Y.; Yin, M.; Mullen, K.; An, C.; Shen, J. Fluorescent nanoparticle delivered dsRNA toward genetic control of insect pests. Adv. Mater. 2013, 25, 4580–4584. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Huang, J.H.; Liu, Y.; Belles, X.; Lee, H.J. Oral delivery of dsRNA lipoplexes to German cockroach protects dsRNA from degradation and induces RNAi response. Pest Manag. Sci. 2017, 73, 960–966. [Google Scholar] [CrossRef] [PubMed]
- Terenius, O.; Papanicolaou, A.; Garbutt, J.S.; Eleftherianos, I.; Huvenne, H.; Kanginakudru, S.; Albrechtsen, M.; An, C.; Aymeric, J.-L.; Barthel, A.; et al. RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 2011, 57, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Haller, S.; Widmer, F.; Siegfried, B.D.; Zhuo, X.; Romeis, J. Responses of two ladybird beetle species (Coleoptera: Coccinellidae) to dietary RNAi. Pest Manag. Sci. 2019, 75, 2652–2662. [Google Scholar] [CrossRef]
- Camargo, R.A.; Barbosa, G.O.; Possignolo, I.P.; Peres, L.E.; Lam, E.; Lima, J.E.; Figueira, A.; Marques-Souza, H. RNA interference as a gene silencing tool to control Tuta absoluta in tomato (Solanum lycopersicum). PeerJ 2016, 4, e2673. [Google Scholar] [CrossRef]
- Ma, Z.; Zhang, Y.; Li, M.; Chao, Z.; Du, X.; Yan, S.; Shen, J. A first greenhouse application of bacteria-expressed and nanocarrier-delivered RNA pesticide for Myzus persicae control. J. Pest Sci. 2023, 96, 181–193. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Chandrashekar, K.; Thakur, N.; Verma, P.C.; Borgio, J.F.; Singh, P.K.; Tuli, R. RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J. Biosci. 2011, 36, 153–161. [Google Scholar] [CrossRef]
- Wu, M.; Dong, Y.; Zhang, Q.; Li, S.; Chang, L.; Loiacono, F.V.; Ruf, S.; Zhang, J.; Bock, R. Efficient control of western flower thrips by plastid-mediated RNA interference. Proc. Natl. Acad. Sci. USA 2022, 119, e2120081119. [Google Scholar] [CrossRef] [PubMed]
- Bensoussan, N.; Dixit, S.; Tabara, M.; Letwin, D.; Milojevic, M.; Antonacci, M.; Jin, P.; Arai, Y.; Bruinsma, K.; Suzuki, T.; et al. Environmental RNA interference in two-spotted spider mite, Tetranychus urticae, reveals dsRNA processing requirements for efficient RNAi response. Sci. Rep. 2020, 10, 19126. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Xu, J.; Palli, R.; Ferguson, J.; Palli, S.R. Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag. Sci. 2011, 67, 175–182. [Google Scholar] [CrossRef]
- Sharif, M.N.; Iqbal, M.S.; Alam, R.; Awan, M.F.; Tariq, M.; Ali, Q.; Nasir, I.A. Silencing of multiple target genes via ingestion of dsRNA and PMRi affects development and survival in Helicoverpa armigera. Sci. Rep. 2022, 12, 10405. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, G.; Asokan, R.; Prasannakumar, N.R.; Kariyanna, B.; Karthi, S.; Alwahibi, M.S.; Elshikh, M.S.; Abdel-Megeed, A.; Ghaith, A.; Senthil-Nathan, S.; et al. RNA Interference Suppression of v-ATPase B and Juvenile Hormone Binding Protein Genes Through Topically Applied dsRNA on Tomato Leaves: Developing Biopesticides to Control the South American Pinworm, Tuta absoluta (Lepidoptera: Gelechiidae). Front. Physiol. 2021, 12, 742871. [Google Scholar] [CrossRef]
- Khan, A.M.; Ashfaq, M.; Khan, A.A.; Naseem, M.T.; Mansoor, S. Evaluation of potential RNA-interference-target genes to control cotton mealybug, Phenacoccus solenopsis (Hemiptera: Pseudococcuidae). Insect Sci. 2018, 25, 778–786. [Google Scholar] [CrossRef]
- Meng, J.; Lei, J.; Davitt, A.; Holt, J.R.; Huang, J.; Gold, R.; Vargo, E.L.; Tarone, A.M.; Zhu-Salzman, K. Suppressing tawny crazy ant (Nylanderia fulva) by RNAi technology. Insect Sci. 2020, 27, 113–121. [Google Scholar] [CrossRef]
- Fu, S.; Liu, Z.; Chen, J.; Sun, G.; Jiang, Y.; Li, M.; Xiong, L.; Chen, S.; Zhou, Y.; Asad, M.; et al. Silencing arginine kinase/integrin β(1) subunit by transgenic plant expressing dsRNA inhibits the development and survival of Plutella xylostella. Pest Manag. Sci. 2020, 76, 1761–1771. [Google Scholar] [CrossRef]
- Liu, F.; Wang, X.D.; Zhao, Y.Y.; Li, Y.J.; Liu, Y.C.; Sun, J. Silencing the HaAK gene by transgenic plant-mediated RNAi impairs larval growth of Helicoverpa armigera. Int. J. Biol. Sci. 2015, 11, 67–74. [Google Scholar] [CrossRef]
- Chen, R.P.; Liu, C.Y.; Shao, H.L.; Zheng, W.W.; Wang, J.X.; Zhao, X.F. Adenylate kinase 2 (AK2) promotes cell proliferation in insect development. BMC Mol. Biol. 2012, 13, 31. [Google Scholar] [CrossRef]
- Jin, H.; Abouzaid, M.; Lin, Y.; Hull, J.J.; Ma, W. Cloning and RNAi-mediated three lethal genes that can be potentially used for Chilo suppressalis (Lepidoptera: Crambidae) management. Pestic. Biochem. Physiol. 2021, 174, 104828. [Google Scholar] [CrossRef] [PubMed]
- Kyre, B.R.; Rodrigues, T.B.; Rieske, L.K. RNA interference and validation of reference genes for gene expression analyses using qPCR in southern pine beetle, Dendroctonus frontalis. Sci. Rep. 2019, 9, 5640. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhang, M.; Li, Y.; He, W.; Li, S.; Zhang, J. Complete protection from Henosepilachna vigintioctopunctata by expressing long double-stranded RNAs in potato plastids. J. Integr. Plant Biol. 2022, 0, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Richtman, N.M.; Zhao, J.Z.; Duncan, K.E.; Niu, X.; Procyk, L.A.; Oneal, M.A.; Kernodle, B.M.; Steimel, J.P.; Crane, V.C.; et al. Discovery of midgut genes for the RNA interference control of corn rootworm. Sci. Rep. 2016, 6, 30542. [Google Scholar] [CrossRef]
- Dhandapani, R.K.; Duan, J.J.; Palli, S.R. Orally delivered dsRNA induces knockdown of target genes and mortality in the Asian long-horned beetle, Anoplophora glabripennis. Arch. Insect Biochem. Physiol. 2020, 104, e21679. [Google Scholar] [CrossRef]
- Singh, S.; Gupta, M.; Pandher, S.; Kaur, G.; Goel, N.; Rathore, P.; Palli, S.R. RNA sequencing, selection of reference genes and demonstration of feeding RNAi in Thrips tabaci (Lind.) (Thysanoptera: Thripidae). BMC Mol. Biol. 2019, 20, 6. [Google Scholar] [CrossRef]
- Velez, A.M.; Fishilevich, E.; Rangasamy, M.; Khajuria, C.; McCaskill, D.G.; Pereira, A.E.; Gandra, P.; Frey, M.L.; Worden, S.E.; Whitlock, S.L.; et al. Control of western corn rootworm via RNAi traits in maize: Lethal and sublethal effects of Sec23 dsRNA. Pest Manag. Sci. 2020, 76, 1500–1512. [Google Scholar] [CrossRef]
- Zhu, J.Q.; Liu, S.; Ma, Y.; Zhang, J.Q.; Qi, H.S.; Wei, Z.J.; Yao, Q.; Zhang, W.Q.; Li, S. Improvement of pest resistance in transgenic tobacco plants expressing dsRNA of an insect-associated gene EcR. PLoS ONE 2012, 7, e38572. [Google Scholar] [CrossRef] [PubMed]
- Malik, H.J.; Raza, A.; Amin, I.; Scheffler, J.A.; Scheffler, B.E.; Brown, J.K.; Mansoor, S. RNAi-mediated mortality of the whitefly through transgenic expression of double-stranded RNA homologous to acetylcholinesterase and ecdysone receptor in tobacco plants. Sci. Rep. 2016, 6, 38469. [Google Scholar] [CrossRef] [PubMed]
- Kottaipalayam-Somasundaram, S.R.; Jacob, J.P.; Aiyar, B.; Merzendorfer, H.; Nambiar-Veetil, M. Chitin metabolism as a potential target for RNAi-based control of the forestry pest Hyblaea puera Cramer (Lepidoptera: Hyblaeidae). Pest Manag. Sci. 2022, 78, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Han, Z.-J. Efficiency of Different Methods for dsRNA Delivery in Cotton Bollworm (Helicoverpa armigera). J. Integr. Agric. 2014, 13, 115–123. [Google Scholar] [CrossRef]
- Xiong, Y.; Zeng, H.; Zhang, Y.; Xu, D.; Qiu, D. Silencing the HaHR3 gene by transgenic plant-mediated RNAi to disrupt Helicoverpa armigera development. Int. J. Biol. Sci. 2013, 9, 370–381. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Zhang, S.Z.; Kulye, M.; Wu, S.R.; Yu, N.T.; Wang, J.H.; Zeng, H.M.; Liu, Z.X. Silencing of molt-regulating transcription factor gene, CiHR3, affects growth and development of sugarcane stem borer, Chilo infuscatellus. J. Insect Sci. 2012, 12, 91. [Google Scholar] [CrossRef]
- Vatanparast, M.; Kazzazi, M.; Mirzaie-Asl, A.; Hosseininaveh, V. RNA interference-mediated knockdown of some genes involved in digestion and development of Helicoverpa armigera. Bull. Entomol. Res. 2017, 107, 777–790. [Google Scholar] [CrossRef]
- Choi, M.Y.; Vander Meer, R.K. Phenotypic Effects of PBAN RNAi Using Oral Delivery of dsRNA to Corn Earworm (Lepidoptera: Noctuidae) and Tobacco Budworm Larvae. J. Econ. Entomol. 2019, 112, 434–439. [Google Scholar] [CrossRef]
- Yang, S.; Zou, Z.; Xin, T.; Cai, S.; Wang, X.; Zhang, H.; Zhong, L.; Xia, B. Knockdown of hexokinase in Diaphorina citri Kuwayama (Hemiptera: Liviidae) by RNAi inhibits chitin synthesis and leads to abnormal phenotypes. Pest Manag. Sci. 2022, 78, 4303–4313. [Google Scholar] [CrossRef]
- Zou, H.; Zhang, B.; Zou, C.; Ma, W.; Zhang, S.; Wang, Z.; Bi, B.; Li, S.; Gao, J.; Zhang, C.; et al. Knockdown of GFAT disrupts chitin synthesis in Hyphantria cunea larvae. Pestic. Biochem. Physiol. 2022, 188, 105245. [Google Scholar] [CrossRef]
- Chang, Y.W.; Wang, Y.C.; Yan, Y.Q.; Xie, H.F.; Yuan, D.R.; Du, Y.Z. RNA Interference of Chitin Synthase 2 Gene in Liriomyza trifolii through Immersion in Double-Stranded RNA. Insects 2022, 13, 832. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.K.R.K.; Rajam, M.V. Targeting chitinase gene of Helicoverpa armigera by host-induced RNA interference confers insect resistance in tobacco and tomato. Plant Mol. Biol. 2016, 90, 281–292. [Google Scholar]
- Shang, F.; Ding, B.Y.; Ye, C.; Yang, L.; Chang, T.Y.; Xie, J.; Tang, L.D.; Niu, J.; Wang, J.J. Evaluation of a cuticle protein gene as a potential RNAi target in aphids. Pest Manag. Sci. 2020, 76, 134–140. [Google Scholar] [CrossRef]
- Yu, H.; Yi, L.; Lu, Z. Silencing of Chitin-Binding Protein with PYPV-Rich Domain Impairs Cuticle and Wing Development in the Asian Citrus Psyllid, Diaphorina citri. Insects 2022, 13, 353. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.H.; Zhang, Q.; Zhu, W.Q.; Shi, Y.; Cao, H.H.; Guo, L.; Chu, D.; Lu, Z.; Liu, T.X. Involvement of Laccase2 in Cuticle Sclerotization of the Whitefly, Bemisia tabaci Middle East-Asia Minor 1. Insects 2022, 13, 471. [Google Scholar] [CrossRef]
- Araujo, R.N.; Santos, A.; Pinto, F.S.; Gontijo, N.F.; Lehane, M.J.; Pereira, M.H. RNA interference of the salivary gland nitrophorin 2 in the triatomine bug Rhodnius prolixus (Hemiptera: Reduviidae) by dsRNA ingestion or injection. Insect Biochem. Mol. Biol. 2006, 36, 683–693. [Google Scholar] [CrossRef]
- Murtaza, S.; Tabassum, B.; Tariq, M.; Riaz, S.; Yousaf, I.; Jabbar, B.; Khan, A.; Samuel, A.O.; Zameer, M.; Nasir, I.A. Silencing a Myzus persicae Macrophage Inhibitory Factor by Plant-Mediated RNAi Induces Enhanced Aphid Mortality Coupled with Boosted RNAi Efficacy in Transgenic Potato Lines. Mol. Biotechnol. 2022, 64, 1152–1163. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, N.; Ma, J.; Huang, L.; Liu, X. Effect of Silencing CYP6B6 of Helicoverpa armigera (Lepidoptera: Noctuidae) on Its Growth, Development, and Insecticide Tolerance. J. Econ. Entomol. 2016, 109, 2506–2516. [Google Scholar] [CrossRef]
- Li, M.; Ma, Z.; Peng, M.; Li, L.; Yin, M.; Yan, S.; Shen, J. A gene and drug co-delivery application helps to solve the short life disadvantage of RNA drug. Nano Today 2022, 43, 101452. [Google Scholar] [CrossRef]
- Ye, C.; Wang, Z.-W.; Sheng, Y.-L.; Wang, Z.-G.; Smagghe, G.; Christiaens, O.; Niu, J.; Wang, J.-J. GNBP1 as a potential RNAi target to enhance the virulence of Beauveria bassiana for aphid control. J. Pest Sci. 2022, 95, 87–100. [Google Scholar] [CrossRef]
- Ran, R.; Li, T.; Liu, X.; Ni, H.; Li, W.; Meng, F. RNA interference-mediated silencing of genes involved in the immune responses of the soybean pod borer Leguminivora glycinivorella (Lepidoptera: Olethreutidae). PeerJ 2018, 6, e4931. [Google Scholar] [CrossRef]
- Zhang, C.; Yan, S.Q.; Shen, B.B.; Ali, S.; Wang, X.M.; Jin, F.L.; Cuthbertson, A.G.S.; Qiu, B.L. RNAi knock-down of the Bemisia tabaci Toll gene (BtToll) increases mortality after challenge with destruxin A. Mol. Immunol. 2017, 88, 164–173. [Google Scholar] [CrossRef]
- Chen, X.; Li, L.; Hu, Q.; Zhang, B.; Wu, W.; Jin, F.; Jiang, J. Expression of dsRNA in recombinant Isaria fumosorosea strain targets the TLR7 gene in Bemisia tabaci. BMC Biotechnol. 2015, 15, 64. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, X.; Bai, R.; Shi, Y.; Tang, Q.; An, S.; Song, Q.; Yan, F. RNA interference of the P450 CYP6CM1 gene has different efficacy in B and Q biotypes of Bemisia tabaci. Pest Manag. Sci. 2015, 71, 1175–1181. [Google Scholar] [CrossRef]
- Qu, X.; Wang, S.; Lin, G.; Li, M.; Shen, J.; Wang, D. The Synergistic Effect of Thiamethoxam and Synapsin dsRNA Targets Neurotransmission to Induce Mortality in Aphis gossypii. Int. J. Mol. Sci. 2022, 23, 9388. [Google Scholar] [CrossRef]
- Allen, M.L.; Walker, W.B., 3rd. Saliva of Lygus lineolaris digests double stranded ribonucleic acids. J. Insect Physiol. 2012, 58, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Song, Y.; Zeng, R. The role of cytochrome P450-mediated detoxification in insect adaptation to xenobiotics. Curr. Opin. Insect Sci. 2021, 43, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Hafeez, M.; Li, X.; Ullah, F.; Zhang, Z.; Zhang, J.; Huang, J.; Fernández-Grandon, G.M.; Khan, M.M.; Siddiqui, J.A.; Chen, L.; et al. Down-Regulation of P450 Genes Enhances Susceptibility to Indoxacarb and Alters Physiology and Development of Fall Armyworm, Spodoptera frugipreda (Lepidoptera: Noctuidae). Front. Physiol. 2022, 13, 884447. [Google Scholar] [CrossRef]
- Li, H.; Guan, R.; Guo, H.; Miao, X. New insights into an RNAi approach for plant defence against piercing-sucking and stem-borer insect pests. Plant Cell Environ. 2015, 38, 2277–2285. [Google Scholar] [CrossRef]
- Asokan, R.; Rebijith, K.B.; Roopa, H.K.; Kumar, N.K. Non-invasive delivery of dsGST is lethal to the sweet potato whitefly, Bemisia tabaci (G.) (Hemiptera: Aleyrodidae). Appl. Biochem. Biotechnol. 2015, 175, 2288–2299. [Google Scholar] [CrossRef]
- Gao, H.; Lin, X.; Yang, B.; Liu, Z. The roles of GSTs in fipronil resistance in Nilaparvata lugens: Over-expression and expression induction. Pestic. Biochem. Physiol. 2021, 177, 104880. [Google Scholar] [CrossRef]
- Yu, X.; Killiny, N. RNA interference of two glutathione S-transferase genes, Diaphorina citri DcGSTe2 and DcGSTd1, increases the susceptibility of Asian citrus psyllid (Hemiptera: Liviidae) to the pesticides fenpropathrin and thiamethoxam. Pest Manag. Sci. 2018, 74, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Shabab, M.; Khan, S.A.; Vogel, H.; Heckel, D.G.; Boland, W. OPDA isomerase GST16 is involved in phytohormone detoxification and insect development. FEBS J. 2014, 281, 2769–2783. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Gupta, G.P.; Rajam, M.V. Silencing of acetylcholinesterase gene of Helicoverpa armigera by siRNA affects larval growth and its life cycle. J. Insect Physiol. 2009, 55, 273–278. [Google Scholar] [CrossRef]
- Sharath Chandra, G.; Asokan, R.; Manamohan, M.; Krishna Kumar, N. Enhancing RNAi by using concatemerized double-stranded RNA. Pest Manag. Sci. 2019, 75, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shang, Q.; Lu, Y.; Zhao, Q.; Gao, X. A transferrin gene associated with development and 2-tridecanone tolerance in Helicoverpa armigera. Insect Mol. Biol. 2015, 24, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Yan, R.; Xu, Z.; Qian, J.; Yu, Y.; Zhu, S.; Wu, H.; Zhu, G.; Chen, M. Silencing of dre4 Contributes to Mortality of Phyllotreta striolata. Insects 2022, 13, 1072. [Google Scholar] [CrossRef]
- Sohail, S.; Tariq, K.; Zheng, W.; Ali, M.W.; Peng, W.; Raza, M.F.; Zhang, H. RNAi-Mediated Knockdown of Tssk1 and Tektin1 Genes Impair Male Fertility in Bactrocera dorsalis. Insects 2019, 10, 164. [Google Scholar] [CrossRef]
- Inberg, A.; Mahak, K. Monsanto Technology LLC and Beeologics. Inc. U.S. Patent 62156751, 3 May 2016. [Google Scholar]
- Xie, X.; Shang, F.; Ding, B.Y.; Yang, L.; Wang, J.J. Assessment of a zinc finger protein gene (MPZC3H10) as potential RNAi target for green peach aphid Myzus persicae control. Pest Manag. Sci. 2022, 78, 4956–4962. [Google Scholar] [CrossRef] [PubMed]
- Raje, K.R.; Peterson, B.F.; Scharf, M.E. Screening of 57 Candidate Double-Stranded RNAs for Insecticidal Activity Against the Pest Termite Reticulitermes flavipes (Isoptera: Rhinotermitidae). J. Econ. Entomol. 2018, 111, 2782–2787. [Google Scholar] [CrossRef]
- Ayari, A.; Rosa-Calatrava, M.; Lancel, S.; Barthelemy, J.; Pizzorno, A.; Mayeuf-Louchart, A.; Baron, M.; Hot, D.; Deruyter, L.; Soulard, D.; et al. Influenza infection rewires energy metabolism and induces browning features in adipose cells and tissues. Commun. Biol. 2020, 3, 237. [Google Scholar] [CrossRef]
- Read, E.B.; Okamura, H.H.; Drubin, D.G. Actin- and tubulin-dependent functions during Saccharomyces cerevisiae mating projection formation. Mol. Biol. Cell 1992, 3, 429–444. [Google Scholar] [CrossRef]
- Kang, J.; Brajanovski, N.; Chan, K.T.; Xuan, J.; Pearson, R.B.; Sanij, E. Ribosomal proteins and human diseases: Molecular mechanisms and targeted therapy. Signal Transduct.Target. Ther. 2021, 6, 323. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Qin, Z.; Liu, W.; Liu, X.; Moussian, B.; Ma, E.; Li, S.; Zhang, J. Nuclear receptor HR3 controls locust molt by regulating chitin synthesis and degradation genes of Locusta migratoria. Insect Biochem. Mol. Biol. 2018, 92, 1–11. [Google Scholar] [CrossRef]
- Kramer, K.J.; Sanburg, L.L.; Kézdy, F.J.; Law, J.H. The Juvenile Hormone Binding Protein in the Hemolymph of Manduca sexta Johannson (Lepidoptera: Sphingidae). Proc. Natl. Acad. Sci. USA 1974, 71, 493–497. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.Y.; Vander Meer, R.K.; Coy, M.; Scharf, M.E. Phenotypic impacts of PBAN RNA interference in an ant, Solenopsis invicta, and a moth, Helicoverpa zea. J. Insect Physiol. 2012, 58, 1159–1165. [Google Scholar] [CrossRef] [PubMed]
- Merzendorfer, H.; Zimoch, L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol. 2003, 206, 4393–4412. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.J.; Xu, K.K.; Yan, X.; Chen, C.X.; Cao, Y.; Meng, Y.L.; Li, C. Functional characterization of chitin deacetylase 1 gene disrupting larval-pupal transition in the drugstore beetle using RNA interference. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2018, 219–220, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Da Lage, J.L. The Amylases of Insects. Int. J. Insect Sci. 2018, 10. [Google Scholar] [CrossRef]
- Feng, K.; Li, W.; Tang, X.; Luo, J.; Tang, F. Termicin silencing enhances the toxicity of Serratia marcescens Bizio (SM1) to Odontotermes formosanus (Shiraki). Pestic. Biochem. Physiol. 2022, 185, 105120. [Google Scholar] [CrossRef]
- Zhang, J.; Ye, C.; Wang, Z.-G.; Ding, B.-Y.; Smagghe, G.; Zhang, Y.; Niu, J.; Wang, J.-J. DsRNAs spray enhanced the virulence of entomopathogenic fungi Beauveria bassiana in aphid control. J. Pest Sci. 2023, 96, 241–251. [Google Scholar] [CrossRef]
- Hay, B.A.; Wassarman, D.A.; Rubin, G.M. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell 1995, 83, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Kong, X.D.; Zhu-Salzman, K.; Qin, Q.M.; Cai, Q.N. The Key Glutathione S-Transferase Family Genes Involved in the Detoxification of Rice Gramine in Brown Planthopper Nilaparvata lugens. Insects 2021, 12, 1055. [Google Scholar] [CrossRef]
- Kishk, A.; Anber, H.A.; AbdEl-Raof, T.K.; El-Sherbeni, A.D.; Hamed, S.; Gowda, S.; Killiny, N. RNA interference of carboxyesterases causes nymph mortality in the Asian citrus psyllid, Diaphorina citri. Arch. Insect Biochem. Physiol. 2017, 94, e21377. [Google Scholar] [CrossRef]
- Sural, T.H.; Peng, S.; Li, B.; Workman, J.L.; Park, P.J.; Kuroda, M.I. The MSL3 chromodomain directs a key targeting step for dosage compensation of the Drosophila melanogaster X chromosome. Nat. Struct. Mol. Biol. 2008, 15, 1318–1325. [Google Scholar] [CrossRef]
- Hoang, B.T.L.; Fletcher, S.J.; Brosnan, C.A.; Ghodke, A.B.; Manzie, N.; Mitter, N. RNAi as a Foliar Spray: Efficiency and Challenges to Field Applications. Int. J. Mol. Sci. 2022, 23, 6639. [Google Scholar] [CrossRef]
- Dubelman, S.; Fischer, J.; Zapata, F.; Huizinga, K.; Jiang, C.; Uffman, J.; Levine, S.; Carson, D. Environmental fate of double-stranded RNA in agricultural soils. PLoS ONE 2014, 9, e93155. [Google Scholar] [CrossRef]
- Fischer, J.R.; Zapata, F.; Dubelman, S.; Mueller, G.M.; Uffman, J.P.; Jiang, C.; Jensen, P.D.; Levine, S.L. Aquatic fate of a double-stranded RNA in a sediment—Water system following an over-water application. Environ. Toxicol. Chem. 2017, 36, 727–734. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.A.; Mickelson, J.; Challender, M.; Moellring, E.; Sult, T.; TeRonde, S.; Walker, C.; Wang, Y.; Maxwell, C.A. Agronomic and compositional assessment of genetically modified DP23211 maize for corn rootworm control. GM Crop. Food 2020, 11, 206–214. [Google Scholar] [CrossRef]
- Rodrigues, T.B.; Mishra, S.; Sridharan, K.; Barnes, E.; Alyokhin, A.; Tuttle, R.; Wimalanathan, K.; Garby, D.; Skizim, N.; Tang, Y.-W.; et al. First Sprayable Double-Stranded RNA-Based Biopesticide Product Targets Proteasome Subunit Beta Type-5 in Colorado Potato Beetle (Leptinotarsa decemlineata). Front. Plant Sci. 2021, 12, 728652. [Google Scholar] [CrossRef]
- Kulkarni, M.M.; Booker, M.; Silver, S.J.; Friedman, A.; Hong, P.; Perrimon, N.; Mathey-Prevot, B. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat. Methods 2006, 3, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Jarosch, A.; Moritz, R.F.A. RNA interference in honeybees: Off-target effects caused by dsRNA. Apidologie 2012, 43, 128–138. [Google Scholar] [CrossRef]
- Boeckman, C.J.; Anderson, J.A.; Linderblood, C.; Olson, T.; Roper, J.; Sturtz, K.; Walker, C.; Woods, R. Environmental risk assessment of the DvSSJ1 dsRNA and the IPD072Aa protein to non-target organisms. GM Crop. Food 2021, 12, 459–478. [Google Scholar] [CrossRef] [PubMed]
| Process | Target | References |
|---|---|---|
| energy metabolism (ATPase) | V-ATPase A, V-ATPase B, V-ATPase D, V-ATPase E, V-ATPase G, ATPase | [6,11,13,17,26,36,40,41,42,43,44,45,46,47,48,49] |
| energy metabolism (others) | arginine kinase, ADP/ATP translocase, adenylate kinase 2, NADH dehydrogenase | [6,41,43,50,51,52,53,54] |
| cytoskeleton related protein | tubulin, actin, myosin, shibire | [6,11,21,26,38,43,44,46,55,56] |
| ribosome | RPL9, RPS13, rpL19, rpL9, rps-14, rpS4, rps10 | [6,13,43,57] |
| proteasome | Rpn7, Rpn3, rpn6, rpn11, Prosα2, pat3, protb, PSMB5, PSMD7, E2 enzyme | [6,13,45,57] |
| SCRT | Snf7, vps2, vps28, | [6,13,44,58,59] |
| COPI | COPβ, αCOP | [6,45,46,50,60] |
| protein trafficking/vesicular transport | SNAP, rab1, rab11, GTPase activator, ADP-ribosylation factor, Rop | [45,56] |
| hormone homeostasis | EcR, ultraspiracle, shadow, HR3, JHE, JHBP, PBAN, bursicon | [47,48,49,61,62,63,64,65,66,67,68] |
| chitin metabolism | trehalase, hexokinase, GFAT, chitin synthase, chitinase, chitin deacetylase 1, cuticular protein, laccase2, knk | [6,33,34,35,36,37,63,69,70,71,72,73,74,75] |
| digestive system | nitrophorin 2, MIF1, SSK, DvSSJ1, DvSSJ2, AMY48, AMY49, KTIs | [57,58,67,76,77,78] |
| immunity | hemocytin, GNBP1, PGRP-LB, Toll, terminicin, serpin2, Ap15782, Ap20844, chaoptin, IAP | [30,58,79,80,81,82,83,84,85] |
| detoxify or pesticide resistance | P450s, GSTs, Ces, EstFE4, acetylcholinesterase, synapsin, transferrin | [20,47,78,86,87,88,89,90,91,92,93,94,95,96] |
| transcription | mRNA capping enzyme, helicase-DNA-binding protein, RNA polymerase II, transcription factor IIB, activating transcription factor, Inr-α, dre4 | [6,45,97] |
| others | Tssk1, Tektin1, MSL3, SRP54, GDPH3, calmodulin, aquaporin, heat shock protein, ZC3H10, apple (ATPase), integrin β1, hexamerin II, GHF9-2 | [6,45,51,54,55,56,57,59,98,99,100,101] |
| RNAi Event | Products | Company | RNAi Gene | Target Pest | Approval |
|---|---|---|---|---|---|
| MON87411 (GM corn) | SmartStax Pro, VorceedTM Enlist®, VT4PRO™… | Bayer, Corteva, Pioneer Hi-Bred | DvSnf7 | D. v. virgifera D. v. zeae D. barberi | 2015–2017 |
| DP23211 (GM corn) | - 1 | DAS | DvSSJ1 | D. v. virgifera | 2021 |
| Ledprona | Calantha | GreenLight | PSMB5 | L. decemlineata | 2021 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Deng, X.; Zhu, Q.; Wu, D.; Zhong, J.; Wen, L.; Yu, X. The dsRNA Delivery, Targeting and Application in Pest Control. Agronomy 2023, 13, 714. https://doi.org/10.3390/agronomy13030714
Lu Y, Deng X, Zhu Q, Wu D, Zhong J, Wen L, Yu X. The dsRNA Delivery, Targeting and Application in Pest Control. Agronomy. 2023; 13(3):714. https://doi.org/10.3390/agronomy13030714
Chicago/Turabian StyleLu, Yuzhen, Xinyue Deng, Qijun Zhu, Denghui Wu, Jielai Zhong, Liang Wen, and Xiaoqiang Yu. 2023. "The dsRNA Delivery, Targeting and Application in Pest Control" Agronomy 13, no. 3: 714. https://doi.org/10.3390/agronomy13030714
APA StyleLu, Y., Deng, X., Zhu, Q., Wu, D., Zhong, J., Wen, L., & Yu, X. (2023). The dsRNA Delivery, Targeting and Application in Pest Control. Agronomy, 13(3), 714. https://doi.org/10.3390/agronomy13030714









