Abstract
Iron (Fe) and (Mn) are essential for the plant but are toxic when in excess. Vacuolar iron transporters (VITs) are involved in plant metal storage and detoxication. In this study, we screened two soybean cultivars (HN51 and SN37) with different responses to iron stress. From HN51 and SN37, we identified a new gene GmVIT1, for which expression is closely related to iron stress response by transcriptomic and quantitative analysis. We obtained GmVIT1 and GmVIT1 promoter from the iron deficiency-tolerant soybean variety Heinong51. Sequence analysis showed that GmVIT1 contained a conserved 170-residue VIT domain and localized at the tonoplast. Moreover, GmVIT1 is expressed in soybean leaves, stems, and roots. The expression of GmVIT1 was significantly induced by excessive Fe/Mn in leaves and stems. GUS assay showed that excess Fe/Mn enhanced GmVIT1 promoter activity. Furthermore, overexpression of GmVIT1 in Arabidopsis seedlings showed reduced phytotoxic effects induced by excess Fe/Mn stress, including yellowing in leaves, decreased chlorophyll content, and accumulated MDA. GmVIT1 overexpression in Arabidopsis showed relatively higher soluble sugar content and SOD, POD, and CAT activity. In addition, the ferric reductase activity in GmVIT1 overexpression in Arabidopsis decreased under excess Fe, while it increased under excess Mn. By integrating all these results, we found that GmVIT1 plays a vital role in plant response to excess Fe/Mn. The results showed that GmVIT1 was worthy of metal homeostasis mechanism research in plants and could be applied in the metal toxic-tolerance improvement in crops.
1. Introduction
Iron (Fe) and manganese (Mn) are essential micronutrients for plants, have redox-active, enzyme-activating functions, and fulfill a structural role in stabilizing proteins [1,2]. Either Fe or Mn deficiency in plants disturbs many biological processes and impairs chlorophyll and organic matter biosynthesis [3]. Although Fe is indispensable for plants, excess accumulation of Fe causes the overproduction of reactive oxygen species (ROS) via the Fenton reaction, which irreversibly impairs cellular and chromatin structure, triggering lipid peroxidation of cellular membrane structures and loss of cellular integrity [4,5,6]. Excess Fe easily appears in waterlogged soils, where soluble Fe concentrations cause a drastic reduction in redox potential. Excessive Fe absorption by plants leads to the appearance of Fe toxicity symptoms called “bronzing”, brown spots spread from the leaf tips to the leaf base [7]. Yield reductions of 10–100% occur in the case of the world’s leading crop species. Fe toxicity is considered one of the most formidable research challenges in rice and wheat cultivation [8,9]. Similarly, plants need Mn in only small quantities and it is toxic in excess. The quantum yield of PSII, CO2 assimilation rate, and stomatal conductance are decreased by Mn toxicity. Furthermore, Mn toxicity generates severe oxidative stress in plant species such as barley [10], cucumber [11], rice [12], and ryegrass [13]. Accumulation of Mn oxides leads to a reduction in shoot biomass, while leaves exhibit toxicity symptoms, including proximal epidermal hypertrophy, and formation of necrotic areas [14]. It has been documented that the uptake of Mn2+ and that of other divalent cations (such as Ca2+, Mg2+, or Fe2+) are closely interrelated, attributed to competition between ions for the absorption site [15,16]. Chlorotic leaves and necrotic spots are the most common symptoms of Mn toxicity [17,18]. One-third of soils in the world are acidic. Mn toxicity is common in poorly drained and acidic soils, and reduces food production [15].
Tight control of Fe and Mn acquisition and translocation is crucial for all plants’ survival and proliferation. As a pivotal intracellular storage organelle, vacuole is essential for regulating metal homeostasis, especially in excess metal detoxification [19]. In addition to the storage role, vacuoles also facilitate the long-distance transport of metals by regulating vacuolar sequestration, which is mainly determined by the interaction between cytoplasmic metal chelators and tonoplast-localized transporters [20]. Transporters at the tonoplast mitigate the toxic effects of excessive metal concentrations on cells by controlling the storage and transport of toxic ions [19]. Some vacuolar metal transporters have been identified, including natural resistance-associated macrophage proteins (NRAMPs) [21,22], metal-tolerance proteins (MTPs) [23], and iron-regulated transporters (IRTs) [24]. Ordinarily, tonoplast transporters have multiple putative trans-membrane domains (TMDs), which carry highly conserved characteristic sequences related to metal selectivity [25]. Vacuolar iron transporters (VITs) are similar to the above proteins. First, VIT genes were identified from Saccharomyces cerevisiae [26,27], and then isolated in plants including Arabidopsis [28], rice [29], rape [30], wheat [31], barley [32], and so on. Recently, VIT was also found as a detoxifier in Plasmodium falciparum [33]. The VIT family proteins contain the DUF125 motif in plants [34]. Phylogenetic analysis revealed that VITs have fewer than four transmembrane domains.
The expression of gene in plant tissues suggests their roles at developmental stages. AtVIT1 in Arabidopsis and OsVIT1, HvVIT1, HvVIT1-2, TaVIT1, TaVIT2, and TaVIT2D in cereals are determined highly expressed in seed embryo and flag leaves [31,32,35,36]. In addition to seeds, VITs are also expressed in the roots and leaves, and nodules [37], indicating that VITs function in all developmental stages of plants. In previous research, VITs were expressed in reproductive organs, while VITs in vegetative organs have received less attention. It was speculated that VITs transport Fe, Mn, Zn, and other metals [37,38,39]. VIT and its homologs can ultimately rescue the growth of Δccc1 yeast under iron toxicity [35,40]. Previous studies found that the VIT family members play a crucial role in excess metal detoxification through the sequestration of Fe from the cytosol to the vacuolar in cells [33,35,40,41]. VIT might participate in Fe accumulation in the endodermal cell layer [42].
VITs are essential genes in plant seeds’ Fe accumulation by regulating metal partitioning between source and absorbing organs. The seed Fe concentration in the nramp3/nramp4 mutant overexpressing AtVTL1, AtVTL2, or AtVTL5 was 50-60% higher than that in non-transformed double mutants or wild-type plants [43]. In rice, OsVIT1 and OsVIT2 mediated the Fe2+ and Mn2+ uptake and transpot in flag leaf [37]. OsVIT2 distributes Fe to the grains by sequestering Fe into vacuoles in the mestome sheath, nodes, and aleurone layer [44]. VITs also play an important role in Fe homeostasis and distribution in plants. In the vit1 mutant of Arabidopsis, Fe diffuses from provascular tissues to the subepidermal cells on the abaxial side of the cotyledon [35]. The constitutive expression of TaVIT2D in vit1 mutant increased the root, leaf, and seed Fe accumulation [36]. AtVIT1 transports Fe into the vacuole for normal seedling development when exposed to excess iron [35]. Excess Fe significantly induces Brassica napa BnMEB2 gene expression in old leaves and roots [30]. BnMEB2 overexpressed plants restored the phenotypes with apparent iron toxicity in roots [30]. In tulip and cornflower petals, Fe ions transported by TgVIT1 and CcVIT combine with anthocyanins in vacuoles to form blue stable chelates [45,46]. In recent years, the application of VIT biofortification has increased significantly [31,47]. These results demonstrate enhancing vacuolar Fe transport in the specific organization as a viable transgenic strategy to biofortify. They could contribute to improved Fe nutrition to help eliminate micronutrient malnutrition in at-risk human populations. However, being a well-known iron storage protein, the detoxification function of excess Fe of VIT has yet to be well-investigated in the plant seedling stage. The underlying mechanism also needs to be elucidated. Further, the detoxification function on excess Mn of VIT is yet to receive notice, nor has it been explicitly studied.
As a major oil crop in the world, soybean is also an iron-sensitive plant. However, soybeans are more sensitive to Fe and Mn stress than other legumes, such as Centrosema pubescens and Phaseolus lathyroide [48,49,50]. Although the reports of GmVTL1 involved nitrogen fixation [40] and GmVTL1a transporting of ferrous Fe from the infected root cell cytosol to the symbiosome [51], research on VIT genes in soybean still needs to be conducted. We found an evolutionarily unique VIT gene that responds to iron deficiency from transcriptomic analysis. The qRT-PCR results showed that iron deficiency and excess iron significantly up-regulated GmVIT1 in HN51, while not significantly in SN37. The results indicated that the GmVIT1 gene is directly related to the different responses to iron stress of HN51 and SN37. Based on the VIT family gene function in the model plants, GmVIT1 might respond to excessive heavy metals such as Fe and Mn and play a role in plant heavy metal detoxification. To prove the function of GmVIT1, we cloned its cds and promoter, analyzed its expression pattern response to excess iron and excess manganese, and explored whether it participated in plant resistance to heavy metal stress by the transgenic method. We hope this work will deepen the understanding of the mechanism of VIT family members in heavy metal stress and provide a candidate gene resource for soybean breeding improvement.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
The Glycine Max L. cv. Heinong 51 (HN51) and Glycine Max L. cv. Suinong 37 (SN37) were supplied by the Crop Tillage and Cultivation Institute, Heilongjiang Academy of Agricultural Sciences. The soybean seeds were soaked in 5% sodium hypochlorite solution for 5 min, disinfected, and rinsed with sterile water. After germination in the dark, soybean seeds were transferred to 1/2 Hoagland nutrient solution. Wild-type Arabidopsis (Arabidopsis thaliana ecotype Columbia-0) seeds were sterilized with 70% (v/v) ethanol, followed by 10% NaClO, and rinsed three times with sterile water, and germinated on one-half strength MS medium supplemented with 0.9% agar plates in an incubator with a light/dark cycle of 16 h/8 h at 23 °C, and 60% relative humidity. Subsequently, the Arabidopsis seedlings were cultured in pots.
2.2. Iron Deficiency Tolerance Detection
The HN51 and SN37 soybean seedlings were cultured in 1/2 Hoagland solution to the trefoil stage and treated with 0 mM Fe-EDTA (−Fe) and 0.1 mM Fe-EDTA (the control) for 16 days. The hydroponic solution was replaced every 4 days. The root length, the lateral root number, and the iron content were detected. We performed a transcriptomic analysis on HN51 and SN37 soybean seedlings exposed to iron deficiency. The soybean samples were dried and ground into powder, and 0.05 g samples were put into the digestion tube, and had 4 mL 65% nitric acid added and digested at 120 °C for 90 min. When temperature dropped to room temperature, 1 mL of 30% hydrogen peroxide was added into the digestive tube and heated at 120 °C for 90 min. After dropping to room temperature, 750 μL of 30% hydrogen peroxide was added and kept until transparent. Finally, ddH2O was used to constant volume to 50 mL. The iron content was determined by an inductively coupled plasma emission spectrometer (PE ICP-OES8000).
2.3. Cloning of GmVIT1 and GmVIT1 Promoter
Total RNA was extracted from HN51 soybean seedlings using a Total RNA Kit II (Omega BioTek, Norcross, GA, USA). The cDNA was synthesized using a TOYOBO kit. Coding sequences of GmVIT1 were amplified from cDNAs. The total DNA of HN51 was extracted using the plant genomic DNA extraction kit (Omega). The GmVIT1 5′-flanking sequence was amplified. The primers are listed in Supplementary Table S1. After purification, the PCR products were sequenced (Sangon Biotech Co., Ltd., Shanghai, China) and analyzed using BLAST (https://www.ncbi.nlm.nih.gov/orffinder/) (accessed on 18 March 2021); the VIT protein sequences from 12 species were downloaded from NCBI (https://www.ncbi.nlm.nih.gov) (accessed on 17 May 2022) and phytozome (https://phytozome-next.jgi.doe.gov/info/Gmax_Wm82_a2_v1) (accessed on 17 May 2022) and analyzed by the MEGA7.0. The analysis of the GmVIT1 promoter was performed by Plant CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (accessed on 20 March 2022).
2.4. Subcellular Localization of GmVIT1
The subcellular localization of GmVIT1 was investigated by 35S::GmVIT1-GFP transiently expressed in onion epidermal cells. The pBI121-35S::GmVIT1-GFP was constructed and transformed into Agrobacterium tumefaciens GV3101 and transformed into the onion epidermal cells. After transformation, cells were incubated in the dark for 2 to 3 days and observed by confocal laser scanning microscopy (TCS SP5 LEICA Microsystems, Germany).
2.5. Gene Expression Analysis
The HN51 and SN37 seedlings were cultured in 1/2 Hoagland solution for 15 days and then treated with 1/2 Hoagland solution with 0.3 mM Fe-EDTA(+Fe), 0 mM of Fe-EDTA(−Fe), and 0.1 mM of Fe-EDTA(CK) for 7 days.To further analyze the relative expression pattern of GmVIT1 in soybean HN51, HN51 seedlings were cultured in 1/2 Hoagland solution for 15 days and then treated with 0.3 mM Fe-EDTA + 0.1 mM of MnCl2 (+Fe), 0.1 mM of Fe-EDTA 0.3 mM MnCl2 (+Mn), and 0.1 mM of Fe-EDTA + 0.1 mM of MnCl2 (CK) for 7 days. Total RNA was isolated from the cell proliferating leaves, the third to fifth leaves, using the RNeasy mini kit (Qiagen, Hilden, Germany), and cDNA was synthesized using the Reverse TraAceaFirst strand cDNA synthesis kit (TOYOBO, Tokyo, Japan). qRT-PCR analysis was carried out using SYBR® Premix Ex TaqTM II (TAKARA, Otsu, Japan) and CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, United States). The relative gene expression was calculated using the ΔΔCt method and was normalized to GmActin. In all experiments, qRT-PCR analyses were performed as triplicates on three different RNA samples isolated independently from each treatment. Primers used in these assays are listed in Supplementary Table S1. The GmVIT1 promoter was cloned into the pBI121 vector to generate a promoter reporter construct GmVIT1pro::β-glucuronidase (GUS). The Agrobacteriummediated transformation of Arabidopsis was performed through the floral dip method. The tissues of the GmVIT1pro::GUS transgenic plants were incubated in GUS solution (Coolaber, SL7160), overnight at 37 °C, and then 95% ethanol solution was used to remove chlorophyll. The stained tissues were monitored using a microscope (EZ4-HD LEICA, Germany).
2.6. Generation and Character Analysis of GmVIT1 Transgenic Arabidopsis
GmVIT1 was inserted into the pBI121 vector (35S::GmVIT1) and introduced into Arabidopsis by Agrobacterium tumefaciens (GV3101)-mediated transformation using the floral dip method. T1 plants were selected by kanamycin and verified by PCR. Established GmVIT1 transgenic Arabidopsis were transferred to soil for subsequent generations by selffertilization in a greenhouse. T3 seedlings were grown in pots and identified by PCR (Figure S1). Three T3 lines and wild-type plants of 25-day-old were treated with 0.3 mM FeEDTA + 0.1 mM MnCl2(+Fe), 0.3 mM MnCl2 + 0.1 mM Fe-EDTA(+Mn), and 0.1 mM Fe-EDTA + 0.1 mM MnCl2 (CK) for 3 days for 7 days. Samples were then observed and photographed. The chlorophyll content was quantified by the acetone method. The soluble sugar and MDA contents were determined according to the experimental method of Dhindsa et al. [52]. The SOD, POD, and CAT activities were measured according to the methods of Shi et al. [53] and Aebi et al. [54]. The ferric iron chelate reductase (FCR) activity was determined using a kit (Geruisi, G0147F).
2.7. Statistical Analysis
Each experiment was repeated in triplicate, and statistical analysis was performed using SPSS statistical software.
3. Results
3.1. The Tolerance of HN51 and SN37 to Fe Deficiency
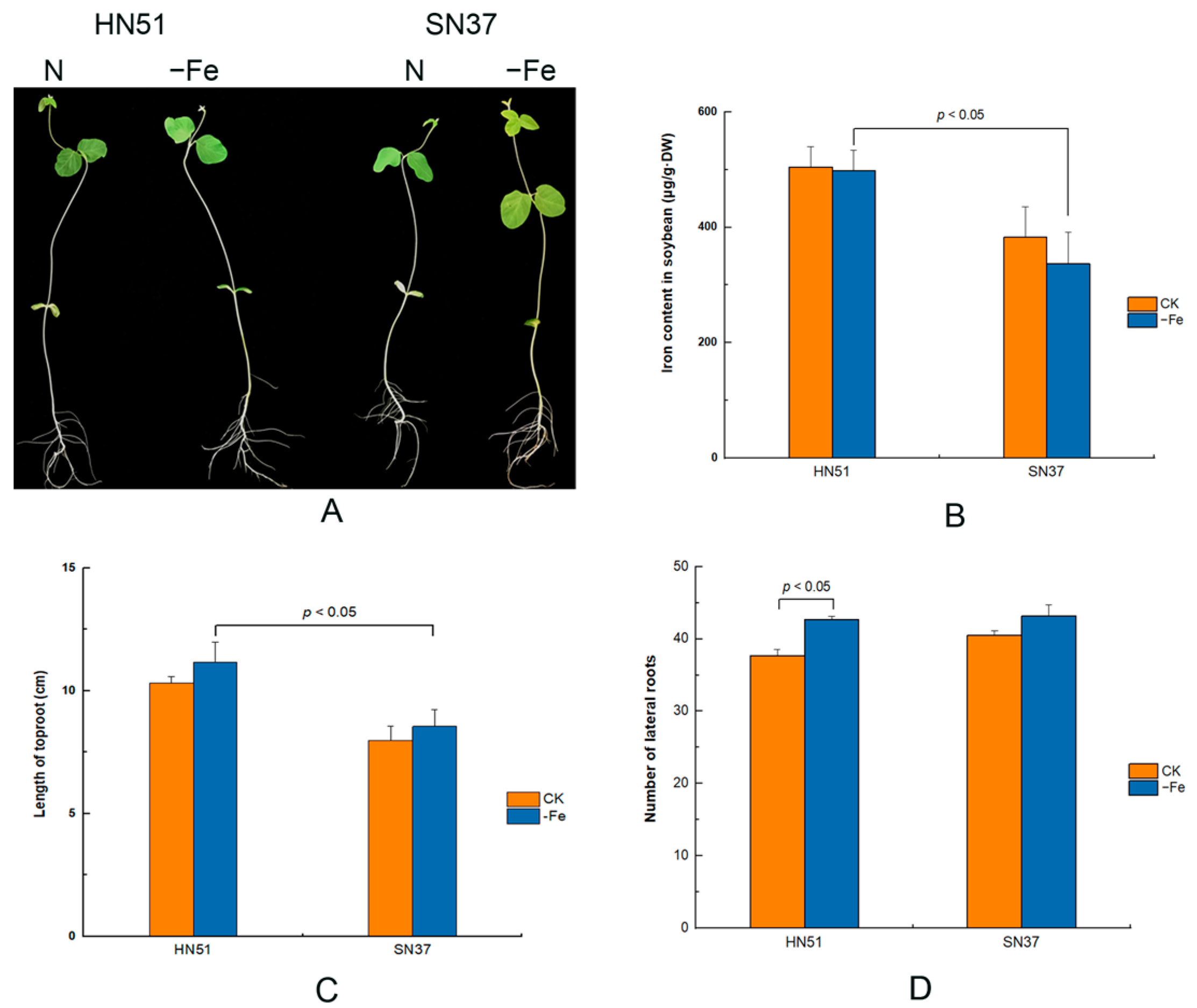
The HN51 and SN37 are two soybean varieties in Northeast China. Under normal conditions, they had no significant difference in their growth states. After 16 days of iron deficiency treatment, the leaves of HN51 soybean showed mild chlorosis compared with SN37. The iron content in HN51 soybean was signifancatly higher than that of SN37 in iron deficiency and iron supply conditions. Compared with SN37, HN51 showed a significantly higher taproot length. These results indicated that HN51 had stronger adaptability and tolerance to iron deficiency (Figure 1).

Figure 1.
The phenotype and total Fe content of HN51 and SN37 under iron deficiency. (A) Phenotypes of HN51 and SN37. (B) Total Fe content in soybean. (C) The taproot length of HN51 and SN37. (D) The lateral root number of HN51 and SN37. The HN51 and SN37 soybean seedlings were treated with 0 mM of Fe-EDTA(−Fe) for 16 days, with 0.1 mM Fe−EDTA as the control. Experiments were repeated three times, and each treatment group contained five samples. The data represent the means of replicates.
3.2. The Expression of GmVITs in Transcriptomic Analysis
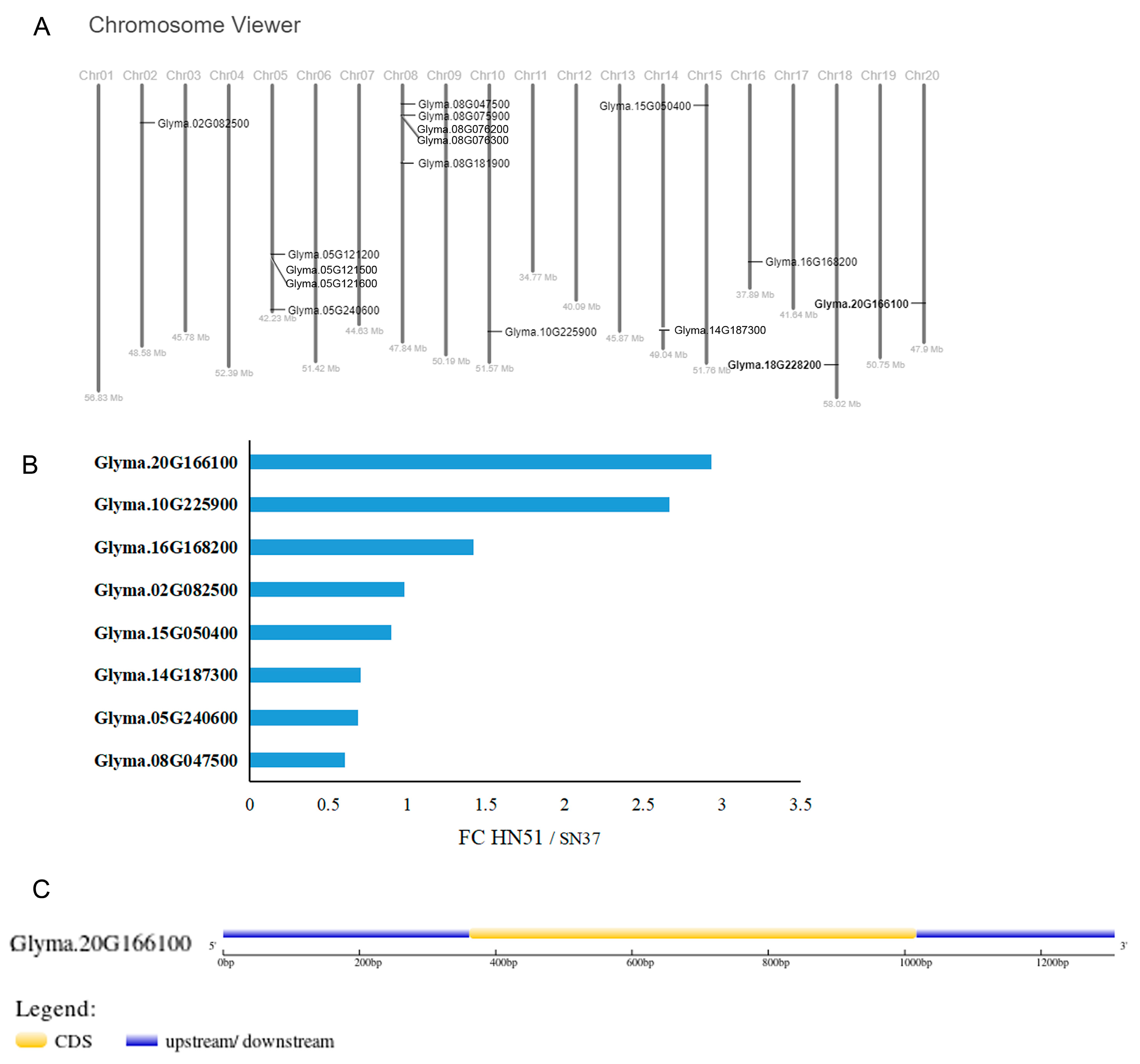
Genome-wide analysis revealed that the soybean genome contains 16 VIT family members. There are tandem repeats of VIT genes on specific chromosomes (Figure 2A). We performed a transcriptomic analysis on HN51 and SN37 soybean seedlings exposed to iron deficiency. The expression of GmVIT1 was significantly higher in HN51 than in SN37. The Fe-responsive VIT family members in soybean were identified from comparative transcriptome data of HN51 and SN37 under iron deficiency. A total of 16 VIT family members were annotated in the transcriptional profile. A total of six VIT genes (Glyma.05G121200, Glyma.05G121500, Glyma.08G076200, Glyma.05G121600, Glyma.08G076300, and Glyma.18G228200) were not expressed, and Glyma.08G075900 was weakly expressed. The transcription of nine VIT genes in soybean (Glyma.05G240600, Glyma.08G047500, Glyma.08G181900, Glyma.15G050400, Glyma.14G187300, Glyma.16G168200, Glyma.02G082500, Glyma.10G225900, and Glyma.20G166100) was detected. Among them, GmVIT1(Glyma.20G166100) showed the highest expression, and its homologs Glyma.10G225900 also showed a higher expression (Figure 2B). The gene structure of GmVIT1 is shown in Figure 2C.

Figure 2.
Screening of soybean GmVIT genes. (A) Chromosome localization of 16 GmVIT genes. (B) The expression of GmVIT genes in HN51 and SN37 under iron deficiency by transcriptomic analysis. (C) The gene structure of GmVIT1.
3.3. GmVIT1 Response to Iron Deficiency and Excess Iron in HN51 and SN37
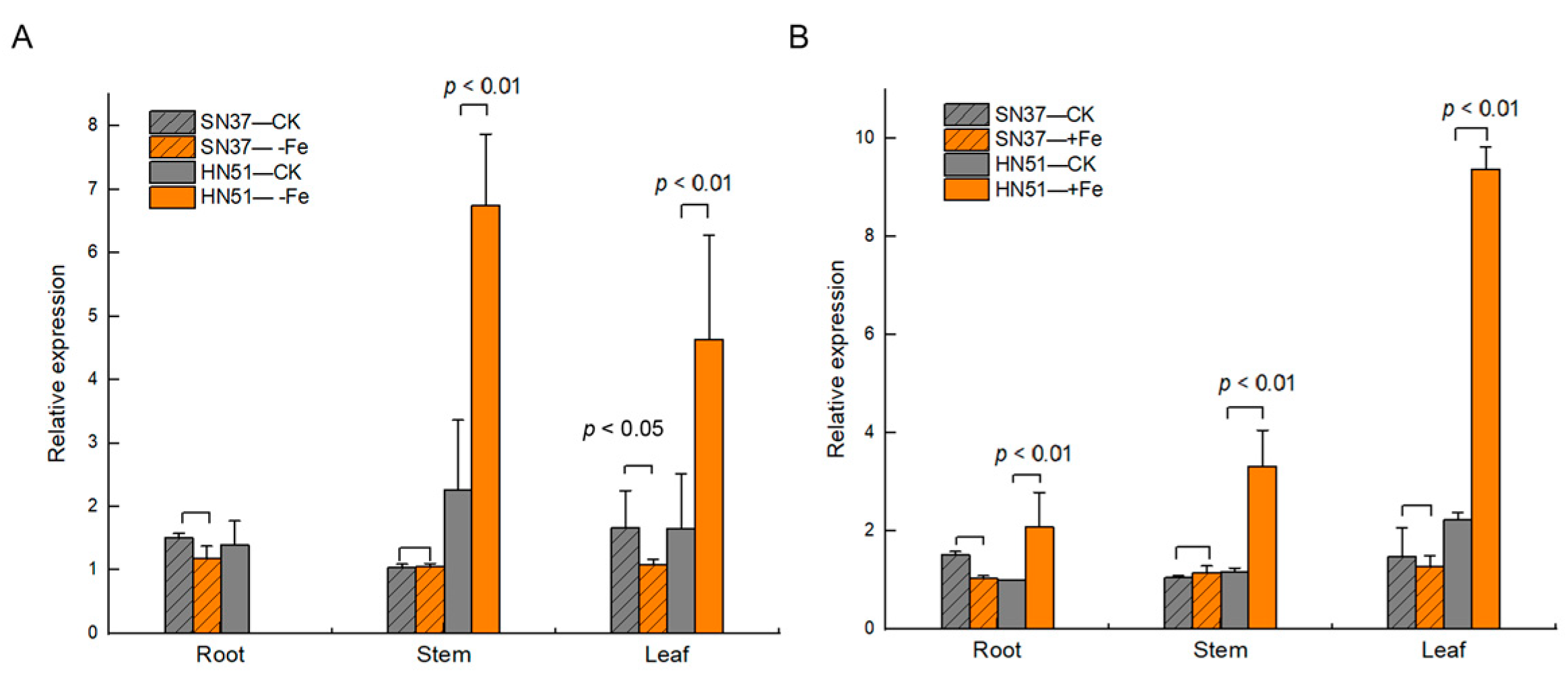
In the root, stems, and leaves of SN37, there were no differences in GmVIT1 expression in −Fe and +Fe treatments, except for the roots under iron deficiency. In HN51 stems and leaves, the expression of GmVIT1 was significantly higher in −Fe and +Fe treatments than that in the control. The expression of GmVIT1 was significantly higher in +Fe treatments than that in the control. The expression of GmVIT1 was not detected in the roots of HN51 under −Fe treatment and showed lower expression than in stems and leaves (Figure 3).

Figure 3.
The expression of GmVIT1 in HN51 and SN37 under iron deficiency (–Fe), excess iron (+Fe), and normal conditions (CK). (A) The expression of GmVIT1 in HN51 and SN37 under iron deficiency (–Fe). (B) The expression of GmVIT1 in HN51 and SN37 under excess iron +Fe: 0.3 mM Fe-EDTA, −Fe: 0 mM of Fe-EDTA, and CK: 0.1 mM of Fe-EDTA. The expression was detected by qRT-PCR. The qRT-PCR experiments were repeated three times, and each treatment group contained five samples. All the data represent the means of three replicates.
3.4. Cloning and Bioinformatics Analysis of GmVIT1
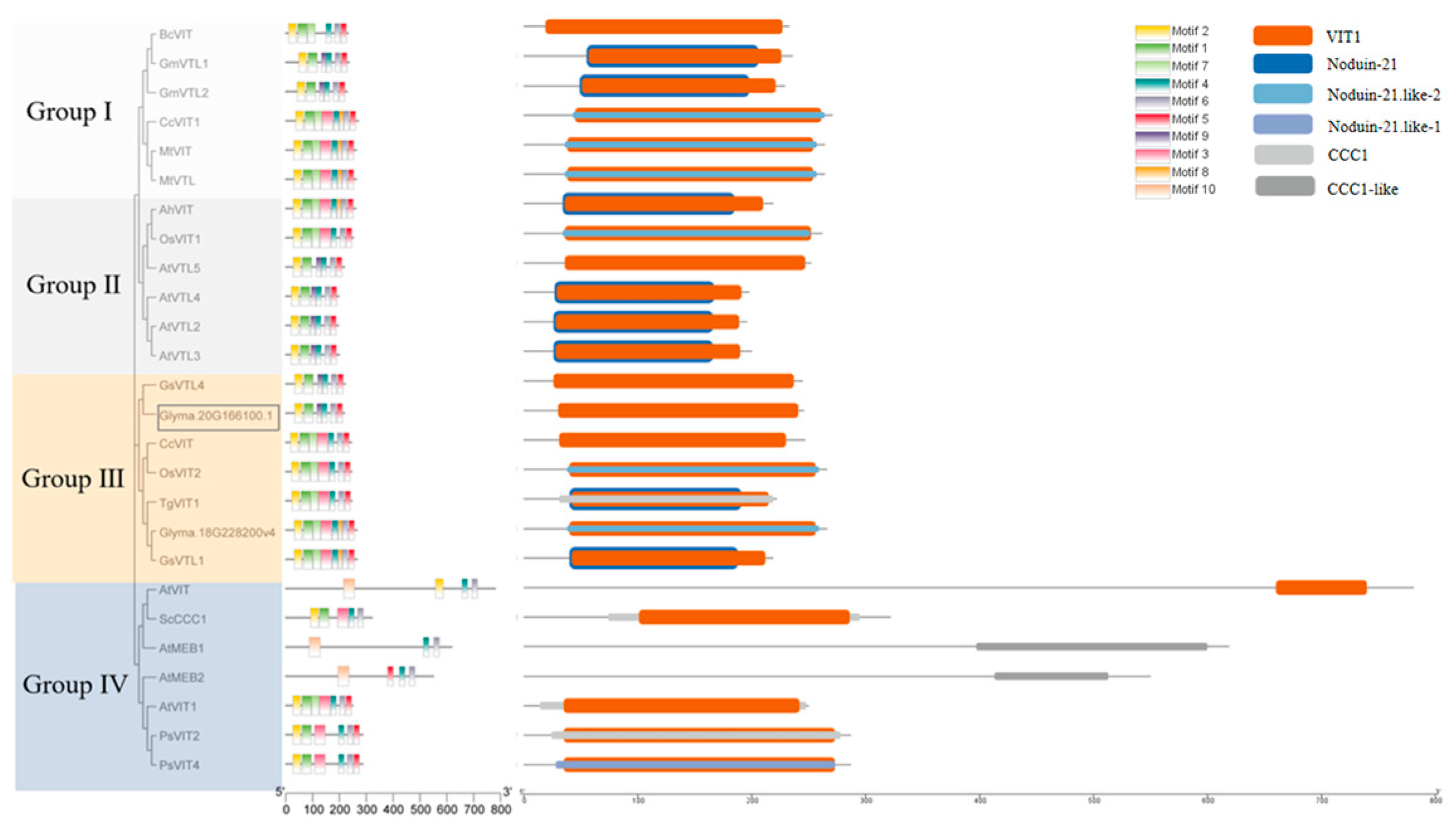
GmVIT1 exists on chromosome 20 and contains an intron (Figure 2A,C). The complete GmVIT1 CDS spanned 660 bp and encoded a 219 amino acid protein with molecular mass of 23,427.16 KD. GmVIT1 has four transmembrane domains and contains a typical VIT domain at 42-212 amino acids (Figure S2 and Figure 4), indicating that it belongs to the VIT family. An evolutionary tree of 26 VIT proteins from 12 species was constructed using MEGA 7.0. GmVIT1 was assigned to group Ⅲ, which contained seven VITs from Glycine max, Glycine soja, Centaurea cyanus, Tulipa gesneriana, and Oryza sativa. GmVIT1 showed a close phylogenetic relationship with Glyma.18G228200v4, GsVIT1, GsVTL4, OsVIT2, CcVIT, and TgVIT1 (Figure 4). GmVIT1 was most close to GsVTL4 from Glycine soja, with a 93.24% similarity in the protein sequence. The other two GmVTLs were assigned to the group and were distantly related to GmVIT1. A total of 10 conserved motifs were identified in VIT proteins using MEME. Among them, six motifs (1, 2, 4, 5, 6, and 9) were distributed in GmVIT1 (Figure 4, Table 1).

Figure 4.
Phylogenetic relationship, motif, and domain composition of the 26 vacuolar iron transporter proteins. Using the Neighbor-Joining method, the phylogenetic tree was constructed with the MEGA 7.0 software. The length of branches corresponds to the degree of divergence. Numbers in the figure represent bootstrap values. Named sequence from Arabidopsis thaliana are AtVIT1 (At2g01770), AtVTL1 (At1g21140), AtVTL2 (At1g76800), AtVTL3 (At3g43630), AtVTL4 (At3g43660), AtVTL5 (At3g25190), from Centaurea cyanus is CcVIT1 (BAO52026), from Glycine max are GmVIT1(Glyma18g228200v4,MN547956.1), GmVTL1 (Glyma.05g121600, NP_001236825), GmVTL2 (Glyma.08g076300, XP_003531056), from Glycine soja are GsVIT1 (D0Y65_013220) and GsVTL1( XP_028233991.1), from Oryza sativa are OsVIT1 (Os04g0463400, BAS89575) and OsVIT2 (Os09g0396900, Q6ERE5.2), from Tulipa gesneriana is TgVIT1 (BAH59029), from Medicago truncatula are MtVTL(Medtr7g076320.1) and MtVIT (XP_024625458.2), from Saccharomyces cerevisiae is ScCCC1(WN66_04396), from Bradyrhizobium cosmicum is BcVIT(FNV92_15665), from Arachis hypogaea is AhVIT (DS421_4g129730), from Cajanus cajan is CcVIT1(XM_020363549.2), from Papaver somniferum are PsVIT2 (XM_026526985.1) and PsVIT4 (XM_026599962.1), and from Tulipa gesneriana is TgVIT1(BAH59029). Different motifs of the VIT proteins are displayed in different-colored boxes. The positions of the structural domain are marked with orange, blue, and grey, respectively.

Table 1.
Details of the VIT protein motifs.
3.5. Subcellular Localization of GmVIT1 Protein
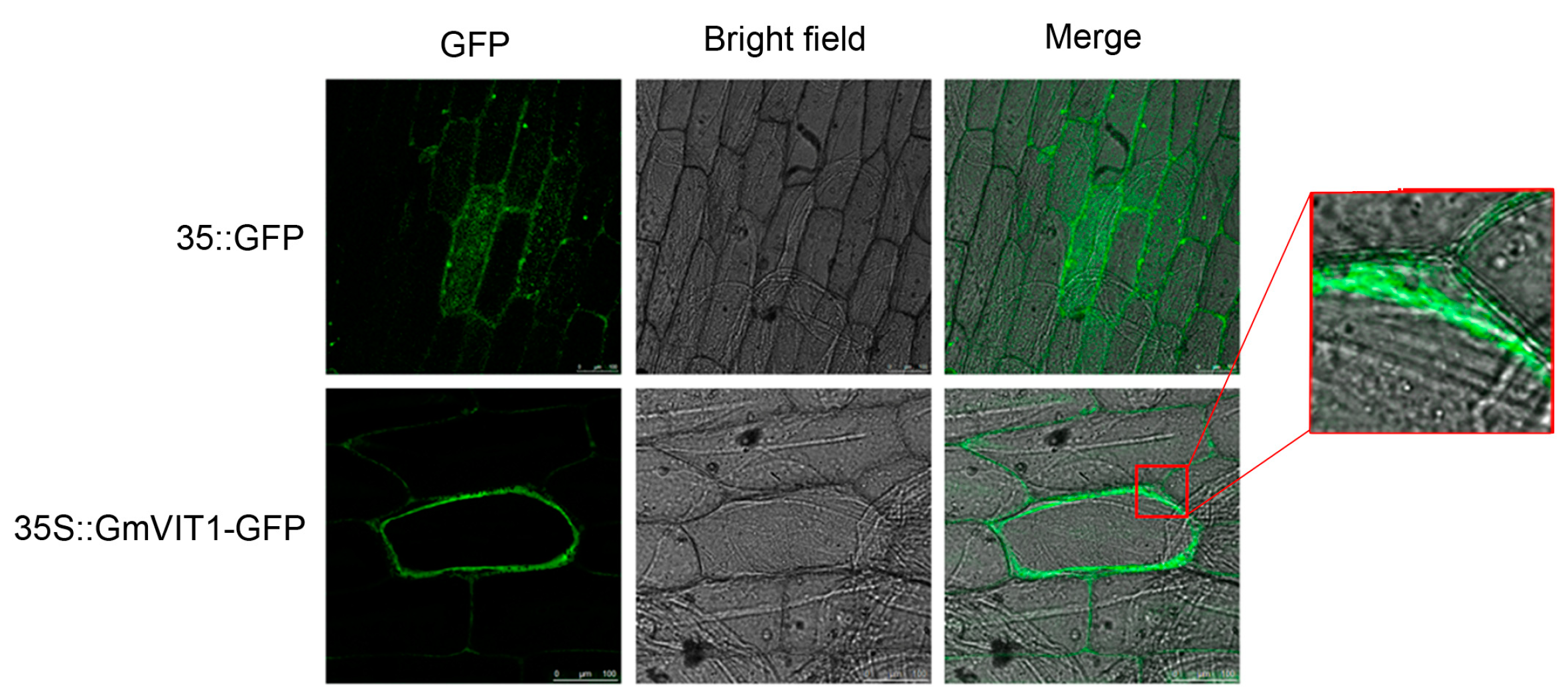
GmVIT1 was fused in frame to a green fluorescent protein (GFP) and driven by a 35S promoter. The 35S::GmVIT1-GFP fluorescence was observed exclusively in the vacuolar membrane, while 35S::GFP fluorescence was found throughout the whole cells (Figure 5). The results showed that the GmVIT1 protein was localized at the vacuolar membrane and may participate in transmembrane transport function as a vacuolar membrane transporter.

Figure 5.
Subcellular localization of 35S::GmVIT1-GFP fusion proteins in onion epidermal cells. GFP: Field of view observed under excitation of green fluorescence; Bright field: Field of view observed without excitation of green fluorescence; Merge: merged green fluorescence and bright-field images. The images were obtained by confocal microscopy. The 35S::GFP was used as a control.
3.6. Expression Patterns of GmVIT1 Gene in Soybean
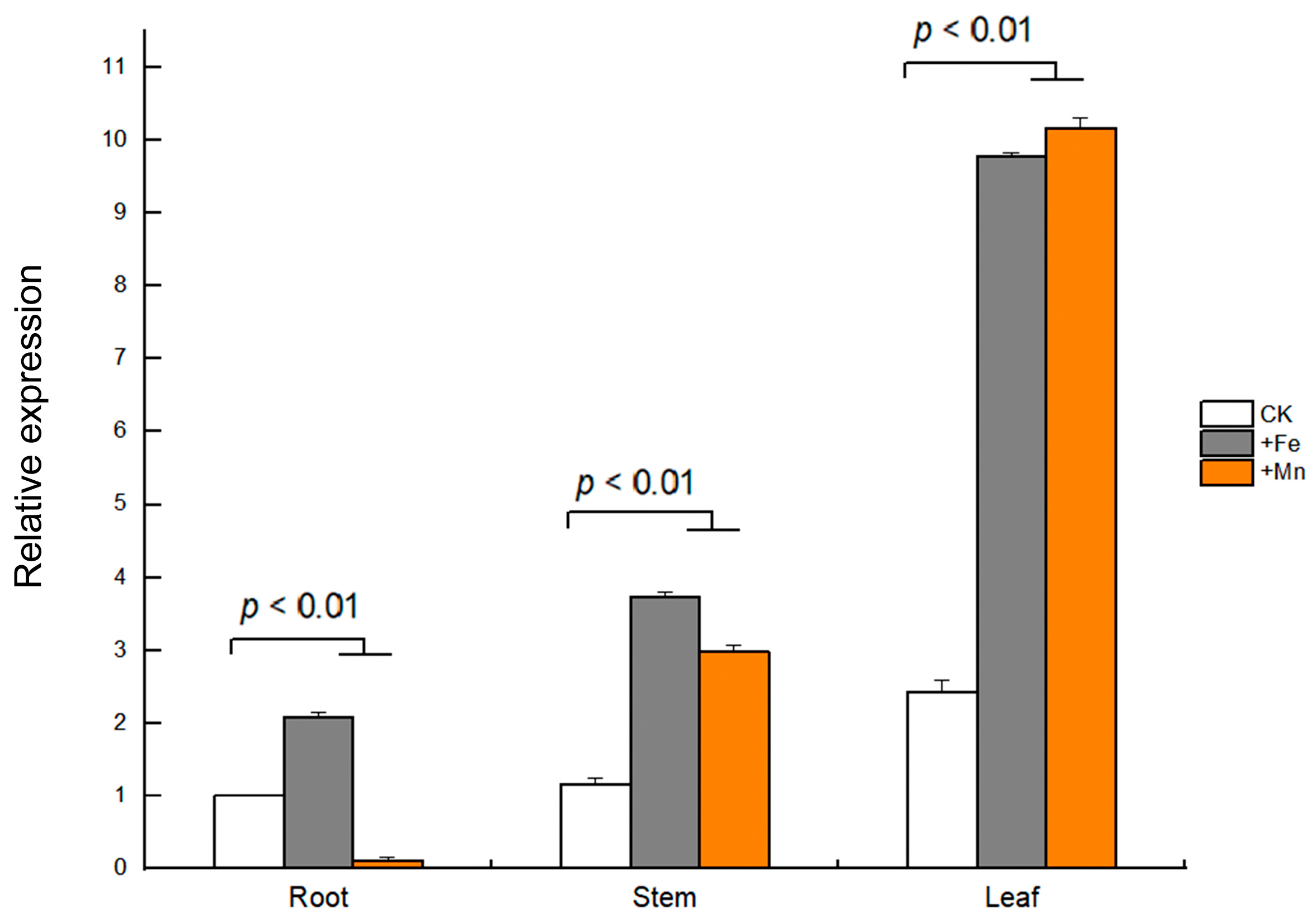
The tissue-specific expression of GmVIT1 in young soybean seedlings was detected by qRT-PCR. The results showed that GmVIT1 was expressed in three tissues, i.e., leaves, stems, and roots. Specifically, the highest expression level of GmVIT1 was in soybean leaves. In the stems and the leaves, the GmVIT1 transcript level was significantly up-regulated (p < 0.01) by +Fe/+Mn stress compared with that in the control. In the root, GmVIT1 was significantly up-regulated (p < 0.01) by +Fe stress while down-regulated (p < 0.01) by +Mn stress. (Figure 6).
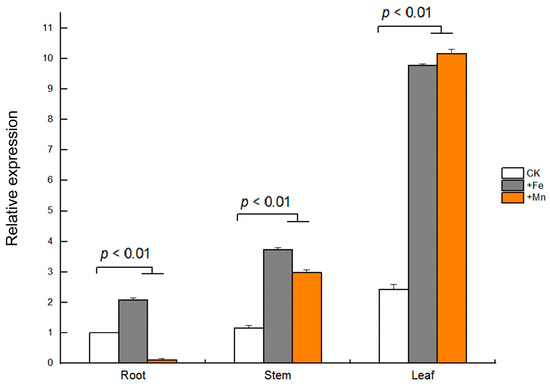
Figure 6.
Relative expression of GmVIT1 in soybean tissues under excessive Fe/Mn treatment. The HN51 seedlings were cultured in 0.3 mM Fe-EDTA + 0.1 mM MnCl2 (+Fe), 0.3 mM MnCl2 + 0.1 mM Fe-EDTA (+Mn), and 0.1 mM Fe-EDTA + 0.1 mM MnCl2 as the control (CK) for 3 days. qRT-PCR analysis of the GmVIT1 transcript in the roots, the stems, and the leaves of the soybean plants was conducted. Experiments were repeated three times, and each group contained five samples. All the data represent the means of three replicates.
3.7. GmVIT1 Promoter Activity in Response to +Fe or +Mn Stress in Arabidopsis
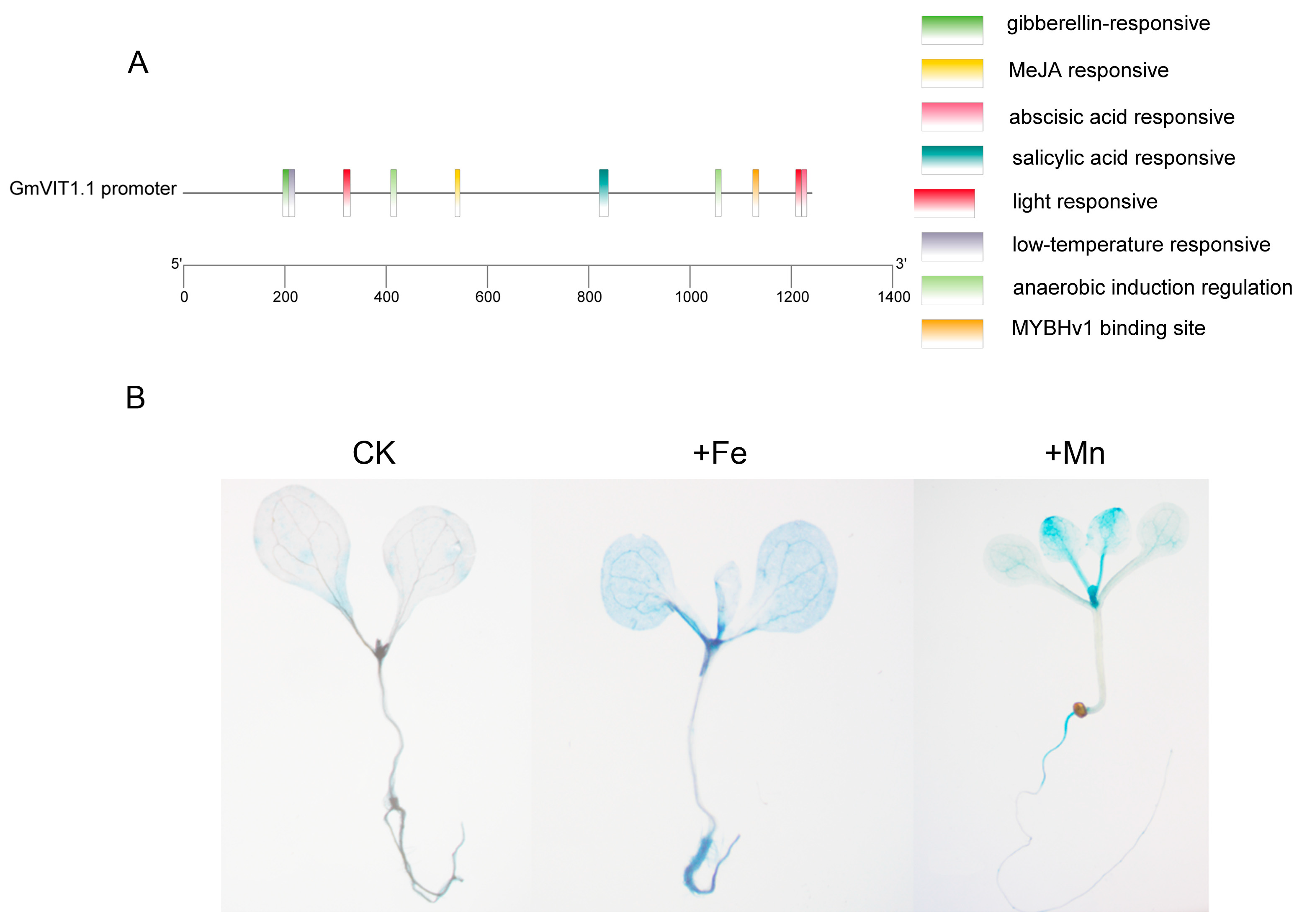
The GmVIT1 promoter contains some cis-elements in response to abscisic acid, gibberellin, salicylic acid, methyl jasmonate, anaerobic, low temperature, and light. MYB recognition sites were also detected in the GmVIT1 promoter (Figure 7A). The slight GUS staining for GmVIT1pro::GUS in roots, stems, and leaves confirmed the widespread and weak expression of GmVIT1 under normal conditions. The +Fe and +Mn stress significantly enhanced GUS activity in Arabidopsis seedlings, confirming the gene expression analysis results. The +Fe stress enhanced GUS activity in the roots, leaves, and petiole of Arabidopsis, especially in old leaf veins. The +Mn stress enhanced GUS activity in the hypocotyl, leaves, and petiole, especially in young leaves (Figure 7B).
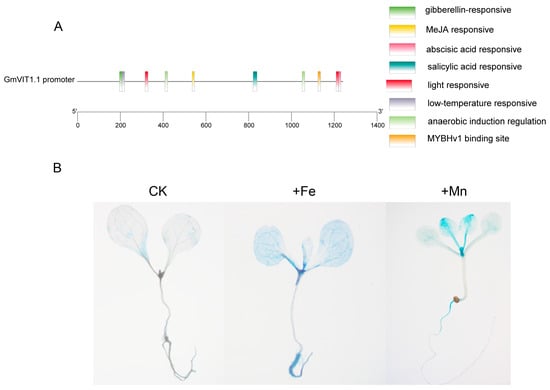
Figure 7.
Cis−elements in the GmVIT1 promoter and histochemical staining of transgenic Arabidopsis harboring the GmVIT1pro::GUS constructs. (A) Predicted positions and functions of cis-elements in the GmVIT1 promoter. (B) GUS staining results of GmVIT1pro::GUS transgenic Arabidopsis under 0.1 mM Fe-EDTA+0.1 mM MnCl2 (CK), 0.3 mM Fe-EDTA + 0.1 mM MnCl2 (+Fe), and 0.3 mM MnCl2 + 0.1 mM Fe-EDTA (+Mn) for 7 days. The seedlings were then stained for detection of β-glucuronidase activity.
3.8. GmVIT1 Transgenic Arabidopsis Exhibit Enhancement of Tolerance to +Fe or +Mn Stress
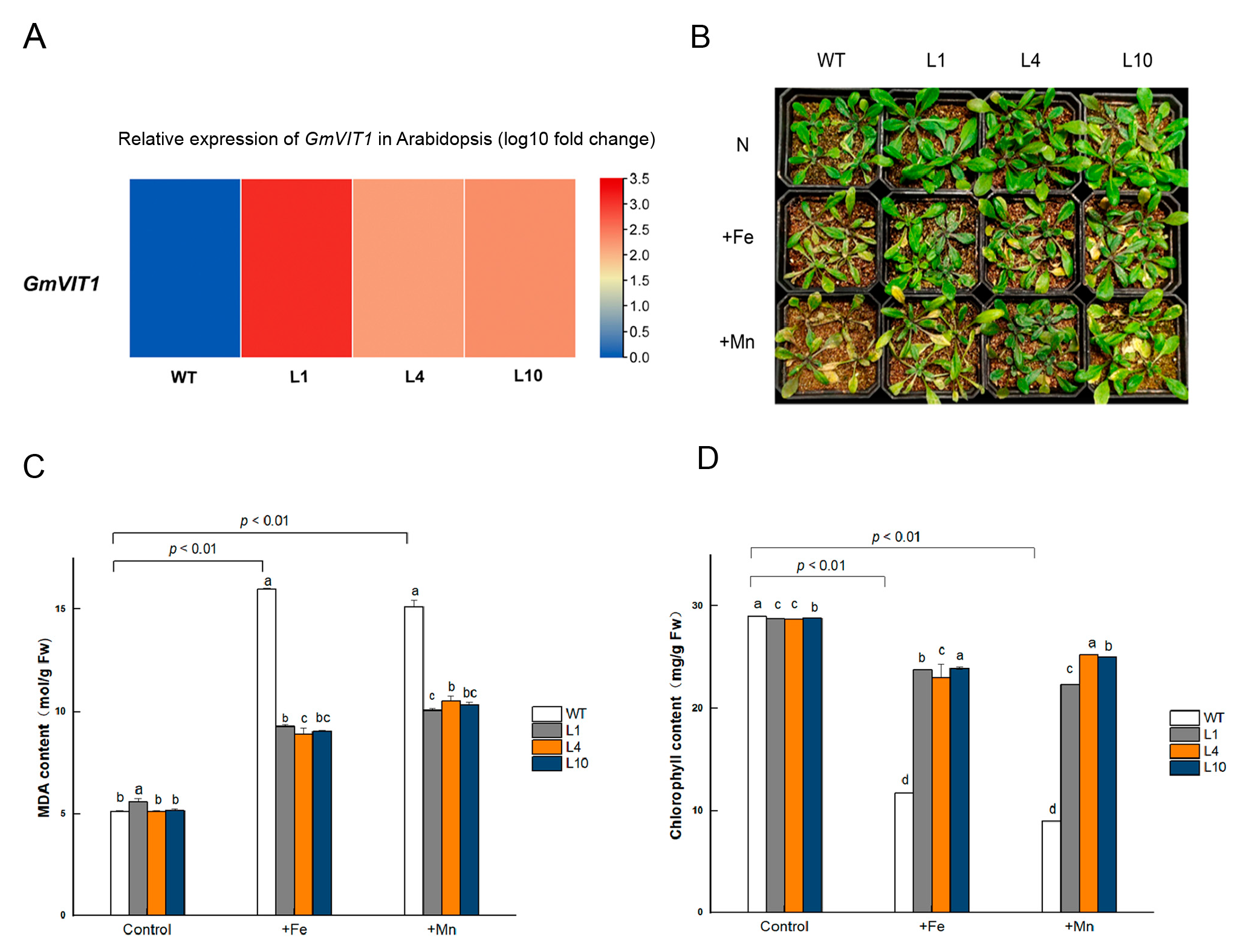
We generated GmVIT1 transgenic Arabidopsis lines and raised T3 lines in this study. The transgenic lines, L1, L4, and L10, which have high expression levels of GmVIT1, were exposed to +Fe and +Mn stress (Figure 8A). GmVIT1 transgenic Arabidopsis lines showed better growth status than wild-type plants. Under +Fe and +Mn stress, both transgenic Arabidopsis and wild-type Arabidopsis showed visible leaf chlorosis compared to non-stress conditions (Figure 8B). The MDA content of transgenic lines and WT increased dramatically under +Fe or +Mn stress (p < 0.01), indicating that +Fe or +Mn resulted in membrane damage of cells in plants. However, MDA content in L1, L4, and L10 were significantly lower than that in the wild-type plants under +Fe or +Mn stress (p < 0.05) (Figure 8C). The MDA results showed that overexpression of GmVIT1 partially rescued damage to the membrane in plant cells. The chlorophyll content in GmVIT1 transgenic Arabidopsis was significantly lower than that in the wild-type plants under normal conditions (p < 0.05). When exposed to +Fe or +Mn stress, chlorophyll content was significantly reduced in transgenic and wild-type Arabidopsis (p < 0.01). The chlorophyll content of L1, L4, and L10 were significantly higher than that in the wild-type plants (p < 0.05) (Figure 8D).
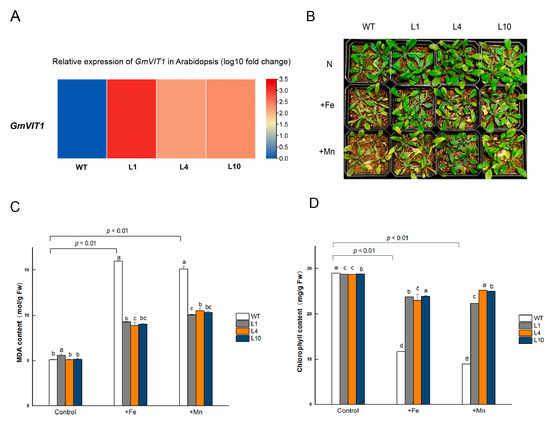
Figure 8.
Phenotype and physiological indicators of GmVIT1 transgenic Arabidopsis and wild-type lines under excess iron or manganese stress. The plants were treated with 0.1 mM Fe-EDTA + 0.1 mM MnCl2 (CK), 0.3 mM Fe-EDTA + 0.1 mM MnCl2 (+Fe), and 0.3 mM MnCl2 + 0.1 mM Fe-EDTA (+Mn) (A) Relative expression levels of the GmVIT1 gene in GmVIT1 transgenic and wild-type Arabidopsis. (B) Growth status of GmVIT1 transgenic lines and wild-type Arabidopsis under +Fe/+Mn stress. (C) The MDA content and (D) the chlorophyll content of GmVIT1 transgenic Arabidopsis and wild-type plants. Experiments were repeated three times, and each group contained five samples. All the data represent the means of three replicates. Lowercase letters represent a statistically significant difference between GmVIT1 transgenic Arabidopsis and wild-type plants under the same condition, p < 0.05.
3.9. GmVIT1 Transgenic Arabidopsis Showed Higher Soluble Sugar Content and Antioxidant Enzyme Activity under +Fe or +Mn Stress
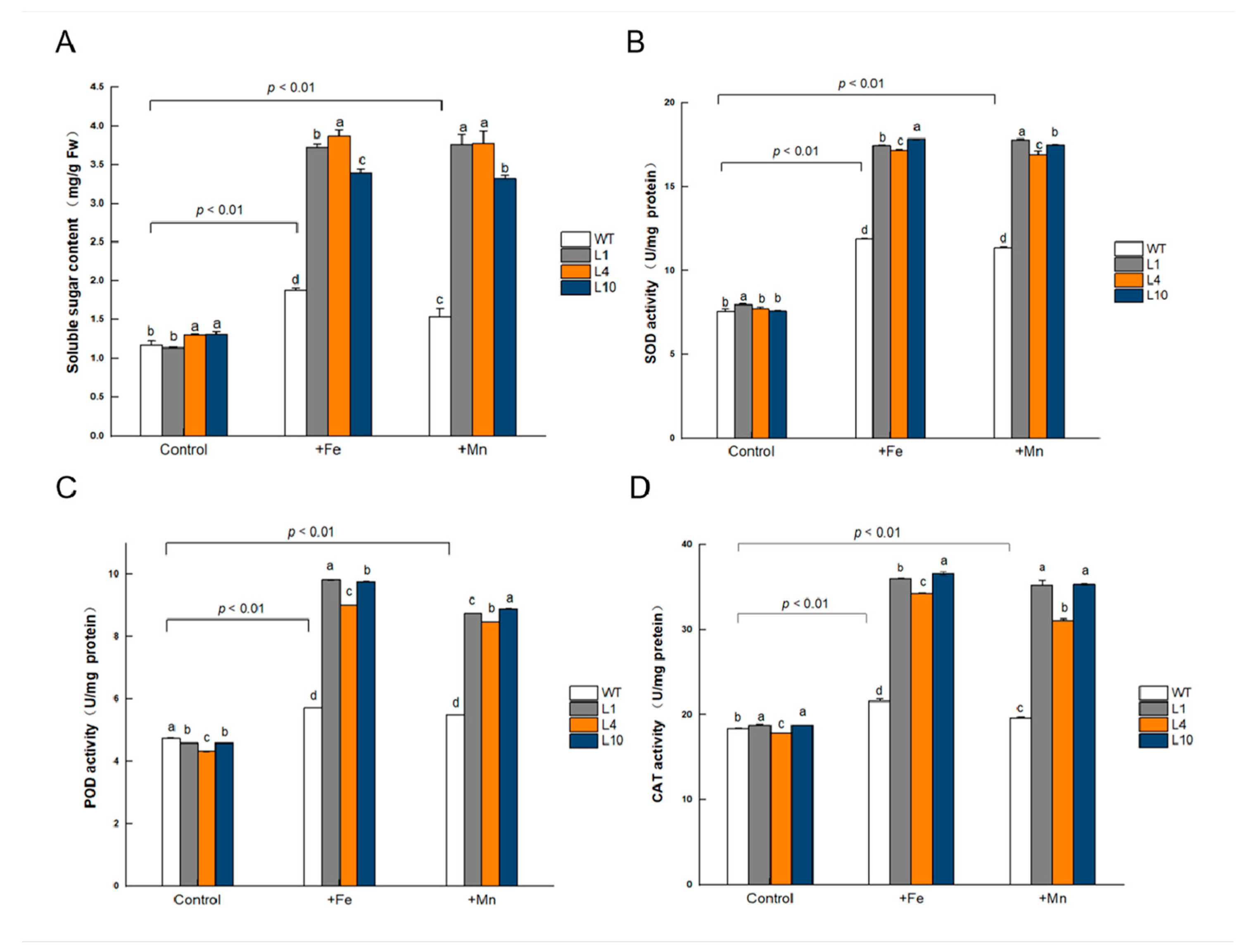
Soluble sugars are important osmolytes that protect plants against abiotic stress. The soluble sugar content showed significant differences (p < 0.05) between GmVIT1 transgenic Arabidopsis and wild-type plants under normal conditions. The +Fe or +Mn stress significantly increased soluble sugar content in all plants (p < 0.01), and the soluble sugar content in GmVIT1 transgenic Arabidopsis was significantly higher than that in the wild-type plants (p < 0.05) (Figure 9A). When grown in normal conditions, the activity of SOD, POD, and CAT showed significant differences (p < 0.05) between GmVIT1 transgenic and wild-type plants. Excess Fe or Mn stress caused a significant increase in SOD, POD, and CAT activities in GmVIT1 transgenic Arabidopsis and wild-type plants (p < 0.01), and the SOD, POD, and CAT activities of GmVIT1 transgenic Arabidopsis were significantly higher than that in the wild-type (p < 0.05) (Figure 9B–D). The results showed that the protective enzyme activity increased in GmVIT1 transgenic Arabidopsis, which could protect plants from oxidative damage.
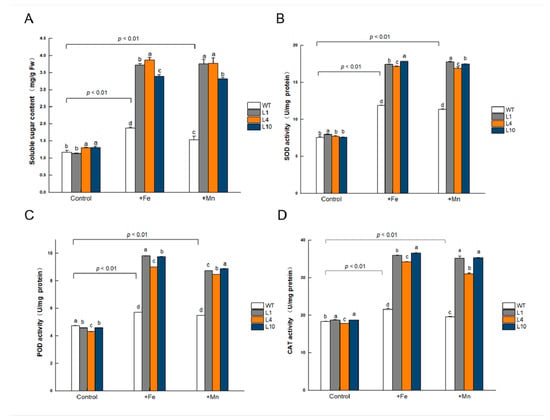
Figure 9.
The physiological index of GmVIT1 transgenic lines and wild-type plants grown under excess iron or manganese. (A) Soluble sugar content. (B) Superoxide dismutase activity. (C) Peroxidase activity. (D) Catalase activity. The plants were grown in 1/2 Hoagland nutrient solution for 14 days and treatedwith 0.1 mM Fe-EDTA + 0.1 mM MnCl2 (CK), 0.3 mM Fe-EDTA + 0.1 mM MnCl2 (+Fe), and 0.3 mM MnCl2 + 0.1 mM Fe-EDTA (+Mn) for 7 days. Experiments were repeated three times, and each group contained five samples. All the data represent the means of three replicates. Lowercase letters represent a statistically significant difference in GmVIT1 transgenic lines and wild-type plants under the same condition, p < 0.05.
3.10. FCR Activity Changed in GmVIT1 Transgenic Plants under +Fe or +Mn Stress
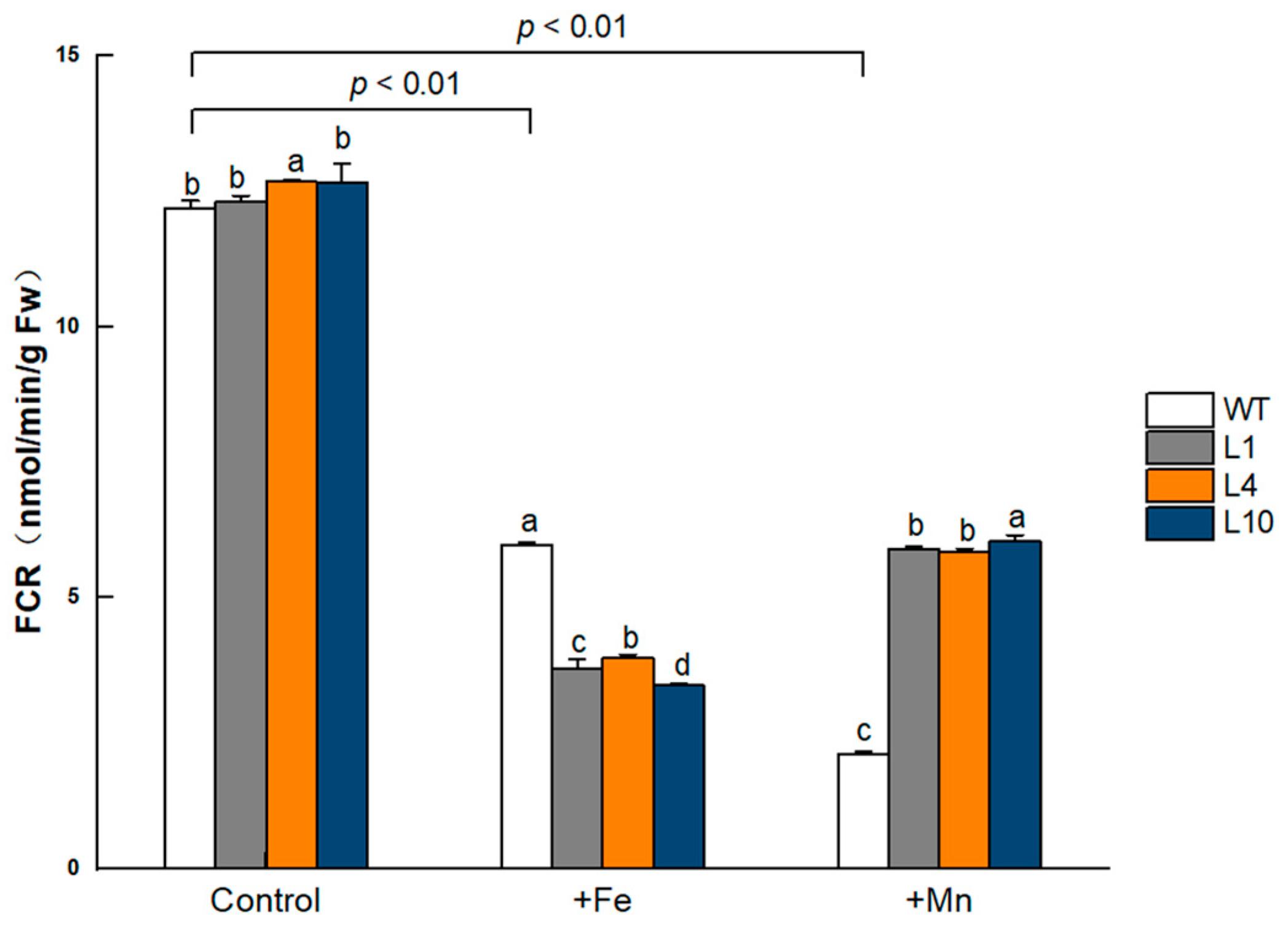
Grown under the normal condition, the FCR activity in GmVIT1 transgenic Arabidopsis roots was significantly higher than that in wild-type plants (p < 0.05). When exposed to excess Fe or Mn, the FCR activity significantly decreased (p < 0.01). The FCR activity in GmVIT1 transgenic Arabidopsis was significantly lower than that in the wild-type plants under +Fe stress (p < 0.05), while significantly higher than that in the wild-type plants under +Mn stress (p < 0.05) (Figure 10). Although the FCR activity in GmVIT1 transgenic plants showed a downward trend, it reflected an opposite response to +Mn and +Fe stress.
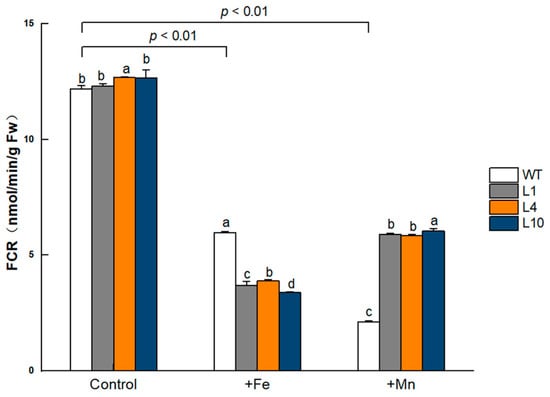
Figure 10.
FCR activity of GmVIT1 transgenic lines and wild-type plants under excess iron or excess manganese stress. Plants were grown in 1/2 Hoagland solution for 14 days and and treatedwith 0.1 mM Fe-EDTA + 0.1 mM MnCl2 (CK), 0.3 mM Fe-EDTA + 0.1 mM MnCl2 (+Fe), and 0.3 mM MnCl2 + 0.1 mM Fe-EDTA (+Mn) for 7 days. Experiments were repeated three times, and each group contained five samples. All the data represent the means of three replicates. Lowercase letters represent a statistically significant difference in GmVIT1 transgenic plants and the wild-type under the same condition, p < 0.05.
4. Discussion
Fe and Mn are redox-active metals and necessary micronutrients for all organisms. When the Fe or Mn concentrations are excessive, free radicals may hinder plant growth [55]. Then, the uptake, compartmentalization, and translocation of Fe and Mn must be strictly regulated in plants. Vacuolar iron transporters have been identified in model plants Arabidopsis, rice, and soybean [56,57]. GmVTL1a is an iron transporter on the symbiosome membrane with an essential function in nitrogen fixation [51], while other soybean VIT functions have not been well-claimed. Here, we cloned GmVIT1 and its promoter from the HN51 soybean and conducted a functional study.
4.1. GmVIT1 Is a Unique Gene in Soybean VIT Family
Most GmVIT genes are present in tandem, and over 61.9 percent of VIT genes are cluster distributed in chromosomes 5 and 8 [34], while GmVIT1 is alone on chromosome 20 in the soybean genome (Figure 2). We found that GmVIT1 has a low similarity with other GmVITs except for Glyma.10G225900. Notably, GmVIT1 shows over 90% similarity to GsVTL1 and GsVTL4 in Glycine soja. Therefore, we believe that GmVIT1 is a relatively primitive VIT gene of soybean from its ancestor Glycine soja. The phylogenetic and motif analysis strongly confirm this conclusion (Figure 4). GmVIT1 contains a conserved VIT domain and four transmembrane domains corresponding to the transport function (Figure S2). GmVIT1, GsVTL1, and GsVTL4 both contain motifs 4, 6, and 9, while other VITs do not contain motif 9 (Figure 4). We believed that GmVIT1 retained in the soybean genome with significance in function during the long-term evolution process. GmVIT1 was localized to the tonoplast, similar to most VIT proteins [40] (Figure 5). Together with structural and phylogenetic results, GmVIT1 is a unique gene and might perform vacuole metal transport function in the plant cell.
4.2. GmVIT1 Was Highly Expressed in Leaves and Induced by Fe/Mn Stress
Metal transporters vary in tissue expression patterns. AtVIT1, BnMEB2, OsVIT1, OsVIT2, TaVIT1, TaVIT2, and TgVIT1 were found to be expressed in mature leaves, flowers, and developing seeds and were involved in the development of propagative organs [28,31,35,36,40,45]. Cao et al. found that soybean VITs were expressed in vegetative and reproductive organs. Almost no expression of GmVTL1 and GmVTL2 was found in the root and leaves of soybean [40]. In this study, we found that GmVIT1 was expressed in the vegetative organs and showed the highest expression level in leaves (Figure 6), consistent with the results of Cao et al. [34]. The results indicated that GmVIT1 might play an important role in soybean leaves.
The transcriptomic analysis revealed that the expression of GmVIT1 was significantly higher in HN51 than that of SN37. The qRT-PCR results showed that iron deficiency and excess iron significantly up-regulated GmVIT1 in HN51, while not significantly in SN37. The results indicated that the GmVIT1 gene is directly related to the different responses to iron stress of HN51 and SN37. The expression of several VITs induced by Fe stress indicated that VITs are involved in the Fe storage of vacuolar to regulate plant Fe homeostasis. The transcript of AtVTL1, AtVTL2, AtVTL5, and BnMEB2 responded more to exogenous Fe [30,43,58]. Fe stress induced the expression of Glyma.16G168200, Glyma08G076300, and Glyma08G075900 in soybean [34]. In this study, GmVIT1 was induced in roots, stems, and leaves by +Fe treatment. Dramatically, the expression of GmVIT1 increased nearly to 9.7-fold in the leaves (Figure 6). The GUS staining verified this result. Therefore, we infer that the GmVIT1 gene may play a more important role in excess Fe response. The unique nature of the leaf makes it prone to the damage of iron overload. Iron is incorporated into iron proteins or transported to cellular organelles, and excess cytosolic iron is sequestered and stored. The up-regulating of VIT1 might help to detoxifying excess Fe [33]. The up-regulation of GmVIT1 in leaves suggests that, during iron excess, GmVIT1 activates vacuolar iron deposition, which appears to be the major mechanism of metal detoxification in soybean. The typical CCC1-like superfamily domains in VIT are also demonstrated for Mn transport from the cytosol to the vacuole [59]. A study has shown that accumulation of TaVTL2 and TaVTL4 in wheat shoots during Mn deprivation [60]. In our results, GmVIT1 was also induced by excessive Mn, suggesting that GmVIT1 might also be involved in detoxifying of excess Mn. Kurt et al. found that VITs were highly transcribed under cold, heat, salt, and drought perturbations [28]. We found abundant stress–response cis-elements present in the GmVIT1 promoter and speculated that GmVIT1 might be regulated by abiotic stress, which remains to be elucidated.
4.3. Overexpression of GmVIT1 Enhances the Tolerance to +Fe/+Mn Stress in Arabidopsis
Leaf chlorosis is one of the most remarkable phenotypes of plants exposed to Fe and Mn toxicity [56,61]. The wild-type Arabidopsis exhibits characteristic chlorosis in leaves under excess Fe in this study. In contrast, GmVIT1 transgenic Arabidopsis old leaves showed mild symptoms, and young leaves showed no symptoms, consistent with less reduction in chlorophyll content. Wheat VIT gene overexpression dramatically increases Fe content in the transgenic plant, resulting in altered partitioning of Fe between source and sink tissues [31]. Overexpression of VIT genes also restored plant growth in iron homeostasis genes mutants such as nramp3/nramp4, vit2, and meb2 [29,30]. These findings implied that VITs transferred cytoplasmic Fe into vacuoles and contribute to Fe homeostasis regulation in plants [44]. In agreement, we detected that GmVIT1 is mainly expressed in leaves and increased plant tolerance to excess Fe. GmVIT1 might involve in iron vacuolar storage and detoxification. In plant tissues, Fe2+ participates in Fenton reactions, catalyzing the generation of hydroxyl radicals (OH-) and other reactive oxygen species (ROS) [62]. The GmVIT1 transgenic plants showed lower MDA and protective enzyme activity, which indicates the ability of resistance to excess Fe enhanced (Figure 8). Previous studies showed that Mn toxicity functions on young leaves more than on old leaves [57]. In the present study, plants exhibited a mild leaf chlorotic phenotype and a better oxidation index under excess Mn. The results supported that the GmVIT1 gene may alleviate Mn toxicity in Arabidopsis. The structural similarity suggests a shared function in Mn-binding and transport of MTP, NRAMP, and VITs [15,57]. It has been discussed previously that VIT1 sequestrates Mn to the vacuole. Expression of Arabidopsis VIT1 also increases the Mn content of yeast cells, but knockout of AtVIT1 does not alter the Mn distribution [35]. OsVIT1 and OsVIT2 may also contribute to sequestrate Mn [40]. TaVIT2 complemented an Mn transporter mutant Dpmr1 yeast and increased barley grain Mn content [31]. Our finding in GmVIT1 indicated that VITs might act as transporters for Mn. Similar to the previous results, GmVIT1 transgenic Arabidopsis improved tolerance to excess Mn. In contrast, some different conclusions were given. The Mn concentration decreased significantly in Osvit2 [29], but a significant change in Mn was observed in brown rice of Osvit1-1 and Osvit2-1 [40]. In our future work, analysis on Fe and Mn contents in GmVIT1 transgenic Arabidopsis under control and stress conditions would demonstrate GmVIT1 gene function. FCR is mainly involved in the catalytic reduction of Fe3+ to Fe2+. The FCR activity was relatively low under sufficient or excess Fe conditions [63]. In this study, the FCR activity of transgenic plants was significantly lower than that of wild-type plants under excess Fe stress, demonstrating that GmVIT1 might reduce the FCR activity (Figure 10). Such a mechanism could be significant and allow plants to adapt to high-iron environments. Metal transporters located in the tonoplast impact on long-distance transport in addition to intracellular metal homeostasis [31]. Wheat VIT1 response to nicotianamine is important for the distribution of Fe [64]. Studies have shown that AtVTL2 expression is regulated by ILR3 [65]. Although GmVIT1 plays a vital role in excess Fe and excess Mn detoxification, the precise molecular mechanism remains to be resolved.
Taken together, the results demonstrated GmVIT1 localized in tonoplast as a vacuole iron transporter family member. GmVIT1 is highly expressed in leaves, and also expressed in stems and roots of soybean. GmVIT1 is induced by iron deficiency and excess Fe and Mn. Overexpression of GmVIT1 confers Arabidopsis tolerance to excess Fe or excess Mn. This study provides new insight into VIT gene fouction and GmVIT1 could be applied in metal toxic tolerance improvement of crops.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy13020384/s1, Figure S1: Seclection of GmVIT1 trangenic Arabidopsis; Figure S2. Transmembrane domains of GmVIT1; Table S1: The primer sequence.
Author Contributions
Y.-D.B. and C.-H.G. designed the experiments and obtained funding for the research. T.L., L.W., L.S. and J.-L.G. performed the experimental analyses. D.-L.G. and T.L. contributed to compiling and analyzing the data and wrote the manuscript. Y.-J.S. conducted statistical analysis. Y.-C.L. and X.-M.Z. participated in the data analysis and supervised the writing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This experiment was funded by the Key Research and Development Project of Heilongjiang Province (No.2022ZX02B05), the National Natural Science Foundation of China (No. U21a20182, 31972507), National Natural Science Foundation of China general Project (No. 31771823).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Andresen, E.; Peiter, E.; Küpper, H. Trace metal metabolism in plants. J. Exp. Bot. 2018, 69, 909–954. [Google Scholar] [CrossRef] [PubMed]
- Nsch, H.R.; Mendel, R.R. Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr. Opin. Plant. Biol. 2009, 12, 259–266. [Google Scholar] [CrossRef]
- Ramirez, L.; Simontacchi, M.; Murgia, I.; Zabaleta, E.; Lamattina, L. Nitric oxide, nitrosyl iron complexes, ferritin and frataxin: A well equipped team to preserve plant iron homeostasis. Plant Sci. 2001, 181, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Nishizawa, N.K. Iron uptake translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 2012, 63, 131–152. [Google Scholar] [CrossRef]
- Briat, J.F.; Ravet, K.; Arnaud, N.; Duc, C.; Boucherez, J.; Touraine, B.; Cellier, F.; Gaymard, F. New insights into ferritin synthesis and function highlight a link between iron homeostasis and oxidative stress in plants. Ann. Bot. 2010, 105, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Dutta, T.; Neelapu, N.; Surekha, C. Iron, Zinc, and Copper Application in Overcoming Environmental Stress. In Protective Chemical Agents in the Amelioration of Plant Abiotic Stress, 1st ed.; Aryadeep, R., Durgesh, K.T., Eds.; Wiley: Noida, India, 2020; Volume 29, pp. 582–596. [Google Scholar]
- Pereira, E.G.; Oliva, M.A.; Rosado-Souza, L.; Mendes, G.C.; Colares, D.S.; Stopato, C.H.; Almeida, A.M. Iron excess affects rice photosynthesis through stomatal and non-stomatal limitations. Plant Sci. 2013, 202, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Becker, M.; Asch, F. Iron toxicity in rice-Conditions and management concepts. J. Plant Nutr. Soil Sci. 2005, 168, 558–573. [Google Scholar] [CrossRef]
- Khabaz, S.H.; Rengel, Z.; Wilson, R.; Setter, T.L. Variation for tolerance to high concentration of ferrous iron (Fe2+) in Australian hexaploid wheat. Euphytica 2010, 172, 275–283. [Google Scholar] [CrossRef]
- Cho, U.H.; Park, J.O. Mercury-induced oxidative stress in tomato seedlings. Plant Sci. 2000, 156, 1–9. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B.; Palma, J.M.; Rodriguez-Ruiz, M. Plant peroxisomes: A nitro-oxidative cocktail. Redox Biol. 2017, 11, 535–542. [Google Scholar] [CrossRef]
- Li, P.; Song, A.L.; Li, Z.J.; Fan, F.L.; Liang, Y.C. Silicon ameliorates manganese toxicity by regulating manganese transport and antioxidant reactions in rice (Oryza sativa L.). Plant Soil 2012, 354, 407–419. [Google Scholar] [CrossRef]
- Ribera, A.E.; Reyes-Díaz, M.M.; Alberdi, M.R.; Alvarez-Cortez, D.A.; Rengel, Z.; Mora, M.D.L.L. Photosynthetic impairment caused by manganese toxicity and associated antioxidative responses in perennial ryegrass. Crop Pasture Sci. 2013, 64, 696–707. [Google Scholar] [CrossRef]
- Santos, E.F.; Santini, K.; Paixão, A.P.; Júnior, E.F.; Lavres, J.; Campos, M.; dos Reis, A.R. Physiological highlights of manganese toxicity symptoms in soybean plants: Mn toxicity responses. Plant Physiol. Biochem. 2017, 56, 1119–1136. [Google Scholar] [CrossRef] [PubMed]
- Alejandro, S.; Höller, S.; Meier, B.; Peiter, E. Manganese in Plants: From Acquisition to Subcellular Allocation. Front. Plant Sci. 2020, 11, 300–323. [Google Scholar] [CrossRef] [PubMed]
- Lovyagina, E.R.; Semin, B.K. Competitive interaction of Mn(II) and Fe(II) cations with the high-affinity Mn-binding site of the photosystem II: Evolutionary aspect. Orig. Life Evol. Biosph. 2022, 52, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.; Mishra, A.K.; Biswal, U.C. Manganese induced peroxidation of thylakoid lipids and changes in chlorophyll-α flfluorescence during aging of cell free chloroplasts in light. Phytochemistry 1987, 26, 3217–3219. [Google Scholar] [CrossRef]
- Hauck, M.; Paul, A.; Gross, S.; Raubuch, M. Manganese toxicity in epiphytic lichens: Chlorophyll degradation and interaction with iron and phosphorus. Environ. Exp. Bot. 2003, 49, 181–191. [Google Scholar] [CrossRef]
- Peng, J.S.; Gong, J.M. Vacuolar sequestration capacity and long-distance metal transport in plants. Front. Plant Sci. 2014, 5, 19–32. [Google Scholar] [CrossRef]
- Agorio, A.; Giraudat, J.; Bianchi, M.W.; Marion, J.; Espagne, C.; Castaings, L.; Lelièvre, F.; Curie, C.; Thomine, S.; Merlot, S. Phosphatidylinositol 3-phosphate-binding protein AtPH1 controls the localization of the metal transporter NRAMP1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 3354–3363. [Google Scholar] [CrossRef]
- Lanquar, V.; Ramos, M.S.; Lelievre, F.; Barbier-Brygoo, H.; Krieger-Liszkay, A.; Kraemer, U.; Thomine, S. Export of vacuolar manganese by AtNRAMP3 and AtNRAMP4 is required for optimal photosynthesis and growth under manganese deficiency. Plant Physiol. 2010, 152, 1986–1999. [Google Scholar] [CrossRef]
- Chang, J.-D.; Huang, S.; Yamaji, N.; Zhang, W.; Ma, J.F.; Zhao, F. OsNRAMP1 transporter contributes to cadmium and manganese uptake in rice. Plant Cell Environ. 2020, 43, 2476–2491. [Google Scholar] [CrossRef] [PubMed]
- Eroglu, S.; Giehl, R.F.H.; Meier, B.; Takahashi, M.; Terada, Y.; Ignatiev, K.; Andresen, E.; Kupper, H.; Peiter, E.; Wiren, N.V. Metal Tolerance Protein 8 Mediates Manganese Homeostasis and Iron Reallocation during Seed Development and Germination. Plant Physiol. 2017, 174, 1633–1647. [Google Scholar] [CrossRef]
- Eckhardt, U.; Marques, A.; Buckhout, T.J. Two iron-regulated cation transporters from tomato complement metal uptake-defificient yeast mutants. Plant Mol. Biol. 2001, 45, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Song, W.Y.; Park, J.; Mendoza-Cozatl, D.J.; Suter-Grotemeyer, M.; Shim, D.; Hortensteiner, S.; Geisler, M.; Weder, B.; Rea, P.A.; Rentsch, D.; et al. Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc. Natl. Acad. Sci. USA 2010, 107, 21187–21192. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, O.S.; Ward, D.M.; Kaplan, J. Ccc1 is a transporter that mediates vacuolar iron storage in yeast. J. Biol. Chem. 2021, 31, 29515–29519. [Google Scholar] [CrossRef] [PubMed]
- Sorribes-Dauden, R.; Peris, D.; Martínez-Pastor, M.T.; Puig, S. Structure and function of the vacuolar Ccc1/VIT1 family of iron transporters and its regulation in fungi. Comput. Struct. Biotechnol. J. 2020, 18, 3712–3722. [Google Scholar] [CrossRef] [PubMed]
- Kurt, F.; Filiz, E. Subcellular iron transport genes in Arabidopsis thaliana: Insights into iron homeostasis. J. BioSci. Biotechnol. 2020, 9, 1–10. [Google Scholar]
- Bashir, K.; Takahashi, R.; Akhtar, S.; Ishimaru, Y.; Nakanishi, H.; Nishizawa, N.K. The knockdown of OsVIT2 and MIT affects iron localization in rice seed. Rice 2013, 6, 31–38. [Google Scholar] [CrossRef]
- Zhu, W.; Zuo, R.; Zhou, R.; Huang, J.; Tang, M.; Cheng, X.; Liu, Y.; Tong, C.; Xiang, Y.; Dong, C.; et al. Vacuolar Iron Transporter BnMEB2 Is Involved in Enhancing Iron Tolerance of Brassica napus. Front. Plant Sci. 2016, 13, 1353–1366. [Google Scholar] [CrossRef]
- Connorton, J.M.; Jones, E.R.; Rodríguez-Ramiro, I.; Fairweather-Tait, S.; Uauy, C.; Balk, J. Wheat Vacuolar Iron Transporter TaVIT2 Transports Fe and Mn and Is Effective for Biofortification. Plant Physiol. 2017, 174, 2434–2444. [Google Scholar] [CrossRef]
- Borg, S.; Brinchpedersen, H.; Tauris, B.; Holm, P.B. Iron Transport, Deposition and Bioavailability in the Wheat and Barley Grain. Plant Soil 2009, 325, 15–24. [Google Scholar] [CrossRef]
- Slavic, K.; Krishna, S.; Lahree, A.; Bouyer, G.; Hanson, K.K.; Vera, I.; Pittman, J.K.; Staines, H.M.; Mota, M.M. A vacuolar iron-transporter homologue acts as a detoxifier in Plasmodium. Nat. Commun. 2016, 7, 10403–10413. [Google Scholar] [CrossRef] [PubMed]
- Cao, J. Molecular Evolution of the Vacuolar Iron Transporter (VIT) Family Genes in 14 Plant Species. Genes 2019, 10, 144. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.A.; Punshon, T.; Lanzirotti, A.; Li, L.T.; Alonso, J.M.; Ecker, J.R.; Kaplan, J.; Guerinot, M.L. Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 2006, 314, 1295–1298. [Google Scholar] [CrossRef] [PubMed]
- Jeet, R.; Singh, S.P.; Tiwari, S.; Pathak, P. Wheat TaVIT2D restores phenotype and mediates iron homeostasis during growth of Arabidopsis thaliana in iron-deficient conditions. Plant Physiol. Rep. 2019, 1, 24–34. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Y.H.; Yi, H.Y.; Gong, J.-M. Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 2012, 72, 400–410. [Google Scholar] [CrossRef]
- Ram, H.; Sardar, S.; Gandass, N. Vacuolar Iron Transporter (Like) proteins: Regulators of cellular iron accumulation in plants. Physiol. Plant. 2021, 171, 823–832. [Google Scholar] [CrossRef]
- Labarbuta, P.; Duckett, K.; Botting, C.H.; Chahrour, O.; Malone, J.; Dalton, J.P.; Law, C.J. Recombinant vacuolar iron transporter family homologue PfVIT from human malaria-causing Plasmodium falciparum is a Fe2+/H+ exchanger. Sci. Rep. 2017, 7, 42850. [Google Scholar] [CrossRef]
- Liu, S.; Liao, L.L.; Nie, M.M.; Peng, W.T.; Zhang, M.S.; Lei, J.N.; Zhong, Y.J.; Liao, H.; Chen, Z.C. A VIT-like transporter facilitates iron transport into nodule symbiosomes for nitrogen fixation in soybean. New Phytol. 2020, 226, 1413–1428. [Google Scholar] [CrossRef]
- Li, L.; Ward, D.M. Iron toxicity in yeast: Transcriptional regulation of the vacuolar iron importer CCC1. Curr. Genet. 2018, 64, 413–416. [Google Scholar] [CrossRef]
- Eroglu, S.; Karaca, N.; Vogel-Mikus, K.; Kavčič, A.; Filiz, E.; Tanyolac, B. The Conservation of VIT1-Dependent Iron Distribution in Seeds. Front. Plant Sci. 2019, 10, 907–921. [Google Scholar] [CrossRef] [PubMed]
- Gollhofer, J.; Timofeev, R.; Lan, P.; Schmidt, W.; Buckhout, T.J. Vacuolar-Iron-Transporter1-Like proteins mediate iron homeostasis in Arabidopsis. PLoS ONE 2014, 9, e110468. [Google Scholar] [CrossRef] [PubMed]
- Che, J.; Yamaji, N.; Ma, J.F. Role of a vacuolar iron transporter OsVIT2 in the distribution of iron to rice grains. New Phytol. 2021, 230, 1049–1062. [Google Scholar] [CrossRef]
- Momonoi, K.; Yoshida, K.; Mano, S.; Takahashi, H.; Nakamori, C.; Shoji, K.; Nitta, A.; Nishimura, M. A vacuolar iron transporter in tulip, TgVit1, is responsible for blue coloration in petal cells through iron accumulation. Plant J. 2009, 59, 437–447. [Google Scholar] [CrossRef]
- Yoshida, K.; Negishi, T. The identification of a vacuolar iron transporter involved in the blue coloration of cornflower petals. Phytochemistry 2013, 94, 60–67. [Google Scholar] [CrossRef]
- Narayanan, N.; Beyene, G.; Chauhan, R.D.; Gaitán-Solis, E.; Grusak, M.A.; Taylor, N.; Anderson, P. Overexpression of Arabidopsis VIT1 increases accumulation of iron in cassava roots and stems. Plant Sci. 2015, 240, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Andrew, C.S.; Hegarty, M.P. Comparative responses to manganese excess of eight tropical and four temperate pasture legume species. Aust. J. Agric. Res. 1969, 20, 687–696. [Google Scholar] [CrossRef]
- Chen, Z.; Sun, L.; Liu, P.; Liu, G.; Tian, J.; Liao, H. Malate synthesis and secretion mediated by a manganese-enhanced malate dehydrogenase confers superior manganese tolerance in Stylosanthes guianensis. Plant Physiol. 2015, 167, 176–188. [Google Scholar] [CrossRef]
- Chen, Z.; Yan, W.; Sun, L.; Tian, J.; Liao, H. Proteomic analysis reveals growth inhibition of soybean roots by manganese toxicity is associated with alteration of cell wall structure and lignification. J. Proteom. 2016, 143, 151–160. [Google Scholar] [CrossRef]
- Brear, E.M.; Bedon, F.; Gavrin, A.; Kryvoruchko, I.S.; Torres-Jerez, I.; Udvardi, M.K.; Day, D.A.; Smith, P.M.C. GmVTL1a is an iron transporter on the symbiosome membrane of soybean with an important role in nitrogen fixation. New Phytol. 2020, 228, 667–681. [Google Scholar] [CrossRef]
- Rajinder, S.; Dhindsa, P.L.; Plumb, D.; David, M. Leaf senescence and lipid peroxidation: Effects of some phytohormones, and scavengers of free radicals and singlet oxygen. Physiol. Plant. 1982, 56, 453–457. [Google Scholar]
- Jun, S.E.; Okushima, Y.; Nam, J.; Umeda, M.; Kim, G.T. Kip-Related Protein 3 Is Required for Control of Endoreduplication in the Shoot Apical Meristem and Leaves of Arabidopsis. Mol. Cells 2013, 35, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Manara, A. Plant Responses to Heavy Metal Toxicity. In Plants and Heavy Metals, 1st ed.; Antonella, F., Ed.; Springer: Dordrecht, The Netherlands, 2012; Volume 3, pp. 27–53. [Google Scholar]
- Aung, M.S.; Masuda, H. How Does Rice Defend Against Excess Iron?: Physiological and Molecular Mechanisms. Front. Plant Sci. 2020, 11, 1102–1110. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xue, Y.; Xie, B.; Zhu, S.; Lu, X.; Liang, C.; Tian, J. Complex gene regulation between young and old soybean leaves in responses to manganese toxicity. Plant Physiol. Biochem. 2020, 155, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Buckhout, T.J.; Yang, T.J.; Schmidt, W. Early iron-deficiency-induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genom. 2009, 10, 147–148. [Google Scholar] [CrossRef] [PubMed]
- Lapinskas, P.J.; Lin, S.J.; Culotta, V.C. The role of the Saccharomyces cerevisiae CCC1 gene in the homeostasis of manganese ions. Mol. Microbiol. 1996, 21, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kaur, G.; Kumar, A.; Meena, V.; Ram, H.; Kaur, J.; Pandey, A.K. Gene Expression Pattern of Vacuolar-Iron Transporter-Like (VTL) Genes in Hexaploid Wheat during Metal Stress. Plants 2020, 9, 229. [Google Scholar] [CrossRef]
- Kisku, G.C.; Kumar, V.; Sahu, P. An Over View of Metal Toxicity in Agricultural Soil and Plants. In Metallic Contamination and Its Toxicity, 1st ed.; Gautam, A., Pathak, C., Eds.; Daya Publishing House: New Delhi, India, 2020; Volume 12, pp. 2451–2456. [Google Scholar]
- Becana, M.; Moran, J.F.; Iturbe-Ormaetxe, I. Iron-dependent oxygen free radical generation in plants subjected to environmental stress: Toxicity and antioxidant protection. Plant Soil 1998, 201, 137–147. [Google Scholar] [CrossRef]
- Durrett, T.P.; Gassmann, W.; Rogers, E.E. The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 2007, 144, 197–205. [Google Scholar] [CrossRef]
- Harrington, S.A.; Connorton, J.M.; Nyangoma, N.I.M.; McNelly, R.; Morgan, Y.M.L.; Aslam, M.F.; Sharp, P.A.; Johnson, A.A.T.; Uauy, C.; Balk, J. A two-gene strategy increases iron and zinc concentrations in wheat flour, improving mineral bioaccessibility. Plant Physiol. 2023, 191, 528–541. [Google Scholar] [CrossRef] [PubMed]
- Tissot, N.; Robe, K.; Gao, F.; Grant-Grant, S.; Boucherez, J.; Bellegarde, F.; Maghiaoui, A.; Marcelin, R.; Izquierdo, E.; Benhamed, M.; et al. Transcriptional integration of the responses to iron availability in Arabidopsis by the bHLH factor ILR3. New Phytol. 2019, 223, 1433–1446. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).