Genetic and Morphological Variation of Belgian Cyperus esculentus L. Clonal Populations and Their Significance for Integrated Management
Abstract
1. Introduction
2. Materials and Methods
2.1. Genetic Clustering of Belgian C. esculentus Clonal Populations Using the Amplified Fragment Length Polymorphism Analysis (AFLP)
2.2. Morphological Clustering of Belgian C. esculentus Clones
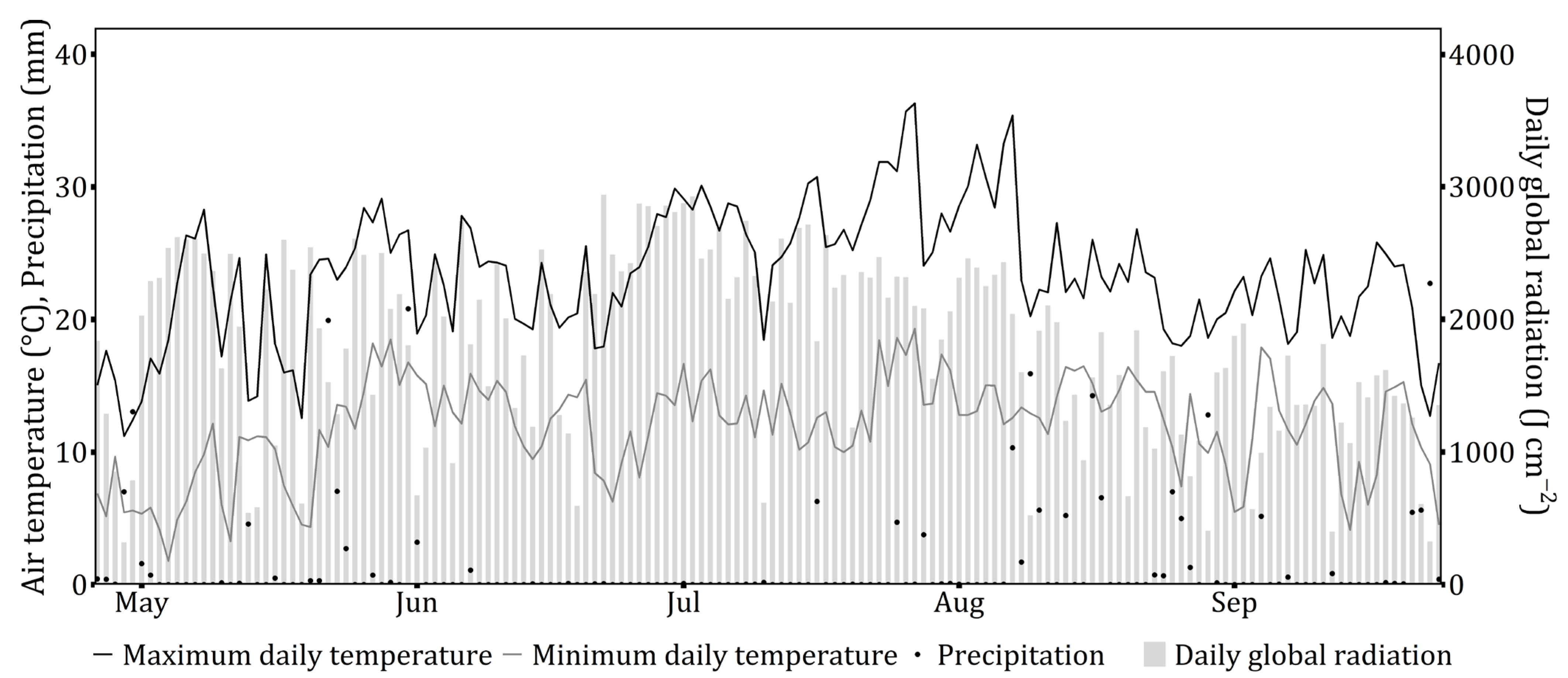
2.3. Data and Statistical Analysis
3. Results
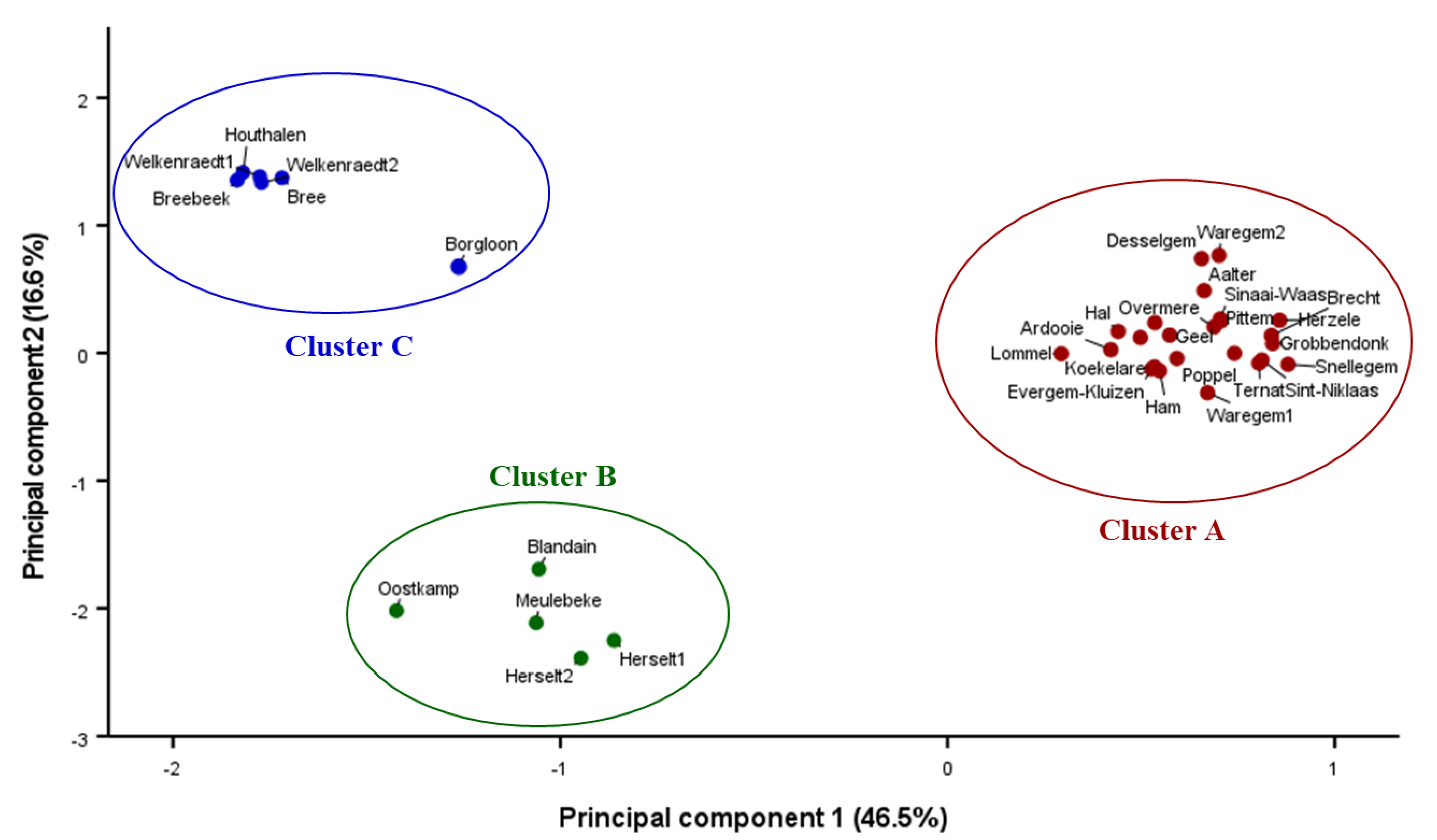
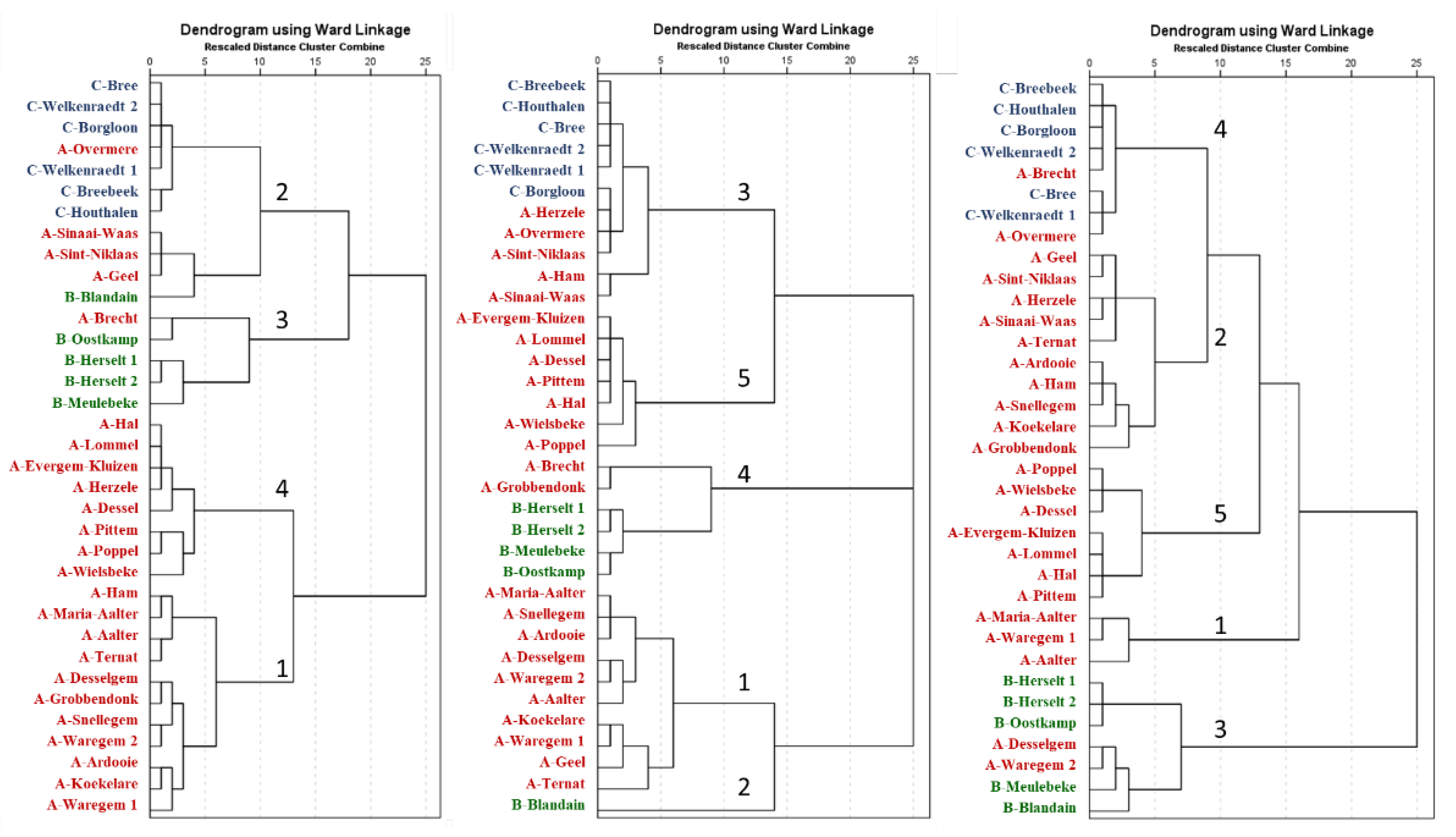
3.1. Genetic Clustering
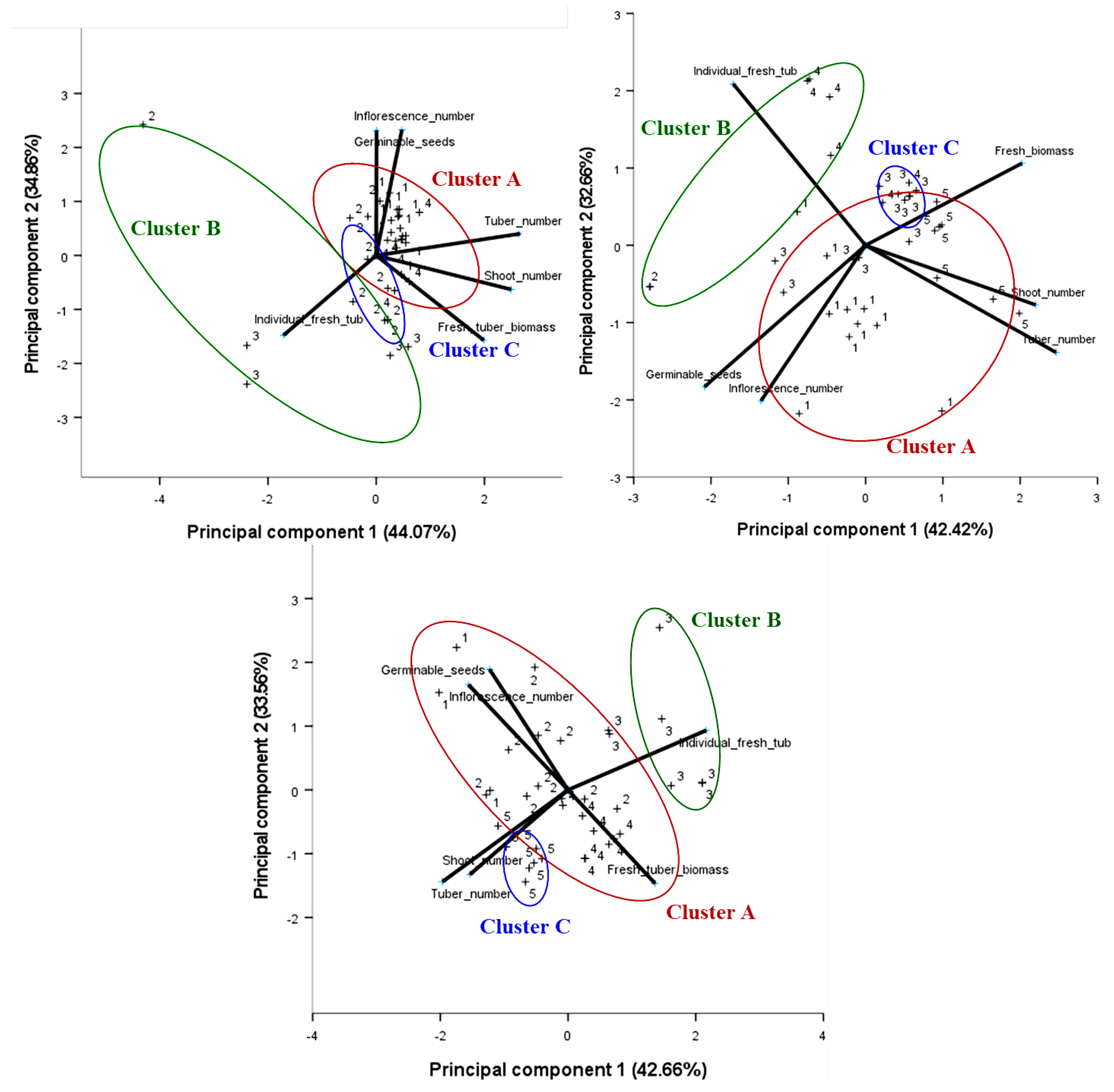
3.2. Morphological Clustering
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Holm, L.R.G.; Plucknett, D.L.; Pancho, J.V.; Herberger, J.P. The World’s Worst Weeds: Distribution and Biology; Krieger Publishing Company: Malabar, FL, USA, 1991; ISBN 978-0-89464-415-3. [Google Scholar]
- Larridon, I.; Reynders, M.; Huygh, W.; Bauters, K.; Van De Putte, K.; Muasya, A.M.; Boeckx, P.; Simpson, D.A.; Vrijdaghs, A.; Goetghebeur, P. Affinities in C3 Cyperus lineages (Cyperaceae) revealed using molecular phylogenetic data and carbon isotope analysis. Bot. J. Linn. Soc. 2011, 167, 19–46. [Google Scholar] [CrossRef]
- Boeckeler, J. Die Cyperaceen des Königlichen herbariums zu Berlin. Linnaea 1870, 36, 287–291. [Google Scholar]
- Ascherson, P.F.A.; Graebner, K.O.R.P.P. Synopsis der Mitteleuropäischen Flora; Verlag von Wilhelm Engelmann: Leipzig, Germany, 1902. [Google Scholar]
- Kükenthal, G. Cyperaceae−Scirpoideae-Cypereae. In Das Pflanzenreich; Engler, A., Diels, L., Eds.; Verlag Wilhelm Engelmann: Leipzig, Germany, 1936; Volume 4. [Google Scholar]
- Schippers, P.; Ter Borg, S.J.; Bos, J.J. A revision of the infraspecific taxonomy of Cyperus esculentus (yellow nutsedge) with an experimentally evaluated character Set. Syst. Bot. 1995, 20, 461. [Google Scholar] [CrossRef]
- De Castro, O.; Gargiulo, R.; Del Guacchio, E.; Caputo, P.; De Luca, P. A molecular survey concerning the origin of Cyperus esculentus (Cyperaceae, Poales): Two sides of the same coin (Weed vs. Crop). Ann. Bot. 2015, 115, 733–745. [Google Scholar] [CrossRef] [PubMed]
- WCSP World Checklist of Selected Plant Families: Royal Botanic Gardens, Kew. Available online: https://wcsp.science.kew.org/prepareChecklist.do?checklist=selected_families%40%40337031220211053975 (accessed on 3 December 2021).
- Tal, A.; Rubin, B. Cyperus esculentus L.—A new weed in Israel. Phytoparasitica 2005, 33, 245–246. [Google Scholar] [CrossRef]
- Follak, S.; Belz, R.; Bohren, C.; De Castro, O.; Del Guacchio, E.; Pascual-Seva, N.; Schwarz, M.; Verloove, F.; Essl, F. Biological flora of Central Europe: Cyperus esculentus L. Perspect. Plant Ecol. Evol. Syst. 2016, 23, 33–51. [Google Scholar] [CrossRef]
- GBIF Global Biodiversity Information Facility: Cyperus esculentus Occurrences. Available online: https://www.gbif.org/occurrence/map?country=BE&taxon_key=2716226 (accessed on 7 January 2022).
- StatBel Kerncijfers Landbouw: Belgische Landbouw in Cijfers: 2021. Available online: https://statbel.fgov.be/nl/nieuws/kerncijfers-landbouw-2021 (accessed on 15 February 2023).
- Sterling, T.M.; Thompson, D.C.; Abbott, L.B. Implications of invasive plant variation for weed management. Weed Technol. 2004, 18, 1319–1324. [Google Scholar] [CrossRef]
- Goolsby, J.A.; De Barro, P.J.; Makinson, J.R.; Pemberton, R.W.; Hartley, D.M.; Frohlich, D.R. Matching the origin of an invasive weed for selection of a herbivore haplotype for a biological control programme. Mol. Ecol. 2005, 15, 287–297. [Google Scholar] [CrossRef]
- Bodo Slotta, T.A. What we know about weeds: Insights from genetic markers. Weed Sci. 2008, 56, 322–326. [Google Scholar] [CrossRef]
- Baker, J.; Hidayat, I.; Preston, C. Molecular tools for understanding distribution and spread of weed genotypes. Crop Prot. 2007, 26, 198–205. [Google Scholar] [CrossRef]
- Ter Borg, S.J.; De Nijs, L.J.; Van Oene, H. Intraspecific Variation of Cyperus esculentus L. in The Netherlands: A Preliminary Report. In 8éme Colloque International des Mauvaises Herbes; Association National pour La Protection des Plantes: Dijon, France, 1988; pp. 181–185. [Google Scholar]
- Li, B.; Shibuya, T.; Yogo, Y.; Hara, T.; Yokozawa, M. Interclonal differences, plasticity and trade-offs of life history traits of Cyperus esculentus in relation to water availability. Plant Species Biol. 2001, 16, 193–207. [Google Scholar] [CrossRef]
- De Cauwer, B.; De Ryck, S.; Claerhout, S.; Biesemans, N.; Reheul, D. Differences in growth and herbicide sensitivity among Cyperus esculentus clones found in Belgian maize fields. Weed Res. 2017, 57, 234–246. [Google Scholar] [CrossRef]
- Horak, M.J.; Holt, J.S.; Ellstrand, N.C. Genetic variation in yellow nutsedge (Cyperus esculentus). Weed Sci. 1987, 35, 506–512. [Google Scholar] [CrossRef]
- Holt, J.S. Genetic variation in life history traits in yellow nutsedge (Cyperus esculentus) from California. Weed Sci. 1994, 42, 378–384. [Google Scholar] [CrossRef]
- Dodet, M.; Petit, R.J.; Gasquez, J. Local spread of the invasive Cyperus esculentus (Cyperaceae) inferred using molecular genetic markers. Weed Res. 2008, 48, 19–27. [Google Scholar] [CrossRef]
- Mulligan, G.A.; Junkins, B.E. Biology of Canadian weeds 17 Cyperus esculentus L. Can. J. Plant Sci. 1976, 56, 339–350. [Google Scholar] [CrossRef]
- De Ryck, S.; Reheul, D.; De Cauwer, B. Impacts of herbicide sequences and vertical tuber distribution on the chemical control of yellow nutsedge (Cyperus esculentus L.). Weed Res. 2021, 61, 454–464. [Google Scholar] [CrossRef]
- DOV Soil Map of Belgium. Available online: https://dov.vlaanderen.be/ (accessed on 5 December 2021).
- Walonmap Géoportail de La Wallonie, Le Site de l’Information Géographique Wallonne! Available online: http://geoportail.wallonie.be/home.html (accessed on 5 December 2021).
- Machery-Nagel Genomic DNA from Plant. Rev. 09 2014. Available online: https://www.takarabio.com/documents/User%20Manual/NucleoSpin%20Plant%20II%20Genomic%20DNA%20Purification%20User%20Manual%20%28PT5028/NucleoSpin%20Plant%20II%20Genomic%20DNA%20Purification%20User%20Manual%20%28PT5028-1%29_Rev_09.pdf (accessed on 15 February 2023).
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; Vandelee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M.; et al. AFLP—A new technique for DNA-fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef]
- De Riek, J.; Dendauw, J.; Mertens, M.; De Loose, M.; Heursel, J.; Van Bockstaele, E. Validation of criteria for the selection of AFLP markers to assess the genetic variation of a breeders’ collection of evergreen azaleas. Theor. Appl. Genet. 1999, 99, 1155–1165. [Google Scholar] [CrossRef]
- Elmer, P. AFLP Plant Mapping Kit Protocol (P/N 402083); Applied Biosystems: Foster City, CA, USA, 1995. [Google Scholar]
- De Riek, J.; Calsyn, E.; Everaert, I.; Van Bockstaele, E.; De Loose, M. AFLP based alternatives for the assessment of distinctness, uniformity and stability of sugar beet varieties. Theor. Appl. Genet. 2001, 103, 1254–1265. [Google Scholar] [CrossRef]
- Vekemans, X. AFLP-SURV, Version 1.0; Laboratoire de Génétique et Ecologie Végétale, Université Libre de Bruxelles: Brussels, Belgium, 2002.
- Horak, M.J.; Holt, J.S. Isozyme variability and breeding systems in populations of yellow nutsedge (Cyperus esculentus). Weed Sci. 1986, 34, 538–543. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; ISBN 3-900051-07-0. [Google Scholar]
- Ward, J.H., Jr. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Tayyar, R.I.; Nguyen, J.H.T.; Holt, J.S. Genetic and morphological analysis of two novel nutsedge biotypes from California. Weed Sci. 2003, 51, 731–739. [Google Scholar] [CrossRef]
- Keller, M.; Krauss, J.; Total, R.; Neuweiler, R. Efficacy of herbicides against yellow nutsedge (Cyperus esculentus) plants originating from seeds. Agroscope 2020, 464, 116–120. [Google Scholar] [CrossRef]
- Okoli, C.A.N.; Shilling, D.G.; Smith, R.L.; Bewick, T.A. Genetic diversity in purple nutsedge (Cyperus rotundus L.) and yellow nutsedge (Cyperus esculentus L.). Biol. Control 1997, 8, 111–118. [Google Scholar] [CrossRef]
- Mueller, M.H.; Valk, A.G. The potential role of ducks in wetland seed dispersal. Wetlands 2002, 22, 170–178. [Google Scholar] [CrossRef]
- Wang, S.; Hou, F.J. Seed bank of livestock dung in the Qilian mountain grassland: A potential resource for vegetation recovery. Rangel. Ecol. Manag. 2021, 78, 90–99. [Google Scholar] [CrossRef]
- Stoller, E.W.; Wax, L.M. Yellow nutsedge shoot emergence and tuber longevity. Weed Sci. 1973, 21, 76–81. [Google Scholar] [CrossRef]
- Verloove, F. Species List | Manual of the Alien Plants of Belgium, Cyperus esculentus. Available online: http://alienplantsbelgium.be/species-list/cyperus-esculentus (accessed on 24 June 2020).
- Follak, S.; Aldrian, U.; Moser, D.; Essl, F. Reconstructing the invasion of Cyperus esculentus in Central Europe. Weed Res. 2015, 55, 289–297. [Google Scholar] [CrossRef]
 for cluster A, green
for cluster A, green  for cluster B, and blue
for cluster B, and blue  for cluster C; see Figure 3). See Table 1 for GPS-coordinates and pedohydrological conditions.
for cluster C; see Figure 3). See Table 1 for GPS-coordinates and pedohydrological conditions.
 for cluster A, green
for cluster A, green  for cluster B, and blue
for cluster B, and blue  for cluster C; see Figure 3). See Table 1 for GPS-coordinates and pedohydrological conditions.
for cluster C; see Figure 3). See Table 1 for GPS-coordinates and pedohydrological conditions.


| Origin | Province | Latitude | Longitude | Soil Texture | Drainage Class * |
|---|---|---|---|---|---|
| Aalter | East Flanders | 51°04′55.90″ N | 3°24′25.10″ E | Loamy sand | d |
| Ardooie | West Flanders | 50°58′25.21″ N | 3°13′53.39″ E | Sandy loam | d |
| Blandain | Hainaut | 50°37′53.60″ N | 3°18′34.80″ E | Loam | b |
| Borgloon | Limburg | 50°49′33.20″ N | 5°23′47.30″ E | Loam | b |
| Brecht | Antwerp | 51°21′30.50″ N | 4°38′48.20″ E | Loamy sand | d |
| Bree | Limburg | 51°10′02.70″ N | 5°38′20.20″ E | Sand | e |
| Breebeek | Limburg | 51°09′26.80″ N | 5°36′29.50″ E | Loamy sand | d |
| Dessel | Antwerp | 51°14′38.30″ N | 5°07′48.40″ E | Sand | d |
| Desselgem | West Flanders | 50°51′56.94″ N | 3°23′22.99″ E | Loamy sand | d |
| Evergem-Kluizen | East Flanders | 51°09′42.30″ N | 3°42′49.50″ E | Loamy sand | d |
| Geel | Antwerp | 51°10′45.50″ N | 4°55′30.80″ E | Sand | c |
| Grobbendonk | Antwerp | 51°09′19.00″ N | 4°44′04.70″ E | Loamy sand | c |
| Hal | Antwerp | 51°26′06.50″ N | 4°46′45.90″ E | Sand | c |
| Ham | Limburg | 51°06′08.70″ N | 5°10′17.00″ E | Sand | c |
| Herselt 1 | Antwerp | 51°04′26.20″ N | 4°53′37.00″ E | Sand | c |
| Herselt 2 | Antwerp | 51°03′59.20″ N | 4°53′28.60″ E | Loamy sand | d |
| Herzele | East Flanders | 50°49′45.80″ N | 3°54′49.50″ E | Loam | b |
| Houthalen | Limburg | 51°02′03.30″ N | 5°23′47.70″ E | Sand | c |
| Koekelare | West Flanders | 51°04′34.50″ N | 2°59′46.87″ E | Loamy sand | d |
| Lommel | Limburg | 51°15′21.20″ N | 5°24′10.20″ E | Sand | c |
| Maria-Aalter | East Flanders | 51°04′54.60″ N | 3°26′26.90″ E | Sand | d |
| Meulebeke | West Flanders | 50°58′09.70″ N | 3°19′13.44″ E | Sandy loam | d |
| Oostkamp | West Flanders | 51°06′46.92″ N | 3°14′41.38″ E | Sand | c |
| Overmere | East Flanders | 51°02′08.80″ N | 3°56′38.60″ E | Sand | c |
| Pittem | West Flanders | 51°00′00.80″ N | 3°13′24.60″ E | Sandy loam | d |
| Poppel | Antwerp | 51°26′24.20″ N | 5°01′18.70″ E | Sand | d |
| Sinaai-Waas | East Flanders | 51°09′06.10″ N | 4°00′10.20″ E | Sand | b |
| Sint-Niklaas | East Flanders | 51°12′07.90″ N | 4°11′04.40″ E | Loamy sand | b |
| Snellegem | West Flanders | 51°09′49.93″ N | 3°07′41.31″ E | Sand | b |
| Ternat | Flemish-Brabant | 50°51′22.60″ N | 4°10′59.50″ E | Silt loam | a |
| Waregem 1 | West Flanders | 50°54′08.96″ N | 3°26′59.83″ E | Loamy sand | d |
| Waregem 2 | West Flanders | 50°52′14.50″ N | 3°22′51.70″ E | Sandy loam | d |
| Welkenraedt 1 | Liège | 50°39′49.10″ N | 5°55′53.40″ E | Loam | d |
| Welkenraedt 2 | Liège | 50°39′20.60″ N | 5°56′18.00″ E | Loam | d |
| Wielsbeke | West Flanders | 50°55′40.48″ N | 3°21′42.25″ E | Sandy loam | c |
| Primer Pair | Number of Markers | Number of Polymorphic Markers | % of Polymorphic Markers | Polymorphic Information Content (PIC) |
|---|---|---|---|---|
| E-AC/M-CAC | 105 | 81 | 77.1 | 0.32 |
| E-AC/M-CAG | 91 | 64 | 70.3 | 0.33 |
| E-AC/M-CAT | 75 | 62 | 82.7 | 0.31 |
| Total | 271 | 207 | 76.4 |
| Variables | Component Loadings | |||||
|---|---|---|---|---|---|---|
| 2018 | 2019 | 2020 | ||||
| PC 1 | PC 2 | PC 1 | PC 2 | PC 1 | PC 2 | |
| Tuber number | 0.950 | 0.144 | 0.948 | 0.175 | −0.769 | −0.563 |
| Shoot number | 0.901 | −0.228 | 0.797 | 0.133 | −0.601 | −0.519 |
| Fresh tuber biomass | 0.719 | −0.568 | 0.406 | −0.443 | 0.534 | −0.574 |
| Individual fresh tuber weight | −0.618 | −0.534 | −0.861 | −0.350 | 0.848 | 0.366 |
| Inflorescence number | 0.172 | 0.842 | −0.276 | 0.851 | −0.610 | 0.645 |
| Number of germinable seeds | 0.002 | 0.838 | −0.170 | 0.932 | −0.481 | 0.740 |
| Cluster | 2018 | 2019 | 2020 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pops./ Cluster | Mean ± SE | Sign. | Pops./ Cluster | Mean ± SE | Sign. | Pops./ Cluster | Mean ± SE | Sign. | ||
| Shoot number | 1 | 11 | 57.9 ± 1.73 | b | 10 | 59.1 ± 4.17 | b | 3 | 72.7 ± 2.49 | b |
| 2 | 11 | 42.4 ± 2.33 | a | 1 | 26.0 ± 0.00 | a | 10 | 61.1 ± 2.12 | ab | |
| 3 | 5 | 46.6 ± 5.18 | ab | 11 | 45.1 ± 3.03 | ab | 7 | 53.0 ± 2.61 | a | |
| 4 | 8 | 78.8 ± 3.94 | c | 6 | 45.6 ± 2.14 | ab | 8 | 57.2 ± 1.75 | a | |
| 5 | - | - | 7 | 90.3 ± 6.46 | c | 7 | 86.2 ± 3.67 | c | ||
| Inflorescence number | 1 | 11 | 10.9 ± 1.02 | b | 10 | 8.9 ± 0.92 | b | 3 | 19.7 ± 1.58 | c |
| 2 | 11 | 4.0 ± 1.02 | a | 1 | 6.0 ± 0.00 | b | 10 | 11.4 ± 1.48 | b | |
| 3 | 5 | 2.7 ± 1.67 | a | 11 | 1.4 ± 0.56 | a | 7 | 6.4 ± 1.85 | ab | |
| 4 | 8 | 5.1 ± 1.21 | a | 6 | 1.9 ± 0.54 | a | 8 | 0.7 ± 0.40 | a | |
| 5 | - | - | 7 | 1.2 ± 0.25 | a | 7 | 5.9 ± 1.32 | ab | ||
| Tuber number | 1 | 11 | 641 ± 26.4 | bc | 10 | 560 ± 52.4 | bc | 3 | 1015 ± 68.2 | bc |
| 2 | 11 | 570 ± 52.2 | ab | 1 | 221 ± 0.0 | a | 10 | 899 ± 59.8 | b | |
| 3 | 5 | 419 ± 92.4 | ab | 11 | 598 ± 28.2 | bc | 7 | 501 ± 42.7 | a | |
| 4 | 8 | 808 ± 62.7 | c | 6 | 359 ± 66.5 | ab | 8 | 932 ± 30.2 | bc | |
| 5 | - | - | 7 | 758 ± 53.9 | c | 7 | 1165 ± 75.9 | c | ||
| Fresh tuber biomass (g) | 1 | 11 | 189.7 ± 7.86 | ab | 10 | 128.5 ± 6.19 | b | 3 | 155.8 ± 5.26 | a |
| 2 | 11 | 150.2 ± 10.38 | ab | 1 | 88.1 ± 0.00 | a | 10 | 162.1 ± 11.58 | a | |
| 3 | 5 | 224.8 ± 28.81 | b | 11 | 124.6 ± 4.70 | b | 7 | 190.7 ± 14.97 | a | |
| 4 | 8 | 200.6 ± 5.44 | b | 6 | 154.2 ± 11.88 | b | 8 | 184.2 ± 5.37 | a | |
| 5 | - | - | 7 | 137.9 ± 3.67 | b | 7 | 188.1 ± 10.82 | a | ||
| Individual fresh tuber weight (g) | 1 | 11 | 0.31 ± 0.021 | a | 10 | 0.25 ± 0.024 | a | 3 | 0.15 ± 0.006 | a |
| 2 | 11 | 0.27 ± 0.014 | a | 1 | 0.40 ± 0.000 | b | 10 | 0.19 ± 0.015 | a | |
| 3 | 5 | 0.59 ± 0.064 | b | 11 | 0.21 ± 0.008 | a | 7 | 0.39 ± 0.034 | b | |
| 4 | 8 | 0.26 ± 0.016 | a | 6 | 0.47 ± 0.051 | b | 8 | 0.20 ± 0.007 | a | |
| 5 | - | - | 7 | 0.19 ± 0.013 | a | 7 | 0.16 ± 0.006 | a | ||
| Number of germinable seeds | 1 | 11 | 1043 ± 161.2 | b | 10 | 2158 ± 235.5 | b | 3 | 6966 ± 2248.4 | b |
| 2 | 11 | 431 ± 139.3 | a | 1 | 7981 ± 0.0 | c | 10 | 1593 ± 398.9 | a | |
| 3 | 5 | 219 ± 101.9 | a | 11 | 566 ± 235.6 | a | 7 | 1731 ± 630.0 | a | |
| 4 | 8 | 479 ± 132.5 | ab | 6 | 260 ± 101.4 | a | 8 | 21 ± 10.9 | a | |
| 5 | - | - | 7 | 555 ± 171.1 | a | 7 | 612 ± 148.8 | a | ||
| Shoot Number | Inflorescence Number | Tuber Number | Fresh Tuber Biomass | Individual Fresh Tuber Weight | Number of Germinable Seeds | |
|---|---|---|---|---|---|---|
| Shoot number | 1.000 | |||||
| Inflorescence number | −0.010 | 1.000 | ||||
| Tuber number | 0.626 ** | −0.106 | 1.000 | |||
| Fresh tuber biomass | 0.235 | −0.239 | 0.163 | 1.000 | ||
| Individual fresh tuber weight | −0.396 * | −0.075 | −0.833 ** | 0.197 | 1.000 | |
| Number of germinable seeds | −0.171 | 0.533 ** | −0.291 | −0.391 * | 0.053 | 1.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Ryck, S.; Reheul, D.; De Riek, J.; De Keyser, E.; De Cauwer, B. Genetic and Morphological Variation of Belgian Cyperus esculentus L. Clonal Populations and Their Significance for Integrated Management. Agronomy 2023, 13, 572. https://doi.org/10.3390/agronomy13020572
De Ryck S, Reheul D, De Riek J, De Keyser E, De Cauwer B. Genetic and Morphological Variation of Belgian Cyperus esculentus L. Clonal Populations and Their Significance for Integrated Management. Agronomy. 2023; 13(2):572. https://doi.org/10.3390/agronomy13020572
Chicago/Turabian StyleDe Ryck, Sander, Dirk Reheul, Jan De Riek, Ellen De Keyser, and Benny De Cauwer. 2023. "Genetic and Morphological Variation of Belgian Cyperus esculentus L. Clonal Populations and Their Significance for Integrated Management" Agronomy 13, no. 2: 572. https://doi.org/10.3390/agronomy13020572
APA StyleDe Ryck, S., Reheul, D., De Riek, J., De Keyser, E., & De Cauwer, B. (2023). Genetic and Morphological Variation of Belgian Cyperus esculentus L. Clonal Populations and Their Significance for Integrated Management. Agronomy, 13(2), 572. https://doi.org/10.3390/agronomy13020572







