Abstract
Soybean (Glycine max (Linn.) Merr.) is a widely-cultivated crop, the yield of which is markedly affected by adverse environmental conditions. Soil salinization, in particular, has led to the degradation of agricultural land, resulting in poor plant growth and decreased crop yields. In plants, serine/threonine protein kinases (STKs) are involved in the plant response to a variety of abiotic stresses. Our previous study identified a transcription factor (GmWRKY20) involved in plant stress resistance, which can directly regulate the expression of GmSTK12. Here, we investigated the effect of the stress-responsive gene GmSTK12 (Glyma.12g198200), which encodes a serine/threonine protein kinase, on soybean salt tolerance. Overall, the overexpression of GmSTK12 (GmSTK12-OE) resulted in increased salt tolerance. Under salt stress, GmSTK12-OE soybeans exhibited significantly increased chlorophyll and proline (PRO) contents; decreased relative electrical conductivity; decreased malondialdehyde (MDA) and superoxide anion (O2−) contents; and increased activities of the antioxidant enzymes catalase (CAT), superoxide dismutase (SOD), and peroxidase (POD). The nitroblue tetrazolium chloride (NBT) staining experiment further confirmed the reduced accumulation of reactive oxygen species (ROS) in GmSTK12-OE soybean leaves. We further determined the Na+ and K+ contents in soybean leaves and roots and found that the Na+ content and Na+/K+ ratio in GmSTK12-OE soybean leaves and roots were significantly lower than those of WT (williams82) soybeans. Furthermore, quantitative real-time PCR (qRT-PCR) analysis revealed that the expression of three SOS pathway genes (GmSOS1, GmSOS2a, and GmSOS2b) was upregulated in GmSTK12-OE soybeans under salt stress. Taken together, the results indicate that GmSTK12 is involved in the mechanism of soybean response to salt stress.
1. Introduction
Soybean (Glycine max (Linn.) Merr.) is one of the most important food crops worldwide, especially in China where it has been cultivated for 5000 years [1]. As ecological and climatic conditions continue to deteriorate in many areas of the world, the soybean crop is becoming progressively more vulnerable to environmental stress. In particular, as the total area and degree of soil salinization continue to grow in many agricultural areas, salt stress is increasingly becoming a threat to soybean yield and quality.
Plant stress can result from biotic or abiotic sources. Biotic stressors include herbivorous insects, microbial pathogens, and intraspecific competitors. Abiotic stressors include drought, extreme temperatures, and salinity, among others. Plant stress alters plant growth, and development and can result in poor health, reductions in yield, and even the death of plants. Plants utilize a variety of mechanisms to respond and adapt to these stressors. A holistic understanding of the molecular mechanisms underpinning plant stress response will aid in efforts to improve crop plant stress tolerance and resistance through breeding and genetic engineering. Such work will be necessary to retain or increase agricultural production in the face of a continuously changing environment.
Stress sensing and signal transduction by protein kinases play a crucial role in the plant response to different stress conditions [2]. Protein kinases such as mitogen-activated protein kinase (MAPK) cascades, receptor-like kinases (RLKs), sucrose nonfermenting1 (SNF1)-related protein kinases (SnRKs), and calcium-dependent protein kinases (CDPKs/CPKs) are essential for regulating plant growth and development, as well as plant responses to stress conditions [3]. Salt stress triggers the accumulation of osmolytes such as proline, polyols, and soluble sugars. These osmolytes regulate intracellular osmotic pressure by reducing the osmotic potential of the cytosolic compartment [4]. Osmolyte accumulation, particularly organic and amino acids, helps to maintain redox homeostasis under salt stress [5]. For example, in sweet potatoes, the β-amylase gene IbBAM1.1 enhances drought and salt stress resistance through regulating ROS homeostasis and osmotic balance [6]. In cotton, salt and alkali stress resistance is enhanced through the regulation of the antioxidant enzyme system, accumulation of osmolytes, and maintenance of ionic balance [7].
Protein kinases (PKs) are widely distributed in both plants and animals, and they are important for growth and development. PKs are classified as either serine/threonine kinases or tyrosine kinases, according to the catalytic substrate [8]. However, the vast majority are Ser/Thr protein kinases. Both types of PKs participate in signal transduction by (1) regulating protein activity through phosphorylation, and (2) signal amplification. Both plant and animal cells can sense signals through cell surface receptors [9,10]. In plants, receptor protein kinases are related to growth and disease resistance and can be classified according to the signals received by the receptors from the environment. Specifically, these receptor protein kinases can be divided into nucleotide binding site NBS-LRR-type and receptor protein kinase PRKs [11,12]. The mode of action of serine/threonine protein kinase is diverse. One class of serine-threonine protein kinases attaches to the helix of the extracellular ligand-binding domain to play a kinase activation role; another class is directly connected to a transmembrane helix without any extracellular domain; and a third class of serine protein kinases can self-phosphorylate serine residues on the activation ring [13,14,15]. Serine/threonine protein kinases have been identified in a variety of plants, including Arabidopsis [16], rice [17], tomato [18], and soybean [19]. Serine/threonine kinase phosphorylation is involved in drought, salt, alkali, and temperature stress signaling. For example, in rice, SAPK4 regulates ionic balance, plant growth, and development under salt stress [20]. Additionally, the gene encoding a serine/threonine protein kinase in Thinopyrum ponticum has been found to increase salt tolerance [21].
The ULK1 kinase-activation loop contains a consensus phosphorylation site at Thr180, which is required for ULK1 autophosphorylation [22]. The central signaling component, SOS2, represents a large family of protein kinases whose catalytic domain is similar to yeast sucrose nonfermenting 1 (SNF1) and mammalian AMP-activated kinase (AMPK). In Arabidopsis, these proteins are generally referred to as SNF1-related kinases (SnRKs). The SnRKs include three members in subfamily 1 (SnRK1s), 10 members in subfamily 2 (SnRK2s), and 25 members in subfamily 3 (SnRK3s) [23]. Calmodulin-binding family proteins, such as calcium-dependent protein kinases or calmodulin-like domain protein kinases (CDPKs), are essential calcium signaling sensor transducers in plants [24].
While the relationship between serine/threonine protein kinase activity and salinity tolerance has been studied in several model plants and crop species, the relationship between GmSTK12 activity and salt tolerance in soybean is poorly characterized. In this study, we verified that GmSTK12-OE improves soybean salt tolerance by enhancing the activity of antioxidant enzymes and maintaining ionic balance. The results of this study will improve our understanding of the mechanism of salt resistance in soybean, particularly the function of serine/threonine protein kinases in Leguminosae plants.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
Soybeans were acquired from the Genetic Engineering Laboratory of the College of Life Sciences, Northeastern Agricultural University, China. GmSTK12-overexpressing (GmSTK12-OE) soybean plants were obtained by Agrobacterium tumefaciens-mediated genetic transformation, with “Williams 82” used as the transformation receptor. Plump and undamaged seeds without lesions were germinated in a greenhouse under the following culture conditions: 25 ± 2 °C; 16 h/8 h light/dark cycle; 20,000 lux light intensity; and 65–75% relative humidity. The seedling-stage salt stress experiment was conducted with 1-week-old seedlings grown hydroponically using Hoagland solution. Experimental seedlings received 2 L of 200 mmol/L NaCl solution. All experiments were conducted with three biological replicates.
2.2. DNA Extraction and PCR Analysis
Total DNA was extracted with a PlantZol kit (TransGen Biotech, Beijing, China). DNA was used as a template for PCR amplification of the bar gene (screening marker gene), using 2×Rapid Taq Master Mix (Vazyme Biotech, Nanjing, China). Positive amplification of the bar gene was used as an indicator of successful genetic transformation.
2.3. RNA Extraction and qRT-PCR Analysis
Total RNA was extracted with an UItrapure RNA kit (CoWin Biotech, Beijing, China). First-strand cDNA was synthesized using EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). Reverse transcription (RT) PCR was performed to amplify the full-length GmSTK12 cDNA. Quantitative real-time PCR was performed in 96-well plates (20 µL reaction volume) using TransStart Top Green qPCR SuperMix (TransGen Biotech, Beijing, China). The relative gene expression levels were calculated using the 2−∆∆Ct method (2−∆∆Ct = [(Ct gene of interest − Ct internal control) sample A-(Ct gene of interest − Ct internal control) sample B]) [25]. Results were plotted as heat maps using TBtools. The soybean GmTUA5 gene was used as the reference gene for normalization. All assays were performed with three independent technical and biological replicates. All primers are listed in Supplementary Table S1.
2.4. Physio-Biochemical Analysis of Plant Materials
The survival rate and number of lateral roots were determined by direct counting. After measuring the fresh weight, the plant material was dried in a 90 °C oven to a constant weight (approximately 30 min) to determine dry weight. Root length and plant height were measured with a ruler. Leaf chlorophyll (Chl) content was determined using a chlorophyll analyzer (Tys-B type). Leaf relative conductivity was measured with a DDS-307 electrical conductivity meter, using the vacuum method. The superoxide dismutase (SOD, nitro-blue tetrazolium method, AKAO001M), catalase (CAT, ammonium molybdate method, AKAO003-2M), and peroxidase (POD, guaiacol method, AKAO005M) activities, as well as the contents of proline (PRO, acidic ninhydrin method, AKAM003M) and malondialdehyde (MDA, TBA (thiobarbiturate) method, AKFA013M), were measured with commercial Boxbio kits (Beijing Boxbio Science & Technology Co., Beijing, China). Each recorded value represented three biological replicates and three technical replicates.
2.5. Determination of Na+ and K+ Contents
A modified method of Gulati et al. [26] was used to extract Na+ and K+ from the samples. Briefly, samples (50 mg) were accurately weighed and placed in glass explosion-proof test tubes. To each test tube was added 10 mL nitrification solution (60 % trichloroacetic acid:concentrated nitric acid:concentrated sulfuric acid, 2:10:1). The samples were extracted in a 90 °C water bath for 5–10 min. The Na+ and K+ contents were measured with a flame photometer (FP6440), with deionized water used as a reference. The standard solution (1 mg/mL) was diluted according to Supplementary Table S2. The experiment was repeated three times per sample.
2.6. Nitroblue Tetrazolium Chloride (NBT) Staining
Soybean leaves were soaked in NBT stain solution, vacuumed with a vacuum pump for 1 h, and developed at room temperature for 24 h in the dark. Leaf pigment was removed by boiling leaves in decolorization solution (acetic acid:glycerol:ethanol, 1:1:3). Subsequently, the stained leaves were observed directly.
2.7. Statistical Analysis
Microsoft Excel 2016 was used to analyze the test results. Statistically significant differences were determined using either a one-way ANOVA or Student’s t-test with GraphPad Prism 9.0. Statistically significant differences were indicated at the p < 0.05 level, very statistically significant differences were indicated at the p < 0.01 level, and extremely significant differences were indicated at the p < 0.001 level.
3. Results
3.1. Overexpression of GmSTK12 Enhances Salt Tolerance of Transgenic Soybean
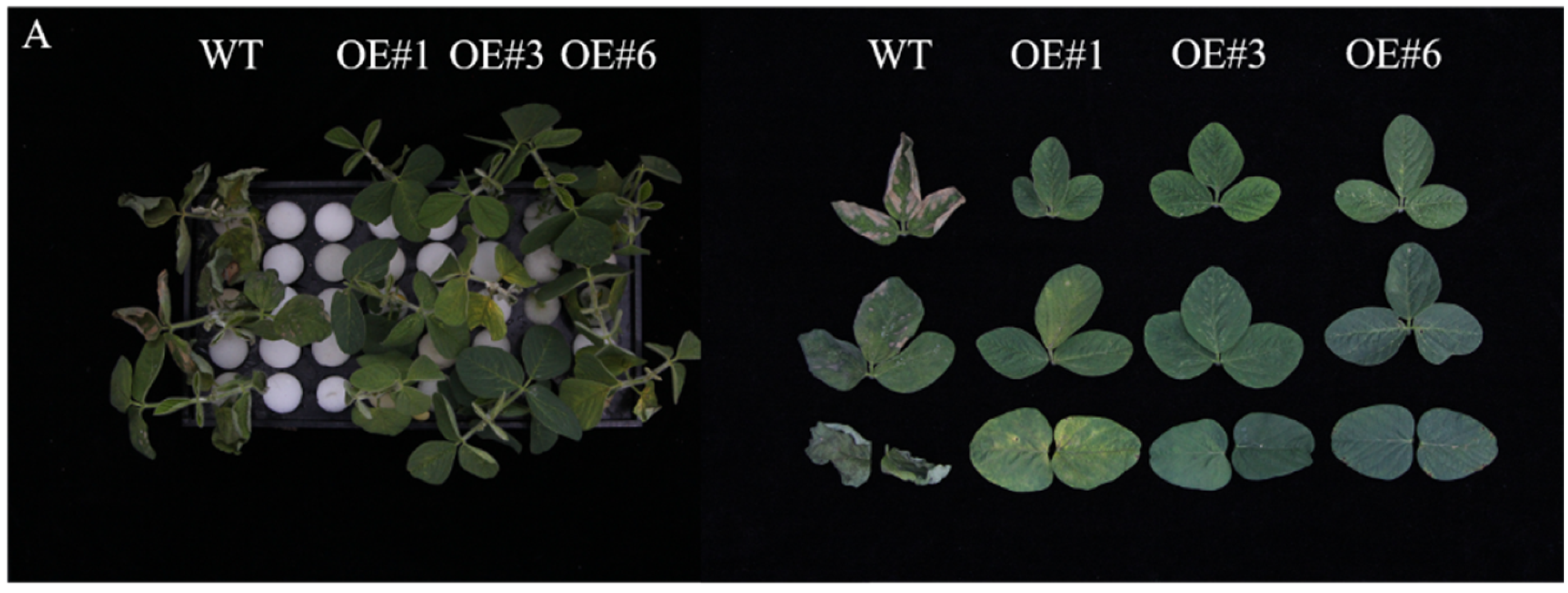
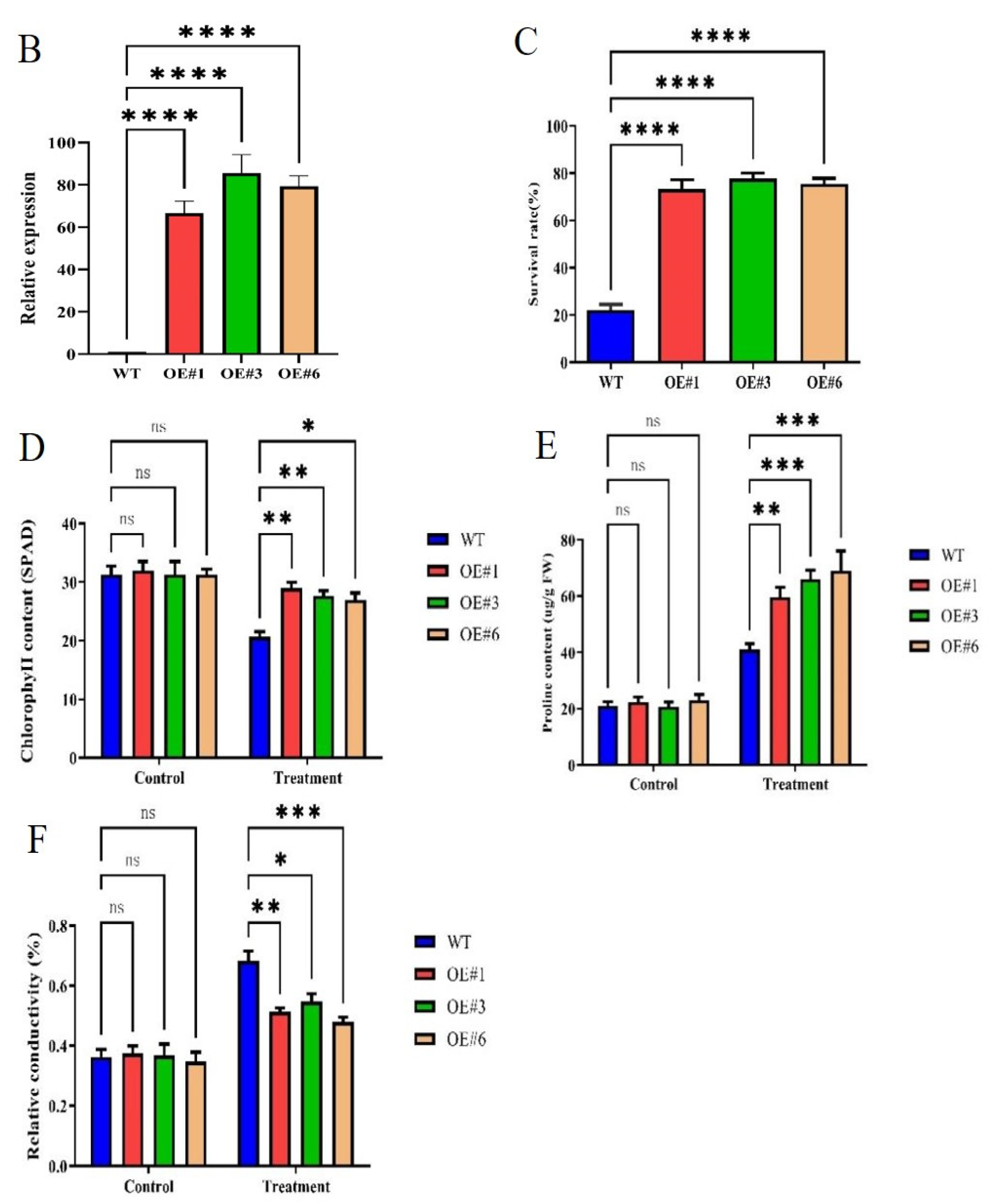
To study the biological function of GmSTK12, overexpression, (OE, lines 1, 3, and 6) lines were treated with Hoagland solution containing 200 mmol/L NaCl solution. Under salt stress, the survival rate and chlorophyll content of soybean seedlings decreased significantly, and the relative electrical conductivity increased significantly (Figure 1). After salt stress treatment, the phenotype of OE seedlings was significantly better than WT seedlings (Figure 1A). Under normal conditions (0mM NaCl), the relative expression of GmSTK12 was significantly (p < 0.05) higher in OE seedlings compared to WT seedlings (Figure 1B). Specifically, after exposure to 200 mmol/L NaCl for 36 h, the survival rate, chlorophyll contents, and proline contents were significantly (p < 0.05) higher in OE seedlings compared to the WT seedlings (Figure 1C–E). The relative conductivity was significantly (p < 0.05) lower in OE seedlings compared to WT seedlings (Figure 1F). Thus, it appears that GmSTK12-OE enhances the salt tolerance of soybean seedlings.

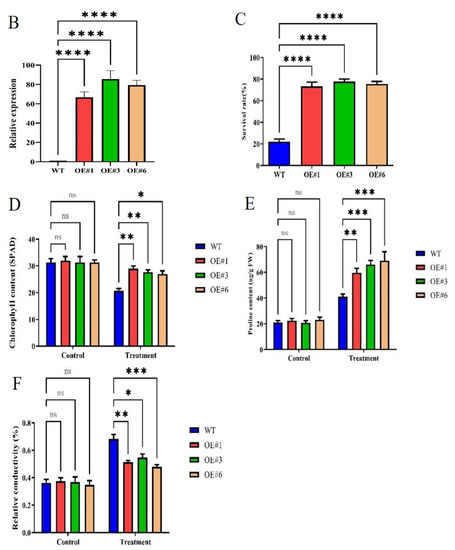
Figure 1.
Phenotype (A), relative expression (B), survival rate (C), chlorophyll content (D), proline content (E), and relative conductivity (F) of control (0 mM NaCl) and salt-treated (200 mM NaCl) soybean seedling leaves after 36 h. Note: A group–group significance analysis was performed, and each treatment (OE#1, OE#3, and OE#6) was compared to WT. All values are presented as the mean ± SD of three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
3.2. Overexpression of GmSTK12 Regulates the Antioxidant Enzyme System under Salt Stress
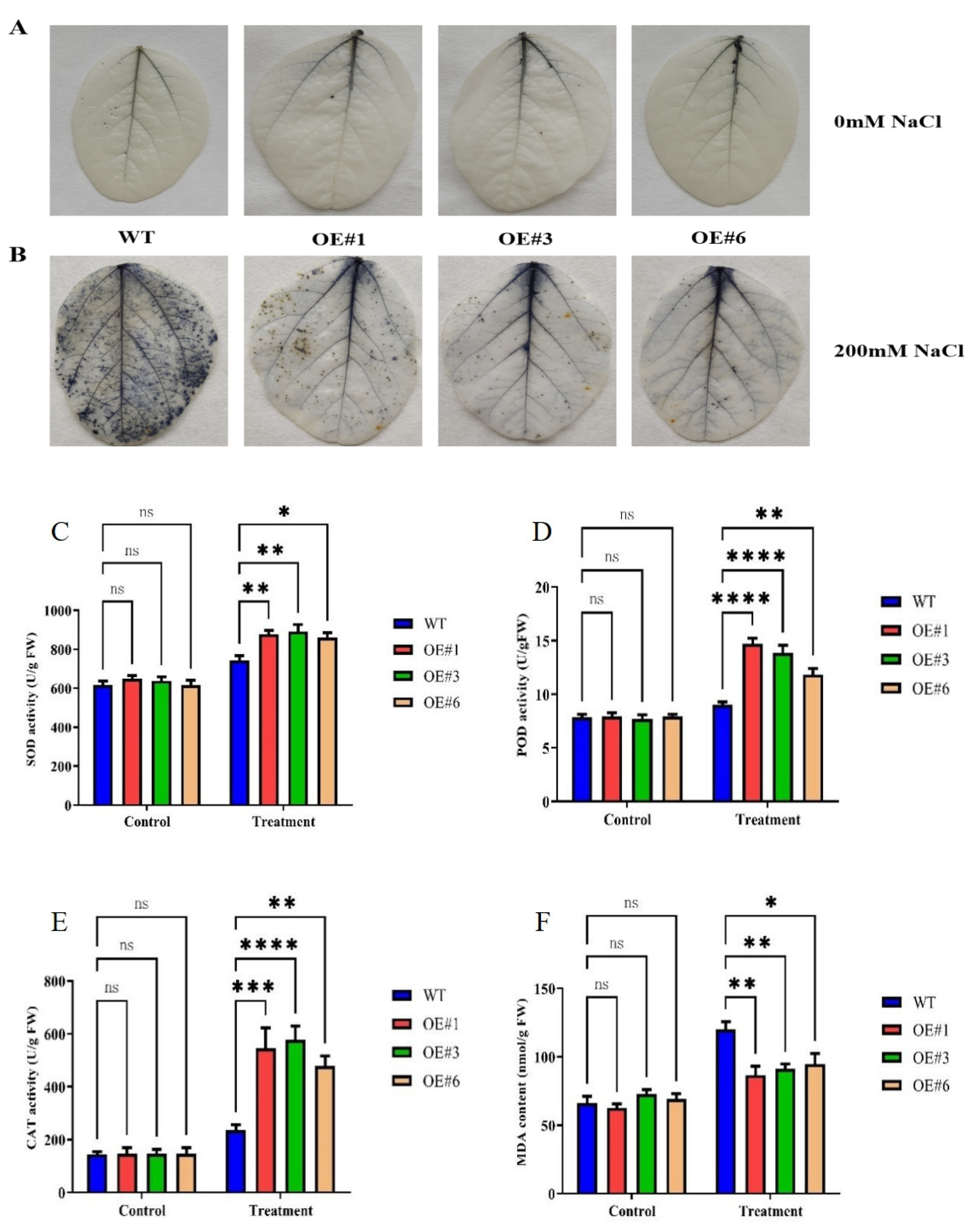
To determine the role of GmSTK12-OE in regulating the antioxidant system, three-week-old salt-stressed soybean leaves were stained with NBT solution, and the activities of SOD, POD, and CAT, as well as the MDA content, were assayed (Figure 2). Under salt stress, a greater quantity of ROS was observed to accumulate in the WT seedlings compared to OE seedlings (Figure 2A,B). Additionally, the activities of SOD, POD, and CAT were significantly higher in the OE seedlings compared to WT seedlings (Figure 2C–E). The MDA content was significantly lower in the OE seedlings compared to WT seedlings (Figure 2F). These results suggest that GmSTK12 improves the antioxidant capacity of transgenic soybeans.
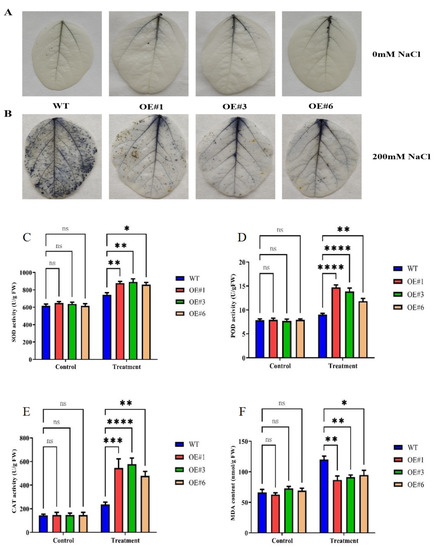
Figure 2.
Phenotype (A,B), SOD activity (C), POD activity (D), CAT activity (E), and MDA content (F) of control (0mM NaCl) and salt-treated (200mM NaCl) soybean seedling leaves after 36 h. Note: A group–group significance analysis was performed, and each treatment (OE#1, OE#3, and OE#6) was compared to WT. All values are presented as the mean ± SD of three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001,**** p < 0.0001.
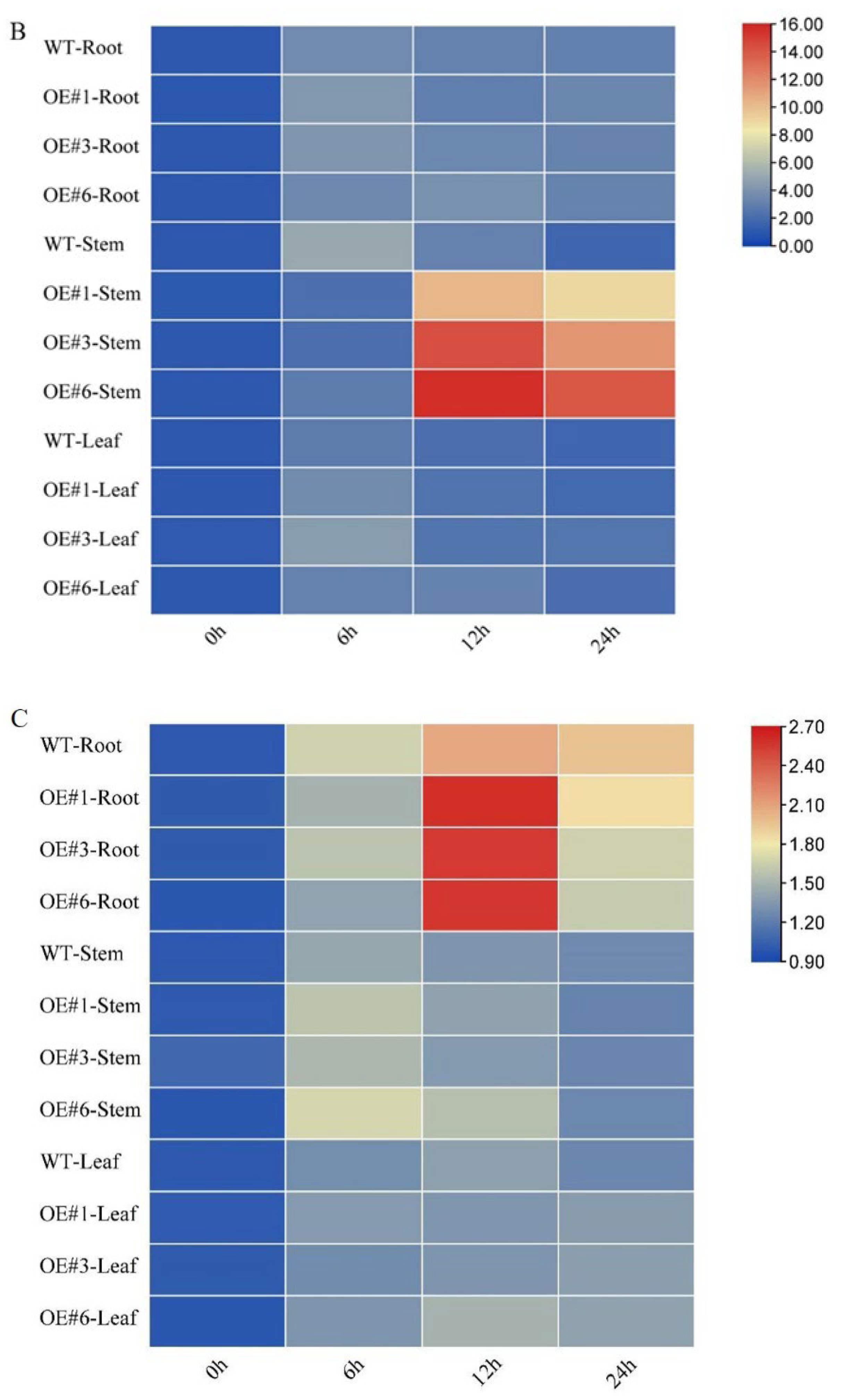
3.3. Overexpression of GmSTK12 Maintains Ionic Balance under Salt Stress
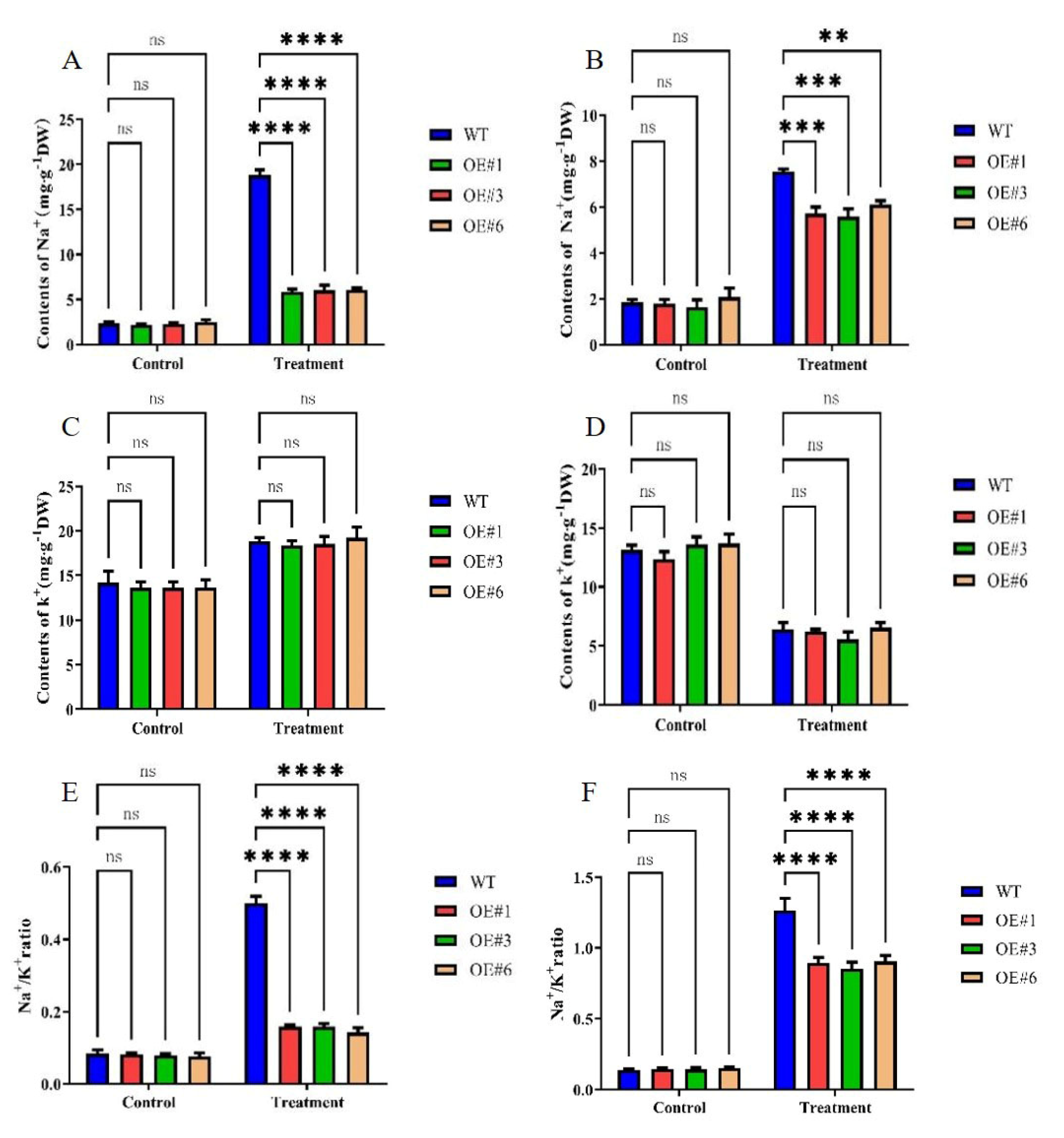
The primary mechanism of salt damage in plants is Na+ toxicity, with the continuous accumulation of Na+ leading to a decrease in K+ content. Thus, the Na+/K+ ratio is an important indicator of the degree of salt damage, and maintaining a suitable Na+/K+ ratio is key to alleviating salt stress. To investigate the ability of GmSTK12-OE soybeans to correct ionic imbalance under salt stress, we determined the Na+ and K+ contents of three-week-old soybean leaves and roots (Figure 3). Although the Na+ content increased in all three groups, less Na+ was accumulated in OE seedlings compared to WT seedlings (Figure 3A,B). The K+ content tended to increase in leaves and decrease in roots, although there was no significant difference between groups either before or after treatment (Figure 3C,D). The leaf and root Na+/K+ ratio was also lower in OE seedlings compared to WT seedlings (Figure 3E,F). These results suggest that GmSTK12 can reduce the accumulation of Na+ in leaves and roots, maintain the balance of Na+ and K+, and improve the salt tolerance of transgenic soybean.
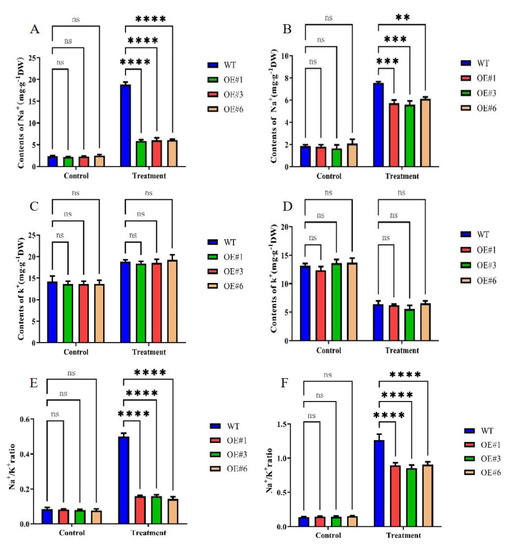
Figure 3.
The Na+ content of leaves (A) and roots (B), the K+ content of leaves (C) and roots (D), and the Na+/K+ ratio of leaves (E) and roots (F) of control (0 mM NaCl) and salt-treated (200 mM NaCl) soybean seedling leaves after 36 h. Note: A group–group significance analysis was performed, and each treatment (OE#1, OE#3, and OE#6) was compared to WT. All values are presented as the mean ± SD of three independent experiments. ** p < 0.01, *** p < 0.001, **** p < 0.0001.
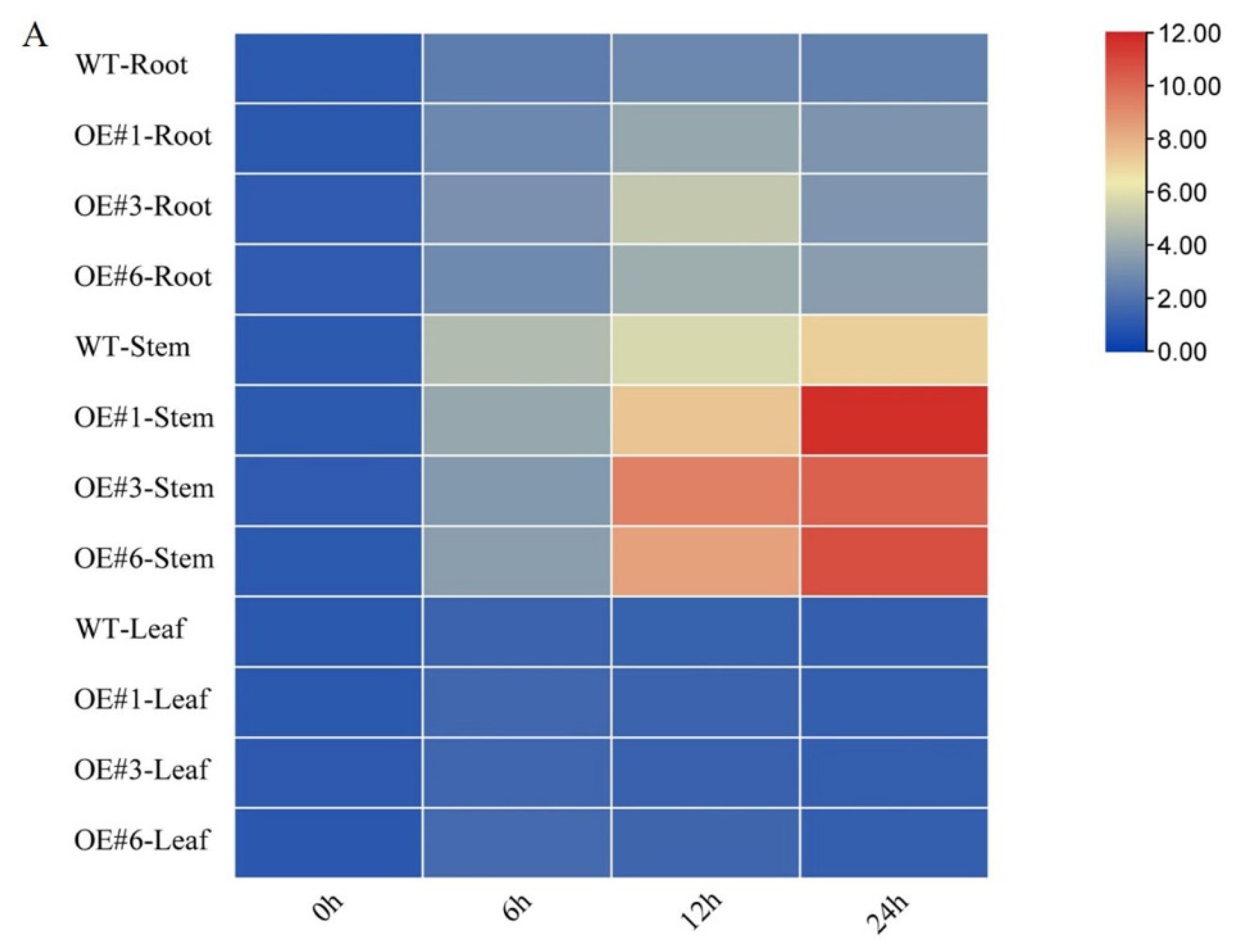
3.4. Effects of GmSTK12 Overexpression on Tissue-Specific SOS Gene Expression
The tissue-specific expression levels of three GmSOS genes were determined by qPCR (Figure 4). Prior to salt treatment, the expression levels of GmSOS1, GmSOS2a, and GmSOS2b were not significantly different between OE and WT seedlings. Six hours after salt treatment, GmSOS1 was significantly upregulated in stem tissues in both OE and WT seedlings, while after 12 and 24 h, GmSOS1 was significantly upregulated only in OE seedlings. Furthermore, GmSOS1 was significantly upregulated in root tissues during the entire experimental period, with no significant differences between OE and WT seedlings (Figure 4A). The expression of GmSOS2a was similar to that of GmSOS1 by 12 h. However, after 24 h, the expression of GmSOS2a decreased, with the OE seedlings exhibiting higher levels of expression than the WT seedlings at 12 and 24 h (Figure 4B). The expression of GmSOS2b was highly tissue-dependent, with the most noticeable expression changes observed in the root. In roots, GmSOS2b expression peaked at 12 h and then decreased, with OE seedlings exhibiting higher levels of expression than WT seedlings at 12 h (Figure 4C). These results suggest that GmSTK12 may affect the regulation of GmSOSs gene expression.
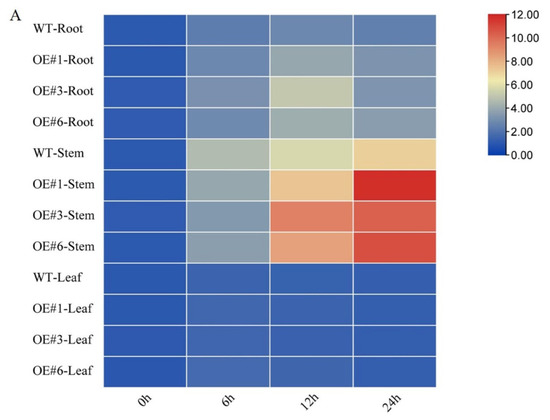
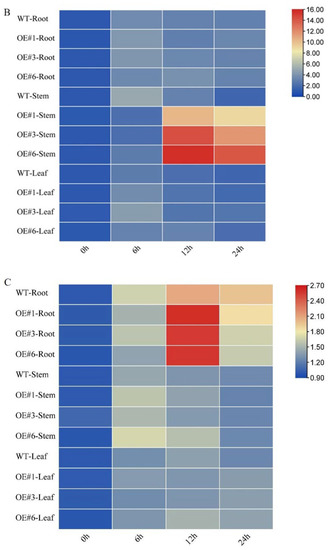
Figure 4.
Tissue-specific relative gene expression of GmSOS1 (A), GmSOS2a (B), and GmSOS2b (C) under normal and salt-stressed conditions. Note: A group–group significance analysis was performed, and each treatment (OE#1, OE#3, and OE#6) was compared to WT. All values are presented as the mean ± SD of three independent experiments.
4. Discussion
Soybean is affected by a number of stressors, including salinity, waterlogging, drought, and others. Salt stress involves both osmotic and ionic stress, in addition to secondary stressors such as nutrient deficiency and oxidative stress [27]. Salt stress inhibits plant growth, impairs physiological and biochemical functioning, and may result in death in severe cases. The primary manifestations of salt stress are cell dehydration, enzymatic activity reduction, and membrane destruction [28]. Here, we found that GmSTK12 improves the salt tolerance of transgenic soybean seedlings by maintaining ionic balance and alleviating osmotic and oxidative stress.
4.1. The Effect of Maintaining Ionic Balance on Plant Salt Tolerance
Salt stress primarily involves Na+, Ca2+, Cl−, SO42−, and other ions generated from the dissociation of sodium and calcium salts. However, these ions cause salt damage and secondary damage to soybean plants only at relatively high concentrations. In order for plants to grow successfully in high-salt environments, plant cells must maintain a relatively low cytosolic sodium concentration [29]. Additionally, the uptake of K+ from saline environments is crucial for the maintenance of intracellular K+/Na+ homeostasis, and helps to reduce Na+ toxicity. Thus, plants respond to elevated Na+ concentrations by maintaining low cytosolic Na+ concentrations and a high cytosolic K+/Na+ ratio. After 36 h of treatment with 200 mM NaCl, the Na+ content in leaves and roots was lower, and Na+/K+ ratio was lower, in OE seedlings compared to WT seedlings (Figure 3). These results may be related to the upregulated expression of GmSOS1, GmSOS2a, and GmSOS2b (Figure 4). Although the content of K+ increased in the leaves and decreased in the roots of both groups, no significant difference was observed between treatments (Figure 3). OE seedlings also exhibited increased survival rate and chlorophyll content, in addition to decreased relative conductivity under salt-stressed conditions (Figure 1). In rice, overexpression of OsCYB5-2 increases OsHAK21-mediated K+ transport and restricts Na+ entry into cells, resulting in improved salt tolerance [30]. In transgenic Lotus corniculatus, overexpression of GmNHX1 results in lower Na+ and K+ contents, a higher K+/Na+ ratio, and enhanced salt tolerance [31]. Overall, our results are consistent with these previous studies.
4.2. The Effect of Regulating the Antioxidant System on Plant Salt Tolerance
As signaling molecules, ROS support cellular proliferation and normal physiological function; therefore, maintenance of a basal level of ROS is essential for life. Salt stress results in an array of adverse effects [32], including the overproduction of ROS. In response to oxidative stress, plants utilize both enzymatic and non-enzymatic antioxidant reactions to maintain normal metabolic functions [33]. The enzymatic antioxidants include superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), ascorbate peroxidase (APX), monodehydroascorbate reductase (MDHAR), glutathione reductase (GR), glutathione peroxidase (GPX), and dehydroascorbate reductase (DHAR) [33]. In soybeans, the expression of GmNHX1 enhances the activity of the antioxidant system, inhibits the accumulation of ROS, limits cadmium (Cd) uptake, and prevents root and cell membrane damage under salt and Cd stress [31]. Here, we found that the activities of SOD, POD, and CAT were higher, and the content of MDA was lower, in OE seedlings compared to WT seedlings under salt stress (Figure 2C–F). NBT staining further confirmed the reduction in ROS accumulation in GmSTK12-OE soybean leaves (Figure 2A,B). Overall, our results are consistent with previous studies [34,35,36]. It appears likely that GmSTK12 enhances the activity of the antioxidant system in soybeans, resulting in decreased membrane lipid damage and oxidative stress, in addition to increased salt tolerance.
4.3. Osmotically Active Substances in Salt-Stressed Plants
Osmotic adjustment is crucial for plant salt tolerance. Osmotic stress results in decreased water absorption by plant roots, which leads to dehydration and wilting [37]. Plants utilize both inorganic ions and organic solutes for osmotic adjustment. Inorganic ions are taken up from the external environment and include K+ and Cl− in microorganisms and halophytes, as well as organic acid potassium in higher plants. Organic solutes are synthesized by plant cells, such as proline, polyols, and sugars, and accumulate under salt stress. These osmolytes participate in the regulation of osmotic pressure by lowering the osmotic potential in the cytosolic compartment [38,39]. Specifically, the properties of proline as an organic osmolyte provide cellular protection against abiotic stresses such as drought and osmotic shock [40,41]. Here, we found that overexpression plants could significantly reduce the accumulation of Na+ cations and maintain a relatively low inorganic salt content (Figure 3A,B). Additionally, overexpression plants accumulated significantly more proline (Figure 1E), a cytoplasmic osmotic pressure regulator, and thus were better able to maintain optimal intracellular osmotic pressure.
5. Conclusions
Under salt-stressed conditions, GmSTK12 overexpression significantly increased the content of proline and the activity of SOD, CAT, and POD, and significantly decreased the contents of MDA and superoxide anion. Additionally, GmSTK12 overexpression significantly increased expression levels of GmSOS1, GmSOS2a, and GmSOS2b, and reduced the Na+ content and Na+/K+ ratio in soybean leaves and roots. Overall, GmSTK12 appears to enhance the osmotic regulation and antioxidant capacity of soybean seedlings. Furthermore, overexpression of GmSTK12 likely influences the soybean response to salt stress.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/agronomy13020613/s1, Table S1: Design of fluorescent quantitative PCR primers; Table S2: Standard solution dilution gradient.
Author Contributions
Conceptualization, Y.L. and S.J.; Data curation, Y.L., Y.C., X.Y., H.Z. (Huimin Zhang) and X.M.; Formal analysis, X.B.; Funding acquisition, X.B.; Investigation, Y.L. and J.Z.; Methodology, Y.L., J.Z., S.J. and X.B.; Project administration, H.Z. (Hong Zhai) and X.B.; Resources, Y.L., Y.C. and X.Y.; Supervision, H.Z. (Huimin Zhang), H.Z. (Hong Zhai) and X.B.; Validation, J.Z., Y.C., X.Y. and S.J.; Visualization, J.Z., Y.C., X.Y. and X.M.; Writing–original draft, Y.L. and S.J.; Writing–review & editing, J.Z. and H.Z. (Hong Zhai). All authors have read and agreed to the published version of the manuscript.
Funding
This research work was funded by the National Natural Science Foundation of China (31971830).
Data Availability Statement
The data presented in this study are available in the Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Goodrich, L.C. The Soybean in China. Science 1941, 93, 183–184. [Google Scholar] [CrossRef]
- Zhu, S.Y.; Yu, X.C.; Wang, X.J.; Zhao, R.; Li, Y.; Fan, R.C.; Shang, Y.; Du, S.Y.; Wang, X.-F.; Wu, F.-Q.; et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 2007, 19, 3019–3060. [Google Scholar] [CrossRef]
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef]
- Li, C.H.; Wang, G.; Zhao, J.L.; Zhang, L.Q.; Ai, L.F.; Han, Y.F.; Sun, D.Y.; Zhang, S.W.; Sun, Y. The Receptor-Like Kinase SIT1 Mediates Salt Sensitivity by Activating MAPK3/6 and Regulating Ethylene Homeostasis in Rice. Plant Cell 2014, 26, 2538–2553. [Google Scholar] [CrossRef]
- Shah, F.A.; Wei, X.; Wang, Q.; Liu, W.; Wang, D.; Yao, Y.; Hu, H.; Chen, X.; Huang, S.; Hou, J.; et al. Karrikin Improves Osmotic and Salt Stress Tolerance via the Regulation of the Redox Homeostasis in the Oil Plant Sapium sebiferum. Front. Plant Sci. 2020, 11, 216. [Google Scholar] [CrossRef]
- Zhu, H.; Yang, X.; Wang, X.; Li, Q.; Guo, J.; Ma, T.; Zhao, C.; Tang, Y.; Qiao, L.; Wang, J.; et al. The sweetpotato β-amylase gene IbBAM1.1 enhances drought and salt stress resistance by regulating ROS homeostasis and osmotic balance. Plant Physiol. Biochem. 2021, 168, 167–176. [Google Scholar] [CrossRef]
- Guo, H.; Huang, Z.; Li, M.; Hou, Z. Growth, ionic homeostasis, and physiological responses of cotton under different salt and alkali stresses. Sci. Rep. 2020, 10, 21844. [Google Scholar] [CrossRef]
- Zhou, P.; Wong, D.; Li, W.; Xie, J.; Av-Gay, Y. Phosphorylation of Mycobacterium tuberculosis protein tyrosine kinase A PtkA by Ser/Thr protein kinases. Biochem. Biophys. Res. Commun. 2015, 467, 421–426. [Google Scholar] [CrossRef]
- Gao, Y.L.; Wang, B.W.; Xu, Z.L.; Li, M.Y.; Song, Z.B.; Li, W.Z.; Li, Y.P. Tobacco serine/threonine protein kinase gene NrSTK enhances black shank resistance. Genet. Mol. Res. 2015, 14, 16415–16424. [Google Scholar] [CrossRef]
- Canova, M.J.; Molle, V. Bacterial serine/threonine protein kinases in host-pathogen interactions. J. Biol. Chem. 2014, 289, 9473–9479. [Google Scholar] [CrossRef]
- Qian, L.H.; Wu, J.Y.; Wang, Y.; Zou, X.; Zhou, G.C.; Sun, X.Q. Genome-Wide Analysis of NBS-LRR Genes From an Early-Diverging Angiosperm Euryale ferox. Front. Genet. 2022, 13, 880071. [Google Scholar] [CrossRef]
- Erickson, J.; Weckwerth, P.; Romeis, T.; Lee, J. What’s new in protein kinase/phosphatase signalling in the control of plant immunity? Essays Biochem. 2022, 66, 621–634. [Google Scholar] [CrossRef]
- Nagarajan, S.N.; Lenoir, C.; Grangeasse, C. Recent advances in bacterial signaling by serine/threonine protein kinases. Trends Microbiol. 2022, 30, 553–566. [Google Scholar] [CrossRef]
- Zhang, W.; Li, L.; Jiang, W.; Zhao, G.; Yang, Y.; Chiao, J. A novel transmembrane serine/threonine protein kinase gene from a rifamycin SV-producing Amycolatopsis mediterranei U32. Eur. J. Biochem. 2000, 267, 3744–3752. [Google Scholar] [CrossRef]
- Janczarek, M.A.-O.; Vinardell, J.A.-O.; Lipa, P.A.-O.; Karaś, M. Hanks-Type Serine/Threonine Protein Kinases and Phosphatases in Bacteria: Roles in Signaling and Adaptation to Various Environments. Int. J. Mol. Sci. 2018, 19, 2872. [Google Scholar] [CrossRef]
- Sheremet Ia, A.; Emets, A.I.; Vissenberg, K.; Verbelen, J.; Blium Ia, B. The effect of inhibitors of serinethreonine protein kinases on Arabidopsis thaliana root morphology and microtubules organization in its cells. Tsitologiia 2010, 52, 389–398. [Google Scholar]
- Heo, J.B.; Lee, Y.M.; Yun, H.R.; Im, C.H.; Lee, Y.S.; Yi, Y.B.; Kwon, C.; Lim, J.; Bahk, J.D. Rice serine/threonine kinase 1 is required for the stimulation of OsNug2 GTPase activity. J. Plant Physiol. 2014, 171, 1601–1608. [Google Scholar] [CrossRef]
- Muhammad, T.; Zhang, J.; Ma, Y.; Li, Y.; Zhang, F.; Zhang, Y.; Liang, Y. Overexpression of a Mitogen-Activated Protein Kinase SlMAPK3 Positively Regulates Tomato Tolerance to Cadmium and Drought Stress. Molecules 2019, 24, 556. [Google Scholar] [CrossRef]
- Sun, X.L.; Yu, Q.Y.; Tang, L.L.; Ji, W.; Bai, X.; Cai, H.; Liu, X.F.; Ding, X.D.; Zhu, Y.M. GsSRK, a G-type lectin S-receptor-like serine/threonine protein kinase, is a positive regulator of plant tolerance to salt stress. J. Plant Physiol. 2013, 170, 505–515. [Google Scholar] [CrossRef]
- Diédhiou, C.J.; Popova, O.V.; Dietz, K.J.; Golldack, D. The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice. BMC Plant Biol. 2008, 8, 49. [Google Scholar] [CrossRef]
- Shen, W.; Gómez-Cadenas, A.; Routly, E.L.; Ho, T.H.; Simmonds, J.A.; Gulick, P.J. The salt stress-inducible protein kinase gene, Esi47, from the salt-tolerant wheatgrass Lophopyrum elongatum is involved in plant hormone signaling. Plant Physiol. 2001, 125, 1429–1441. [Google Scholar] [CrossRef]
- Bach, M.; Larance, M.; James, D.E.; Ramm, G. The serine/threonine kinase ULK1 is a target of multiple phosphorylation events. Biochem. J. 2011, 440, 283–291. [Google Scholar] [CrossRef]
- Hrabak, E.M.; Chan, C.W.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R.; et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef]
- Harper, J.F.; Breton, G.; Harmon, A. Decoding Ca(2+) signals through plant protein kinases. Annu. Rev. Plant Biol. 2004, 55, 263–288. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Gulati, A.; Jaiwal, P.K. Comparative salt responses of callus cultures of Vigna radiata L. Wilczek to various osmotic and ionic stresses. J. Plant Physiol. 1993, 141, 120–124. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Blumwald, E. Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 2000, 12, 431–434. [Google Scholar] [CrossRef]
- Song, T.; Shi, Y.; Shen, L.; Cao, C.; Shen, Y.; Jing, W.; Tian, Q.; Lin, F.; Li, W.; Zhang, W. An endoplasmic reticulum-localized cytochrome b(5) regulates high-affinity K(+) transport in response to salt stress in rice. Proc. Natl. Acad. Sci. USA 2021, 118, e2114347118. [Google Scholar] [CrossRef]
- Yang, L.; Han, Y.; Wu, D.; Yong, W.; Liu, M.; Wang, S.; Liu, W.; Lu, M.; Wei, Y.; Sun, J. Salt and cadmium stress tolerance caused by overexpression of the Glycine Max Na(+)/H(+) Antiporter (GmNHX1) gene in duckweed (Lemna turionifera 5511). Aquat. Toxicol. 2017, 192, 127–135. [Google Scholar] [CrossRef]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Ketehouli, T.; Zhou, Y.G.; Dai, S.Y.; Carther, K.F.I.; Sun, D.Q.; Li, Y.; Nguyen, Q.V.H.; Xu, H.; Wang, F.W.; Liu, W.C.; et al. A soybean calcineurin B-like protein-interacting protein kinase, GmPKS4, regulates plant responses to salt and alkali stresses. J. Plant Physiol. 2021, 256, 153331. [Google Scholar] [CrossRef]
- Qu, Y.; Guan, R.; Yu, L.; Berkowitz, O.; David, R.; Whelan, J.; Ford, M.; Wege, S.; Qiu, L.; Gilliham, M. Enhanced reactive oxygen detoxification occurs in salt-stressed soybean roots expressing GmSALT3. Physiol. Plant 2022, 174, e13709. [Google Scholar] [CrossRef]
- Hoang, X.L.T.; Chuong, N.N.; Hoa, T.T.K.; Doan, H.; Van, P.H.P.; Trang, L.D.M.; Huyen, P.N.T.; Le, D.T.; Tran, L.P.; Thao, N.P. The Drought-Mediated Soybean GmNAC085 Functions as a Positive Regulator of Plant Response to Salinity. Int. J. Mol. Sci. 2021, 22, 8986. [Google Scholar] [CrossRef]
- Cardoso, A.A.; Brodribb, T.J.; Kane, C.N.; DaMatta, F.M.; McAdam, S.A. Osmotic adjustment and hormonal regulation of stomatal responses to vapour pressure deficit in sunflower. AoB Plants 2020, 12, plaa025. [Google Scholar] [CrossRef]
- Cui, Y.N.; Li, X.T.; Yuan, J.Z.; Wang, F.Z.; Guo, H.; Xia, Z.R.; Wang, S.; Ma, Q. Chloride is beneficial for growth of the xerophyte Pugionium cornutum by enhancing osmotic adjustment capacity under salt and drought stresses. J. Exp. Bot. 2020, 71, 4215–4231. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Liu, M.A.-O.; Zhou, H.A.-O.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- Szabados, L.; Savouré, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef]
- Jin, T.; Sun, Y.; Shan, Z.; He, J.; Wang, N.; Gai, J.; Li, Y.A.-O. Natural variation in the promoter of GsERD15B affects salt tolerance in soybean. Plant Biotechnol. J. 2021, 19, 1155–1169. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).