Abstract
A fruiting body of a basidiomycete fungus was discovered growing on chopped Ficus nitida tree trunks in the student housing on the Assiut University campus during the course of this inquiry and a normal collecting operation in the Assiut Governorate, Egypt. Following the growth of the basidioma’s inner tissue on PDA, fungal mycelial growth was achieved. Internal transcribed spacer region (ITS) sequencing has allowed for the identification of the fungus as Tomophagus colossus. On the dry weight basis, chemical analysis of T. colossus AUMC 14536 basidioma revealed that it contains 28.81% carbohydrates, 25.34% crude fats, 23.44% crude fibers, 20.64% crude proteins, and 3.02% ash, in addition to potassium, phosphorus, calcium, selenium, iron, and zinc (133.59, 114.46, 6.27, 3.08, 1.28, and 0.73 mg/100 g dry weight, respectively). The total phenolic compounds (39.26 mg/g) and total flavonoids (5.62 mg/g) were also evaluated. The basidioma extract’s antioxidant activity was assessed as %DPPH radical scavenging activity with an IC50 of 4.15 µg/mL compared with a 1.89 µg/mL IC50 of ascorbic acid. In solid-state fermentation (SSF), the fungus could ferment broad bean straw, palm leaf hay, rice husk, rice straw, sugarcane bagasse, and wheat bran to produce endoglucanase, exoglucanase, laccase, pectinase, and xylanase in substantial amounts. Specific activity exhibited the highest values for endoglucanase (81.48 U/mg), exoglucanase (114.35 U/mg), pectinase (81.94 U/mg), and xylanase (70.18 U/mg) on the rice husk, while the peak of laccase activity (94.27 U/mg) was gained on bean straw. This is the first assessment of the organism’s nutritional value, amino acid content, antioxidant activity, and enzymatic capabilities in Egypt.
Keywords:
amino acids; antioxidant; biodiversity; cytotoxicity; Ficus; ITS; phenolic; phylogeny; polyporaceae 1. Introduction
The family Polyporaceae Fr. ex Corda includes polypore fungi that have considerable economic significance. Many species cause significant annual crop losses in many tropical countries [1], and some are of vital importance for medical uses, especially in East Asia [2,3]. Ganoderma species are a major group of Polyporaceae [1,2,3], which develop double-walled basidiospores that differentiate them from other members of this family. Eventually, the term “Ganoderma” was used to describe the Costa Rican species Tomophagus colossus (Fr.) [4], formerly known as Polyporus colossus Fr. [5]. Many scientists [6,7,8,9,10] recognized Tomophagus colossus as a synonym for Ganoderma colossus (Fr.) C.F. Baker, but many others [6,7,8,9] rejected the genus Tomophagus. The exceptionally dense and pale context of the genus Tomophagus, which dries to a soft and light consistency, led to its recognition as distinct from Ganoderma. Additionally, recent molecular phylogenetic analyses using internal transcribed spacers (ITS) of ribosomal DNA repeats [11] and nearly complete mitochondrial small-subunit ribosomal DNA sequences [12] led to the recognition of Tomophagus as a distinct genus.
Although rare, reports of this species have been found in all of the tropics, with the exception of East Africa [8,13]. The first record of T. colossus in Vietnam dates back to Patouillard [14]. Patouillard mistakenly identified the species as Ganoderma obokense, which was subsequently synonymized with G. colossus [6,7,8,15]. Vietnam is the only place where this endangered species has been found again [4]. In Egypt, the Suez Canal University’s Faculty of Agriculture’s ornamental farm was the source of the first isolation of T. colossus [16].
The members of Polyporaceae have been shown to be pharmacologically active, and their medicinal use has been studied [17,18]. Several new lanostan triterpenes and lactones (colossolactones) from cultivated T. colossus Murrill have been identified in recent studies [19,20]. As a consequence, the current study was created to provide T. colossus, which has been connected, for the first time, with the Ficus nitida tree in Assiut, Egypt. The fungal basidioma underwent chemical investigation, including the assessments of macro- and micronutrients’ content, antioxidant activity, amino acid content, and cytotoxicity, as well as the exploitation of some agricultural wastes for enzymes synthesis. Finding this species in Egypt for the first time may have implications for the creation of new bioactive compounds for medical applications and/or extracellular cocktail enzymes that may find application in numerous other industrial domains.
2. Materials and Methods
2.1. Collection and Isolation of Fungal Material
In close proximity to the student residence buildings (55QG + QHF) at Assiut University, Assiut Governorate, Egypt, fruiting bodies on Ficus nitida were found. After being severed, the fungal basidiomata were promptly transported to the Assiut University Mycological Center’s mycological laboratory. A tiny portion of the inner tissue of the basidioma was excised under sterile conditions and plated on Petri dishes containing potato dextrose agar (PDA) [21]. The plates were then left to develop fungal growth in an incubation period of 10 days at 25 °C. The fungus’s pure culture was isolated on PDA plates and preserved as AUMC 14536, frozen mycelia stored at −86 °C and lyophilized cultures, in the culture collection of the Assiut University Mycological Center (AUMC).
2.2. Molecular Identification of the Fungal Sample
The DNA extraction from the fungus was performed following the method described by Moubasher et al. [22], in which 0.2 g of 7-day-old fungal mycelia grown on PDA was grinded and transferred to a 1.5 mL microfuge tube. The isolation of the DNA was performed by CTAB method (800 μL CTAB buffer composed of 3% CTAB, 1.4 M/L NaCl, 0.2% Mercaptoethanol, 20 mM EDTA, 100 mM Tris-HCl pH 8.0, and 1% PVP-40). The PCR reaction was carried out using the universal primers ITS1 and ITS4 [23] and SolGent EF-Taq, as described by Al-Bedak and Moubasher [24] and Al-Bedak et al. [25].
2.3. Alignments and Phylogenetic Analyses
DNASTAR (version 5.05) was used to produce the contiguous sequence of Tomophagus sp. In this study. From GenBank, sequences of the closest species in the genera Ganoderma and Tomophagus, as well as those of the accessible-type specimens, were retrieved. As an outgroup, Favolus subtropicus BJFC Cui4292 was employed. Using MAFFT (version 6.861b) and the default settings, the sequences of Tomophagus sp. AUMC 14536 and those retrieved from GenBank were aligned together [26]. Alignment gaps and parsimony uninformative characters were optimized by BMGE [27]. Maximum-likelihood (ML) and maximum-parsimony (MP) phylogenetic analyses were performed using MEGA X version 10.2.6 [28]. The robustness of the most parsimonious trees was evaluated by 1000 bootstrap replications [29]. The best optimal model of nucleotide substitution for the ML analyses was determined using Akaike information criterion (AIC), as implemented in Modeltest 3.7 [30]. The phylogenetic tree was drawn and visualized using MEGA X (version 10.2.6) [28], and the resulting phylogenetic tree was edited and saved as TIF file [25].
2.4. Macro- and Micronutrient Analyses of Tomophagus colossus Basidioma
All chemical analyses were performed at the Central Laboratories Unit, Faculty of Agriculture, Assiut University, Assiut, Egypt. Prior to chemical analyses, T. colossus basidioma was oven-dried at 50 ± 1 °C for 36 h and then grinded to fine powder. Nutrient composition of the T. colossus basidioma was determined using the standard methods of the association of official analytical chemists’ procedures (AOAC). All values were expressed in mg/100 g dry weight. Triplicate samples were used for moisture content in a hot-air-circulating oven. Ash was determined by incineration (550 °C) of the known weight of the sample in a muffle furnace (Method No 930.05). Crude fat was determined by exhaustively extracting a known weight of the sample in petroleum ether (boiling point, 40 °C to 60 °C) using a Soxhlet extractor (Method No 930.09). Protein (N × 6.25) was determined by the Kjeldahl method (Method No 978.04). Crude fiber was determined after digesting a known weight of fat-free sample in refluxing 1.25% sulfuric acid and 1.25% sodium hydroxide (Method No. 930.10). Mineral content was determined using the atomic absorption amanitas (Perkin–Elmer, Model 3300, Encino, CA, USA). Total phenolic compounds was determined by the Folin–Ciocalteau reagent method using Gallic acid as standard [31,32]. The total flavonoids were determined using the technique described by [33]. The carbohydrate content (%) was determined by difference. The addition of all the percentages of moisture content (MC), fat content (FC), ash content (AC), crude fiber (CF), and crude protein (CP) was subtracted from 100% according to Equation (1)
%Carbohydrate = 100 − (%MC + %FC + %AC + %CF + %CP)
2.5. Antioxidant Activity of Tomophagus colossus’s Basidioma
This test was carried out at Nawah Center for Scientific Services (www.nawah-scientific.com, accessed on 16 September 2023), Al-Moqattam, Cairo, Egypt. The antioxidant activity was determined by the colorimetric technique [34,35] using the radical scavenging activity of a freshly made methanol solution containing 0.004% (w/v) of 2, 2′-Diphenyl-1-picrylhydrazyl (DPPH). A total of 1 mL of this solution was added to 3 mL of various concentrations (3.9, 7.8, 15.62, 31.25, 62.5, 125, 250, 500, and 1000 µg/mL) of T. colossus extract in ethanol. The mixture was vigorously mixed and left to stand for 30 min at room temperature (30 °C). UV–VIS Milton Roy spectrophotometer (Milton Roy, Spectronic 1201; Markham, ON, Canada) was used to determine the absorbance at 517 nm. The experiment was carried out in triplicate, with ascorbic acid serving as the reference chemical. Using a log dosage inhibition curve, the sample’s IC 50 value—the amount of sample needed to block 50% of the DPPH free radical—was determined. Higher levels of free radical activity were indicated by the reaction mixture’s lower absorbance. Estimates were made of the DPPH radical’s percentage of inhibition [36] according to Equation (2).
where Ac = absorbance of the control at zero time, and At = absorbance of the antioxidant at 1 h.
2.6. Amino Acids Content of T. colossus Basidioma
2.6.1. Preparation of Standard Stock Solution
A stock solution of 17 amino acids, namely aspartic acid, glutamic acid, threonine, serine, proline, glycine, alanine, cystine, valine, methionine, isoleucine, leucine, tyrosine, phenylalanine, histidine, lysine, and arginine, was prepared at a concentration of 2.5 µmol/mL, except Cystine, which was prepared at 1.25 µmol/mL.
2.6.2. Sample Hydrolysis
Glass ampoules were filled with approximately 30 mg of the defatted material. Nitrogen gas was introduced to the samples along with 7.0 mL of 6 M HCl to remove any remaining oxygen. The glass ampoules were sealed using a Bunsen flame and baked for 22 h at 105 ± 5 °C. Humins were eliminated by filtering the contents after the ampoule was allowed to cool. After that, the filtrate was vacuum-sealed and dried at 40 °C in a rotating evaporator. Before being used, the residue was dissolved in 5 mL of pH 2.0 acetate buffer and kept in a deep freezer.
2.6.3. Analysis of Amino Acids Content
About 2.5 g of the basidioma sample were weighed into the extraction thimble, and fat was extracted with a chloroform/methanol (2:1, v/v) mixture using Soxhlet apparatus (AOAC, 2005). The extraction lasted for 5–6 h. The stock solution was diluted by 60 µL in 1.5 mL vial with buffer and filtered by 0.22 µm syringe filter. Afterwards, Sykam Amino Acid Analyzer (Sykam GmbH, Eresing, Germany) subjected 100 µL of this solution to analysis. The amino acid analyzer was equipped with Solvent Delivery System S 2100 (Quaternary pump with flow rate of 0.01–10.00 mL/min and maximum pressure up to 400 bar), Autosampler S 5200, Amino Acid Reaction Module S4300 (with built-in dual-filter photometer between 440 and 570 nm with constant signal output and signal summary option) and Refrigerated Reagent Organizer S 4130.
2.7. Cytotoxicity Test
This test was conducted at Nawah Scientific Inc., Moqattam, Cairo, Egypt. Oral epithelial cells (OECs) were used in this test, and the methodology described by [37,38] was followed. The cells were kept alive in DMEM medium at 37 °C in a humidified atmosphere of 5% (v/v) CO2. The DMEM was supplemented with 100 mg/mL streptomycin, 100 U/mL penicillin, and 10% heat-inactivated fetal bovine serum. The sulforhodamine B (SRB) assay was used for cell density determination, based on the measurement of cellular protein content [39]. Aliquots of 100 µL cell suspension (contained 5 × 103 cells/mL) were grown in complete medium in 96-well plates for 24 h. To treat the cells, 100 µL of media with various concentrations of T. colossus fruiting body’s extract was utilized. After 72 h of extract exposure, cells were fixed by replacing the medium with 150 µL of 10% TCA and incubated at 4 °C for 60 min. After removing the TCA solution, the cells were washed five times with distilled water. Aliquots of a 70 µL LSRB solution (0.4% w/v) were added, and the mixture was incubated in a dark atmosphere for 10 min at room temperature. The plates were then washed three times with 1.0% acetic acid before being allowed to air dry overnight. To dissolve the protein-bound SRB dye, 150 µL of 10 mM TRIS was added. The absorbance was then measured at 540 nm using a BMG LABTECH®- FLUOstar Omega microplate reader (Ortenberg, Germany) [37].
2.8. Cell Viability
Cell viability was calculated as a ratio to the reference values. The scores were compared using the intraclass correlation coefficient (ICC) and limits of agreement statistics [40,41]. As a descriptive measure of agreement, the data on limits of agreement were also employed. The dose–response curve was used to obtain the IC50, and Equation (3) was used to determine the relative viability percentage [42].
2.9. Solid-State Fermentation of Some Lignocellulosic Wastes by T. colossus AUMC 14536
2.9.1. Substrate Pretreatment
Six different agricultural wastes, including bean straw (BS), palm leaf hay (PLH), rice husk (RH), rice straw (RS), sugarcane bagasse (SB), and wheat bran (WB), were chosen for solid-state fermentation (SSF). Every substrate was purchased from a public market in the governorates of Assiut and New Valley. The samples were cleaned with distilled water before being used. They were then uniformly weighted after being oven dried at 50 °C.
2.9.2. Enzyme Production in SSF
Each 250 mL Erlenmeyer flask in the triplicate set included 5 g of agricultural waste. Each was given 10 mL of the fermentation medium, which contains the following ingredients (g/L): oat spelt xylan, 1.0; NaNO3, 2.0; K2HPO4, 1.0; KCl, 0.5; MgSO4·7H2O, 0.5; FeSO4, 0.01; ZnSO4, 0.01; and CuSO4, 0.005. At the end, the pH was set to 7.0. The flasks were injected with 1.0 mL of spore suspension containing 1.5 × 108 spores derived from a 7-day-old culture of Tomophagus isolate AUMC 14536 after being autoclaved (121 °C for 20 min). The inoculated flasks underwent a 14-day static incubation period at 30 °C. After the incubation, centrifugation (10,000 rpm for 10 min at 4 °C) produced the cell-free supernatants, which were then used as crude enzymes.
2.9.3. Enzyme Assay and Protein Estimation
Using the substrates carboxymethyl cellulose (CMC), microcrystalline cellulose (MCC), pectin, and oat spelts xylan, the estimation of endoglucanase, exoglucanase, pectinase, and xylanase was performed. Prepared in a 50 mM Na-citrate buffer (pH 5.0), the reaction mixture contained 0.5 mL of crude enzyme and 0.5 mL of the aforementioned substrates at a 1.0% concentration. The reaction was carried out at 50 °C in a water bath for 20 min and stopped by introducing 2 mL of 3, 5, Dinitrosalicylic acid (DNS) reagent. The quantity of reducing sugars released was measured using glucose or xylose calibration curves [43]. When measuring the activity of the laccase enzyme at 450 nm, the reddish brown color that resulted from laccase’s oxidation of guaiacol was employed [44]. The reaction mixture contained 1.0 mL of 2 mM guaiacol, 1.0 mL of the fungal supernatant, and 3.0 mL of 10 mM sodium acetate buffer (pH 5), and the reaction was carried out at 30 °C for 15 min [45]. Protein content was estimated by Lowry’s method, using bovine serum albumin as standard [46]. Calculations of sugar concentration and enzyme activity were conducted according to Al-Bedak et al. [25] and Al-Kolaibe et al. [47].
3. Results
3.1. Molecular Identification of the Tomophagus Isolate
Based on a megablast search in the NCBI database using ITS sequence of Tomophagus isolate AUMC 14536, the closest hits are Tomophagus colossus cultivar Sai Gon ((GenBank accession number MG650109; identities = 614/614 (100%); Gaps = 0/614 (0%)) and T. colossus isolate A164FB2 ((GenBank accession number OQ558872; identities = 613/614 (99.83%); Gaps = 0/614 (0%)). The taxonomic status of Tomophagus sp. in this investigation was ascertained using a phylogenetic analysis of the ITS dataset. There were 24 species in the final batch of ITS data. Out of the 614 characters in the maximum-parsimony dataset, 509 could be aligned without any doubt, 151 were variable characters that were parsimony-uninformative, and 101 were counted as parsimony-informative. For nucleotide substitution, the Kimura 2-parameter and Gamma distribution (K2 + G) model was ideal. Four trees were produced via maximum-parsimony analysis, and the most parsimonious one was given; it had a tree length of 432 steps, a consistency index of 0.548387, a retention index of 0.736752, and a composite index of 0.404025. Figure 1 displays the single tree that the maximum-likelihood analysis produced (log likelihood = 2964.62).
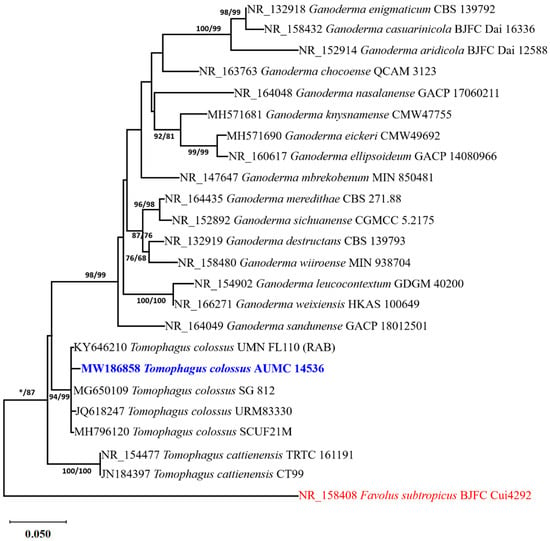
Figure 1.
The most parsimonious phylogenetic tree generated from ML/MP analysis using a heuristic search (1000 replications) of ITS sequence of T. colossus AUMC 14536 (in blue color) compared with other closely similar ITS sequences belonging to Tomophagus and Ganoderma in GenBank. Bootstrap support values for ML/MP ≥ 50% are indicated near the respective nodes. Bootstrap support values less than 50% are signed with (*). The tree is rooted to Favolus subtropicus BJFC Cui4292 as outgroup (in red color).
The relationships between the Tomophagus sp. AUMC 14536 and other Ganoderma and Tomophagus species were described by the phylogenetic result. A very strong bootstrap support value of 94% ML/99% MP is endorsed by the strain found within the T. colossus clade at the same branch as T. colossus SG 812 (100% similarity), T. colossus URM 83330 (99.84% similarity), T. colossus SCUF21M (99.83%), and T. colossus UMNFL 110 (RAB) (99.51% similarity). This led to the identification of the strain under study as T. colossus (Figure 1).
3.2. Brief Description of T. colossus AUMC 14536
Pileus thick, 8–15 (−25) cm, spongy, floccose, light in weight when dried, sessile, or with a very short cylindrical stipe. Pilear surface glabrous, opaque in appearance, yellowish in young specimens, brownish in older basidiocarps, thin, readily broken when cracked or pressed (Figure 2A–C). Chlamydospores dark-brown, thick-walled, 10–12 × 7–10 µm (Figure 2D). Basidiospores abundant, verrucose, ovate, truncate or rounded at the apex, yellowish, thick-walled, 13–18 × 8–12 µm (Figure 2E). Colonies on PDA attaining a diameter of 75 mm after 7 days at 28.0 ± 2.0 °C, centrally brown, floccose; margin entire, wide, white; reverse brown beneath the colony center, pale outwards (Figure 2F). Chlamydospores formed on PDA culture, globose, double-walled, pigmented, ornamented with abundant papillate projections on the surface (Figure 2G–I).
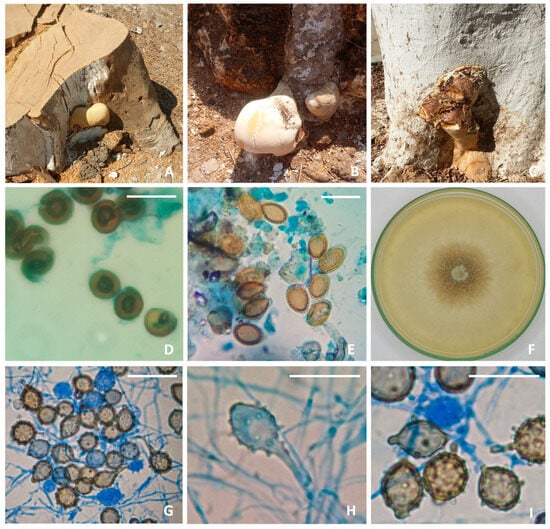
Figure 2.
T. colossus AUMC 14536 (A–C), Ficus nitida-associated basidioma (D), dark-colored, thick-walled chlamydospores observed in the context (E), verrucose basidiospores (F), seven-day-old colony on PDA at 25 °C (G–I), globose, double-walled, pigmented, ornamented chlamydospores with abundant papillate projections (scale bars = 20 µm).
3.3. Chemical Analyses of T. colossus’s Basidioma
Tomophagus colossus AUMC 14536 basidioma’s chemical analysis showed that, based on dry weight, crude fats and carbohydrates (28.81 and 25.34%, respectively) make up the majority of the basidioma, followed by crude fibers (23.44%) and crude proteins (20.64%), as well as ash content (3.02%) (Table 1). We assessed potassium, phosphorus, calcium, iron, zinc, and selenium. The highest quantities of calcium (6.27 mg/100 g) were followed by potassium (133.59 mg/100 g) and phosphorus (114.46 mg/100 g). Iron and selenium levels were moderate (3.08 and 1.28 mg/100 g, respectively) at the same time. Moreover, it contains the least zinc (0.73 mg/100 g). The concentrations of total flavonoids and total phenolic compounds were 562 and 3926 mg/100 g, respectively (Table 1).

Table 1.
Chemical Composition of T. colossus AUMC 14536’s Basidioma.
3.4. Antioxidant Activity of T. colossus AUMC 14536 Biomass
A DPPH test was used to further investigate the biomass of T. colossus AUMC 14536’s capacity to scavenge hydrogen, and the free radical scavenging activity was reported as an IC50 value. The results indicate that DPPH activity increased with increasing sample concentration (Figure 3A). The extract of T. colossus AUMC 14536 basidioma had an IC50 value of 4.15 µg/mL, which was higher than the ascorbic acid activity value of 1.89 µg/mL (Figure 3B).
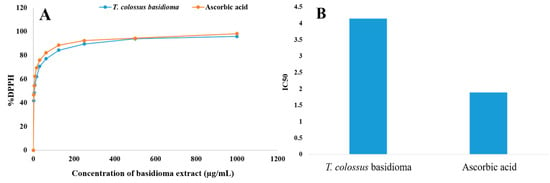
Figure 3.
(A) Antioxidant activity (% DPPH) and (B) IC50 of the T. colossus basidioma extract compared with standard ascorbic acid.
3.5. Analysis of Amino Acids Content
When the T. colossus AUMC 14536 basidioma extract was analyzed for amino acid content, 16 amino acids were identified (Figure 4). It was shown that arginine (14.396 mg/100 mg dry weight), glutamic acid (6.801), alanine (5.556), histidine (4.558), and aspartic acid (2.727) were the most common. From 0.282 (methionine) to 1.945 (leucine), the remaining amino acids are listed in Table 2.
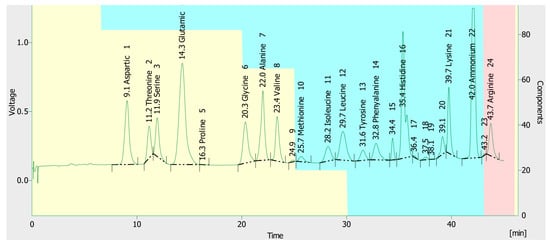
Figure 4.
Amino acid profile of the T. colossus AUMC 14536’s basidioma.

Table 2.
Amino acids profile of T. colossus AUMC 14536’s basidioma.
3.6. Cytotoxicity
The percentage of cell viability was used to illustrate the cytotoxic effects of T. colossus AUMC 14536’s basidioma biomass on the OECs. The SRB assays’ negative control reactions had intraclass correlation coefficient values (ICC) of 0.997, 0.985, 0.974, 0.968, and 0.960 at different concentrations of 0.01, 0.1, 1.0, 10.0, and 100 µg/mL, respectively. The IC50 was found to be 4.15 µg/mL, and the cell viability varied across all tested concentrations, ranging from 95.997% to 99.714 (Figure 5).

Figure 5.
Dose–response curve for evaluation of cytotoxicity of T. colossus AUMC 14536’s basidioma on OECs using SRB assays.
3.7. Cell Wall Hydrolyzing Enzymatic Potential of T. colossus AUMC 14536 in SSF
The lignocellulosic wastes utilized in SSF could potentially be fermented by T. colossus AUMC 14536, which could result in the production of large quantities of all the enzymes that were identified (endoglucanase, exoglucanase, laccase, pectinase, and xylanase). The highest specific activity was obtained by the rice husk waste for endoglucanase (81.48 U/mg), exoglucanase (114.35 U/mg), pectinase (81.94 U/mg), and xylanase (70.18 U/mg). Laccase production was boosted by bean straw and wheat bran, with maximal specific activities of 94.27 and 91.7 U/mg, respectively (Figure 6).
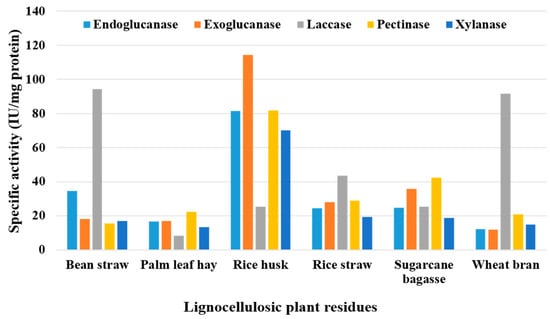
Figure 6.
Specific activity of endoglucanase, exoglucanase, laccase, pectinase, and xylanase produced by T. colossus AUMC 14536 in SSF at 30 °C.
4. Discussion
The members of the Polyporaceae are identified by their characteristic double-walled basidiospores, which were clearly observed in T. colossus in this study. The existence of chlamydospores is a significant feature for species identification, as evidenced by molecular phylogenetic research. Chlamydospores are found in the basidiomata of just a few Ganoderma species, including G. subamboinense (Henn.) Bazzalo and J.E. Wright and G. weberianum (Bres. and Henn. ex Sacc.) Steyaert [12,48]. The macro- and micromorphological characteristics of Ganoderma are very variable, with over 250 species reported [8]. As a result, it is the most difficult genus to classify among order Polyporales [11,49]. The variety of Ganoderma morphological features encouraged several taxonomists to look into chemical and molecular methods for differentiating Ganoderma species. Based on morphological features and molecular phylogeny, the strain AUMC 14536 was identified as T. colossus in this study, which was associated with the Ficus nitida tree for the first time in Egypt and worldwide. Tomophagus colossus has been isolated from the Faculty of Agriculture Ornamental farm at Suez Canal University, Egypt [16], but this is the first time that its nutritional value, amino acids content, antioxidant activity, and cytotoxicity, as well as its enzymatic capabilities, have been assessed.
High levels of carbohydrates, fats, fibers, and proteins were found in the chemical analysis of T. colossus basidioma used in this study. Additionally, high levels of potassium, phosphorus, calcium, selenium, iron, and zinc were found, suggesting that T. colossus biomass has a high nutritional value as a nutritional supplement. Polypore fungi such as Ganoderma have a long history of usage in traditional medicine in several Asian countries to promote health and longevity. It has also been widely employed in a variety of medical applications and nutritional supplements to prevent and treat a wide range of illnesses [50]. Regarding this, Triterpenoids (Colossolactones A, B, C, D, E, F, and G) and colossolactones I, II, III, IV, V, VI, and VIII, as well as Ganomycin B, Ganomycin I, farnesyl hydroquinone, Ganomycin I, Ganomycin B, schisanlactone A, Colossolactone VII, Ergosterol, and Ganorbiformin A, were extracted from Vietnamese T. colossus [19,20,51,52]. Conversely, polysaccharides, triterpenoids, nucleosides, sterols, fatty acids, protein, and alkaloids are only a few of the bioactive compounds discovered and extracted from many Polypore fungi [53,54,55,56,57,58,59,60,61,62,63,64].
Considerable levels of flavonoids (562 mg/100 g) and phenolic compounds (3926 mg/100 g) were detected in the biomass of T. colossus basidioma in this investigation, as well as strong antioxidant activity with IC50 of 4.15 µg/mL. Regarding this, Hericium erinaceus, Ganoderma lucidum, and Agrocybe aegerita fruiting bodies have total phenolic content values of 17.10, 28.11, and 16.05 mg/g, respectively. However, the total flavonoid content in Hericium erinaceus, Ganoderma lucidum, and Agrocybe aegerita exhibits values of 445.6, 627.7, and 393.9 μg/g, respectively [65]. Both Ganoderma carnosum Pat. and Ganoderma pfeifferi Bres. yielded significant amounts of flavonoids (10.06 ± 0.51 and 2.71 ± 0.24 mg/g) and phenolic compounds (43.28 ± 3.83 and 22.81 ± 0.68 mg/g) [66]. However, the existence of flavonoids in some mushroom species is uncertain, because these species lack the chalcone isomerase enzymes necessary for the biosynthesis of flavonoids [67]. Despite not being the primary component of edible and medicinal mushrooms, phenolic compounds have been shown to exhibit a number of advantageous properties, including antioxidant [68,69] and enzyme inhibition [70], which appears to be closely associated with the anti-inflammatory qualities of mushrooms [71].
Enzymatic potential by mushrooms and filamentous fungi has been shown to be the most efficient, cost-effective, environmentally friendly, and, in most situations, high-yielding method [72,73,74]. In the present study, T. colossus AUMC 14536 showed good growth on all agricultural wastes (BS, PLH, RH, RS, SB, and WB) used in SSF, and produced high levels of endoglucanase, exoglucanase, laccase, pectinase, and xylanase enzymes. These findings bode well for the development of a practical application for the treatment of lingo-cellulosic plant wastes, as well as for the bioremediation of damaged ecosystems produced by the burning of such residues. Regarding this, no research on T. colossus enzyme synthesis in SSF has been disclosed. Some Ganoderma strains isolated from the base of the trees and decayed wood in Havana, Cuba [75], and Pakistan [76] have been used to produce laccases and peroxidases. In liquid media and solid-state cultures incorporating wood chips from Eucalyptus globulus or Drimys winteri, some strains of Ganoderma australe (Fr.) Pat. (A464 and A272) generated laccase and manganese peroxidase activity [77]. Two strains of Ganoderma multipileum Ding Hou produced a maximum laccase activity of 1355.5 ± 8.8 U/L utilizing guaiacol as substrate [78]. Two strains of Ganoderma (GASI3.4, and CCB209) produced lignin peroxidase, manganese peroxidase, and laccase on wheat bran [79].
5. Conclusions
In addition to using T. colossus for enzyme synthesis and lignocellulosic residue consumption, this work is a first for Egypt, since it is the first to isolate the fungus in relation to Ficus nitida trees. It was discovered that the basidiomata biomass had strong antioxidant activity, along with high concentrations of minerals, phenolic compounds, total lipids, total proteins, total fibers, carbohydrates, and amino acid content. Due to its enzymatic properties, this species is proposed to be accepted here for potential use in the manufacturing of animal feed, as well as in the prevention and treatment of various medical conditions. This requires special cultivation techniques, extracts, and commercial formulations. Although T. colossus seems promising, more investigation and credible scientific evidence are needed to confirm Tomophagus’s safe and effective use as a dietary supplement.
Author Contributions
All authors participated equally in data analysis, authoring, and revising the article. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Researchers Supporting Project number (RSP2023R364), King Saud University, Riyadh, Saudi Arabia.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare that there are no potential conflicts of interest regarding the publication of this paper.
References
- Flood, J.; Bridge, P.; Holderness, M. Ganoderma Diseases of Perennial Crops; CABI: Wallingford, UK, 2000. [Google Scholar]
- Jong, S.; Birmingham, J. Medicinal benefits of the mushroom Ganoderma. Adv. Appl. Microbiol. 1992, 37, 101–134. [Google Scholar]
- Lin, Z.B.; Zhang, H.N. Anti-tumor and immunoregulatory activities of Ganoderma lucidum and its possible mechanisms. Acta Pharmacol. Sin. 2004, 25, 1387–1395. [Google Scholar] [PubMed]
- Le, X.T.; Nguyen Le, Q.H.; Pham, N.D.; Duong, V.H.; Dentinger, B.T.; Moncalvo, J.-M. Tomophagus cattienensis sp. nov., a new Ganodermataceae species from Vietnam: Evidence from morphology and ITS DNA barcodes. Mycol. Prog. 2012, 11, 775–780. [Google Scholar]
- Fries, E.M. Novae symbolae mycologicae, in peregrinis terris a botanicis danicis collectae. In Nova Acta Regia Societis Scientiarum Upsaliensis, Ser. 3; 1851; pp. 17–136. Available online: https://cir.nii.ac.jp/crid/1130000795112975744 (accessed on 16 October 2023).
- Furtado, J.S. Ganoderma colossum and the status of Tomophagus. Mycologia 1965, 57, 979–984. [Google Scholar] [CrossRef]
- Steyaert, R. Species of Ganoderma and related genera mainly of the Bogor and Leiden Herbaria. Persoonia-Mol. Phylogeny Evol. Fungi 1972, 7, 55–118. [Google Scholar]
- Ryvarden, L. Synopsis Fungi, Genera of Polypores: Nomenclature and Taxonomy; Fungiflora A/S: Oslo, Norway, 1991; Volume 5, pp. 1–363. [Google Scholar]
- Corner, E.J.H. Amauroderma and Ganoderma; Cramer: Berlin, Germany, 1983. [Google Scholar]
- Wu, S.-H.; Zhang, X. The finding of three Ganodermataceae species in Taiwan. Collect. Res. 2003, 16, 61–66. [Google Scholar]
- Moncalvo, J.; Ryvarden, L. A nomenclatural study of the Ganodermataceae Donk; Fungiflora: Oslo, Norway, 1997; pp. 107–114. [Google Scholar]
- Hong, S.G.; Jung, H.S. Phylogenetic analysis of Ganoderma based on nearly complete mitochondrial small-subunit ribosomal DNA sequences. Mycologia 2004, 96, 742–755. [Google Scholar] [CrossRef]
- Ofodile, L.N.; Uma, N.; Kokubun, T.; Grayer, R.J.; Ogundipe, O.; Simmons, M.S. Antimicrobial colossolactones from a Nigerian polypore Ganoderma colossum (Fr.) CF Baker. Int. J. Med. Mushrooms 2005, 7, 437–438. [Google Scholar] [CrossRef]
- Patoullard, N. Contributions à la flore mycologique du Tonkin. J. Bot. 1897, 11, 367–370. [Google Scholar]
- Ryvarden, L. Neotropical polypores: Part 1: Introduction, Ganodermataceae & Hymenochaetaceae; Fungiflora: Oslo, Norway, 2004; p. 277. [Google Scholar]
- Abdel-Azeem, A.; Nafady, N. New records on the genus Tomophagus and Battarrea for mycobiota of Egypt. Curr. Res. Environ. Appl. Mycol. 2019, 9, 77–84. [Google Scholar] [CrossRef]
- Dzubak, P.; Hajduch, M.; Vydra, D.; Hustova, A.; Kvasnica, M.; Biedermann, D.; Markova, L.; Urban, M.; Sarek, J. Pharmacological activities of natural triterpenoids and their therapeutic implications. Nat. Prod. Rep. 2006, 23, 394–411. [Google Scholar] [CrossRef]
- Liby, K.T.; Yore, M.M.; Sporn, M.B. Triterpenoids and rexinoids as multifunctional agents for the prevention and treatment of cancer. Nat. Rev. Cancer 2007, 7, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Kleinwächter, P.; Anh, N.; Kiet, T.T.; Schlegel, B.; Dahse, H.-M.; Härtl, A.; Gräfe, U. Colossolactones, new triterpenoid metabolites from a Vietnamese mushroom Ganoderma Colossum. J. Nat. Prod. 2001, 64, 236–239. [Google Scholar]
- El Dine, R.S.; El Halawany, A.M.; Ma, C.-M.; Hattori, M. Anti-HIV-1 protease activity of lanostane triterpenes from the vietnamese mushroom Ganoderma Colossum. J. Nat. Prod. 2008, 71, 1022–1026. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.; Onions, A.H. The Preservation and Maintenance of Living Fungi; CAB International: Egham, UK, 1994; p. 122. [Google Scholar]
- Moubasher, A.H.; Ismail, M.A.; Al-Bedak, O.A.; Mohamed, R.A. Ramophialophora chlamydospora, a new species from an alkaline lake ofWadi-El-Natron, Egypt. Asian J. Mycol. 2019, 2, 110–117. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: Cambridge, MA, USA, 1990; Volume 18, pp. 315–322. [Google Scholar]
- Al-Bedak, O.A.; Moubasher, A.H. Aspergillus gaarensis, a new addition to section Circumdati from soil of Lake El-Gaar in Wadi-El-Natron, Egypt. Stud. Fungi 2020, 5, 59–65. [Google Scholar] [CrossRef]
- Al-Bedak, O.A.; Teama, E.; Ali, E.; Said, M.; Shalaby, E.; Moharram, Z. Impact of fumigation with phosphine on viability of wheat grains stored for six months at two levels of moisture content, in addition to description of four new records of associated fungi and assessment of their potential for enzymatic production. J. Basic Appl. Mycol. 2020, 11, 77–97. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Criscuolo, A.; Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010, 10, 210. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. Modeltest: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.Y.; Lim, T.T.; Tee, J.J. Antioxidant properties of guava fruit: Comparison with some local fruits. Sunway Acad. J. 2006, 3, 9–20. [Google Scholar]
- Maurya, S.; Singh, D. Quantitative analysis of total phenolic content in Adhatoda vasica Nees extracts. Int. J. PharmTech Res. 2010, 2, 2403–2406. [Google Scholar]
- Zhishen, J.; Mengcheng, T.; Jianming, W. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999, 64, 555–559. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.-E.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT-Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Ramadan, A.M.; Shehata, R.M.; El-Sheikh, H.H.; Ameen, F.; Stephenson, S.L.; Zidan, S.A.; Al-Bedak, O.A. Exploitation of Sugarcane Bagasse and Environmentally Sustainable Production, Purification, Characterization, and Application of Lovastatin by Aspergillus terreus AUMC 15760 under Solid-State Conditions. Molecules 2023, 28, 4048. [Google Scholar] [CrossRef]
- Yen, G.C.; Duh, P.D. Scavenging effect of methanolic extracts of peanut hulls on free-radical and active-oxygen species. J. Agric. Food Chem. 1994, 42, 629–632. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New colorimetric cytotoxicity assay for anticancer-drug screening. JNCI J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef]
- Allam, R.M.; Al-Abd, A.M.; Khedr, A.; Sharaf, O.A.; Nofal, S.M.; Khalifa, A.E.; Mosli, H.A.; Abdel-Naim, A.B. Fingolimod interrupts the cross talk between estrogen metabolism and sphingolipid metabolism within prostate cancer cells. Toxicol. Lett. 2018, 291, 77–85. [Google Scholar] [CrossRef]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986, 327, 307–310. [Google Scholar] [CrossRef]
- Deyo, R.A.; Diehr, P.; Patrick, D.L. Reproducibility and responsiveness of health status measures statistics and strategies for evaluation. Control. Clin. Trials 1991, 12, S142–S158. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.B.; Nielsen, S.E.; Berg, K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J. Immunol. Methods 1989, 119, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Kalra, K.; Chauhan, R.; Shavez, M.; Sachdeva, S. Isolation of Laccase Producing Trichoderma Spp. and Effect. Int. J. ChemTech Res. 2013, 5, 2229–2235. [Google Scholar]
- Abd El Monssef, R.A.; Hassan, E.A.; Ramadan, E.M. Production of laccase enzyme for their potential application to decolorize fungal pigments on aging paper and parchment. Ann. Agric. Sci. 2016, 61, 145–154. [Google Scholar] [CrossRef]
- Lowry, O.; Rosebrough, N.; Farr, A.; Randall, R. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- AL-Kolaibe, A.M.; Moharram, A.M.; Al-Bedak, O.A. Worthwhile enzyme production and eco-friendly bioconversion of three agricultural residues by Aspergillus curvatus and Aspergillus gaarensis, promising enzyme-producers isolated from extreme environment. J. Basic Appl. Mycol. 2021, 12, 1–14. [Google Scholar]
- Douanla-Meli, C.; Langer, E. Ganoderma carocalcareus sp. nov., with crumbly-friable context parasite to saprobe on Anthocleista nobilis and its phylogenetic relationship in G. resinaceum group. Mycol. Prog. 2009, 8, 145–155. [Google Scholar] [CrossRef]
- Ryvarden, L. Type studies in the Polyporaceae 17. Species described by WA Murrill. Mycotaxon 1995, 23, 169–198. [Google Scholar]
- Hapuarachchi, K.; Cheng, C.R.; Wen, T.C.; Jeewon, R.; Kakumyan, P. Mycosphere Essays 20: Therapeutic potential of Ganoderma species: Insights into its use as traditional medicine. Mycosphere 2017, 8, 1653–1694. [Google Scholar] [CrossRef]
- El Dine, R.S.; El Halawany, A.M.; Ma, C.M.; Hattori, M. Inhibition of the dimerization and active site of HIV-1 protease by secondary metabolites from the Vietnamese mushroom Ganoderma Colossum. J. Nat. Prod. 2009, 72, 2019–2023. [Google Scholar] [CrossRef] [PubMed]
- Isaka, M.; Chinthanom, P.; Kongthong, S.; Srichomthong, K.; Choeyklin, R. Lanostane triterpenes from cultures of the Basidiomycete Ganoderma orbiforme BCC 22324. Phytochemistry 2013, 87, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Paterson, R.R.M. Ganoderma–a therapeutic fungal biofactory. Phytochemistry 2006, 67, 1985–2001. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.-C.; Yang, Z.-L.; Cui, B.-K.; Yu, C.-J.; Zhou, L.-W. Species diversity and utilization of medicinal mushrooms and fungi in China. Int. J. Med. Mushrooms 2009, 11, 287–302. [Google Scholar] [CrossRef]
- Cheng, C.R.; Yang, M.; Wu, Z.Y.; Wang, Y.; Zeng, F.; Wu, W.Y.; Guan, S.H.; Guo, D.A. Fragmentation pathways of oxygenated tetracyclic triterpenoids and their application in the qualitative analysis of Ganoderma lucidum by multistage tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2011, 25, 1323–1335. [Google Scholar] [CrossRef]
- Cao, Y.; Yuan, H.-S. Ganoderma mutabile sp. nov. from southwestern China based on morphological and molecular data. Mycol. Prog. 2013, 12, 121–126. [Google Scholar] [CrossRef]
- De Silva, D.D.; Rapior, S.; Fons, F.; Bahkali, A.H.; Hyde, K.D. Medicinal mushrooms in supportive cancer therapies: An approach to anti-cancer effects and putative mechanisms of action. Fungal Divers. 2012, 55, 1–35. [Google Scholar] [CrossRef]
- De Silva, D.D.; Rapior, S.; Hyde, K.D.; Bahkali, A.H. Medicinal mushrooms in prevention and control of diabetes mellitus. Fungal Divers. 2012, 56, 1–29. [Google Scholar] [CrossRef]
- De Silva, D.D.; Rapior, S.; Sudarman, E.; Stadler, M.; Xu, J.; Aisyah Alias, S.; Hyde, K.D. Bioactive metabolites from macrofungi: Ethnopharmacology, biological activities and chemistry. Fungal Divers. 2013, 62, 1–40. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, Z.; Cheng, C.; Qing, F. Synthesis and Antitumor Activity Evaluation of γ-Monofluorinated and γ, γ-Difluorinated Goniothalamin Analogues. Chin. J. Chem. 2013, 31, 805–812. [Google Scholar] [CrossRef]
- Hapuarachchi, K.; Wen, T.; Jeewon, R.; Wu, X.; Kang, J.; Hyde, K. Mycosphere Essays 7. Ganoderma lucidum-are the beneficial anti-cancer properties substantiated? Mycosphere 2016, 7, 305–332. [Google Scholar]
- Baby, S.; Johnson, A.J.; Govindan, B. Secondary metabolites from Ganoderma. Phytochemistry 2015, 114, 66–101. [Google Scholar] [CrossRef]
- Bishop, K.S.; Kao, C.H.; Xu, Y.; Glucina, M.P.; Paterson, R.R.M.; Ferguson, L.R. From 2000 years of Ganoderma lucidum to recent developments in nutraceuticals. Phytochemistry 2015, 114, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-H.; Lin, Y.-S.; Lin, K.-L.; Lu, Y.-L.; Chen, C.-H.; Chien, M.-Y.; Shang, H.-F.; Lin, S.-Y.; Hou, W.-C. Effects of hot-water extracts from Ganoderma lucidum residues and solid-state fermentation residues on prebiotic and immune-stimulatory activities in vitro and the powdered residues used as broiler feed additives in vivo. Bot. Stud. 2015, 56, 17. [Google Scholar] [CrossRef]
- Gąsecka, M.; Mleczek, M.; Siwulski, M.; Niedzielski, P.; Kozak, L. Phenolic and flavonoid content in Hericium erinaceus, Ganoderma lucidum, and Agrocybe aegerita under selenium addition. Acta Aliment. 2016, 45, 300–308. [Google Scholar] [CrossRef]
- Yalcin, O.U.; Sarikurkcu, C.; Cengiz, M.; Gungor, H.; Gungor, H.; Ćavar Zeljković, S. Ganoderma carnosum and Ganoderma pfeifferi: Metal concentration, phenolic content, and biological activity. Mycologia 2020, 112, 1–8. [Google Scholar] [CrossRef]
- Gil-Ramírez, A.; Pavo-Caballero, C.; Baeza, E.; Baenas, N.; Garcia-Viguera, C.; Marín, F.R.; Soler-Rivas, C. Mushrooms do not contain flavonoids. J. Funct. Foods 2016, 25, 1–13. [Google Scholar] [CrossRef]
- Ferreira, I.C.; Barros, L.; Abreu, R. Antioxidants in wild mushrooms. Curr. Med. Chem. 2009, 16, 1543–1560. [Google Scholar] [CrossRef]
- Palacios, I.; Lozano, M.; Moro, C.; D’Arrigo, M.; Rostagno, M.A.; Martínez, J.A.; García-Lafuente, A.; Guillamón, E.; Villares, A. Antioxidant properties of phenolic compounds occurring in edible mushrooms. Food Chem. 2011, 128, 674–678. [Google Scholar] [CrossRef]
- Stanikunaite, R.; Khan, S.I.; Trappe, J.M.; Ross, S.A. Cyclooxygenase-2 inhibitory and antioxidant compounds from the truffle Elaphomyces granulatus. Phytother. Res. 2009, 23, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Muszyńska, B.; Grzywacz-Kisielewska, A.; Kala, K.; Gdula-Argasińska, J. Anti-inflammatory properties of edible mushrooms: A review. Food Chem. 2018, 243, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Tapwal, A.; Pandey, S.; Borah, R.K.; Borah, D.; Borgohain, J. Macro-fungal diversity and nutrient content of some edible mushrooms of Nagaland, India. Nusant. Biosci. 2013, 5, 1–7. [Google Scholar] [CrossRef]
- Ghazala, I.; Haddar, A.; Romdhane, M.B.; Ellouz-Chaanouni, S. Screening and molecular identification of new microbial strains for production of enzymes of biotechnological interest. Braz. Arch. Biol. Technol. 2016, 59, 1–12. [Google Scholar] [CrossRef]
- Karthikeyan, V.; Ragunathan, R.; Johney, J.; Kabesh, K. Production, optimization and purification of laccase produced by Pleurotus ostreatus MH591763. Res. Rev. A J. Microbiol. Virol. 2019, 9, 56–64. [Google Scholar]
- Torres-Farradá, G.; Manzano Leon, A.M.; Rineau, F.; Ledo Alonso, L.L.; Sánchez-López, M.I.; Thijs, S.; Colpaert, J.; Ramos-Leal, M.; Guerra, G.; Vangronsveld, J. Diversity of ligninolytic enzymes and their genes in strains of the genus Ganoderma: Applicable for biodegradation of xenobiotic compounds? Front. Microbiol. 2017, 8, 898. [Google Scholar] [CrossRef]
- Aftab, A.; Siddique, I.; Aftab, M.; Yousaf, Z.; Chaudhry, F.A. Wood degrading mushrooms potentially strong towards laccase biosynthesis in pakistan. Wood Res. 2020, 65, 809–818. [Google Scholar] [CrossRef]
- Elissetche, J.-P.; Ferraz, A.; Freer, J.; Rodriguez, J. Enzymes produced by Ganoderma australe growing on wood and in submerged cultures. World J. Microbiol. Biotechnol. 2007, 23, 429–434. [Google Scholar] [CrossRef]
- Alshiekheid, M.A.; Umar, A.; Ameen, F.; Alyahya, S.A.; Dufosse, L. Biodegradation of chromium by laccase action of Ganoderma Multipileum. J. King Saud Univ. Sci. 2023, 35, 102948. [Google Scholar] [CrossRef]
- de Souza Silva, C.M.M.; De Melo, I.S.; De Oliveira, P.R. Ligninolytic enzyme production by Ganoderma spp. Enzym. Microb. Technol. 2005, 37, 324–329. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).