Genome-Wide Identification and Analysis of the MAPK and MAPKK Gene Families in Potato (Solanum tuberosum L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of MAPK and MAPKK Genes in Potato
2.2. Proteins Sequence and Phylogenetic Analysis of StMAPK and StMAPKK
2.3. Sequence Analysis of StMAPK and StMAPKK Genes
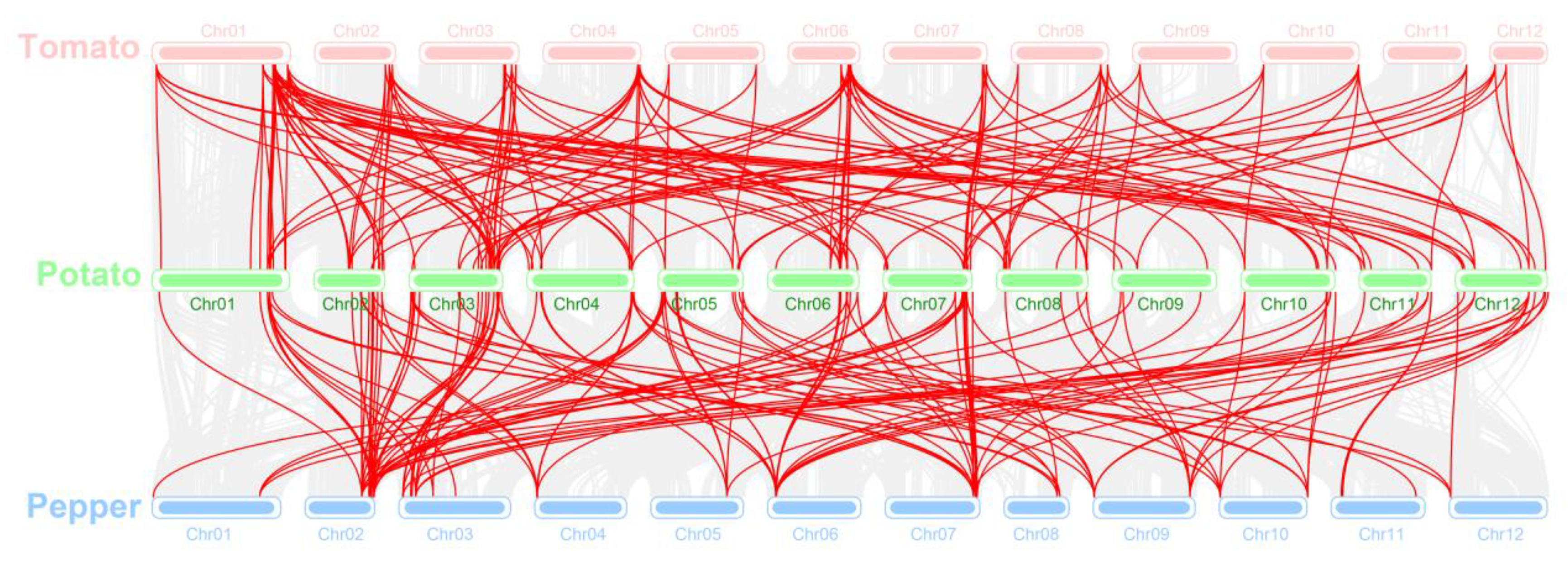
2.4. Synteny Analysis and Chromosomal Localization
2.5. StMAPKs and StMAPKKs Protein Characterization
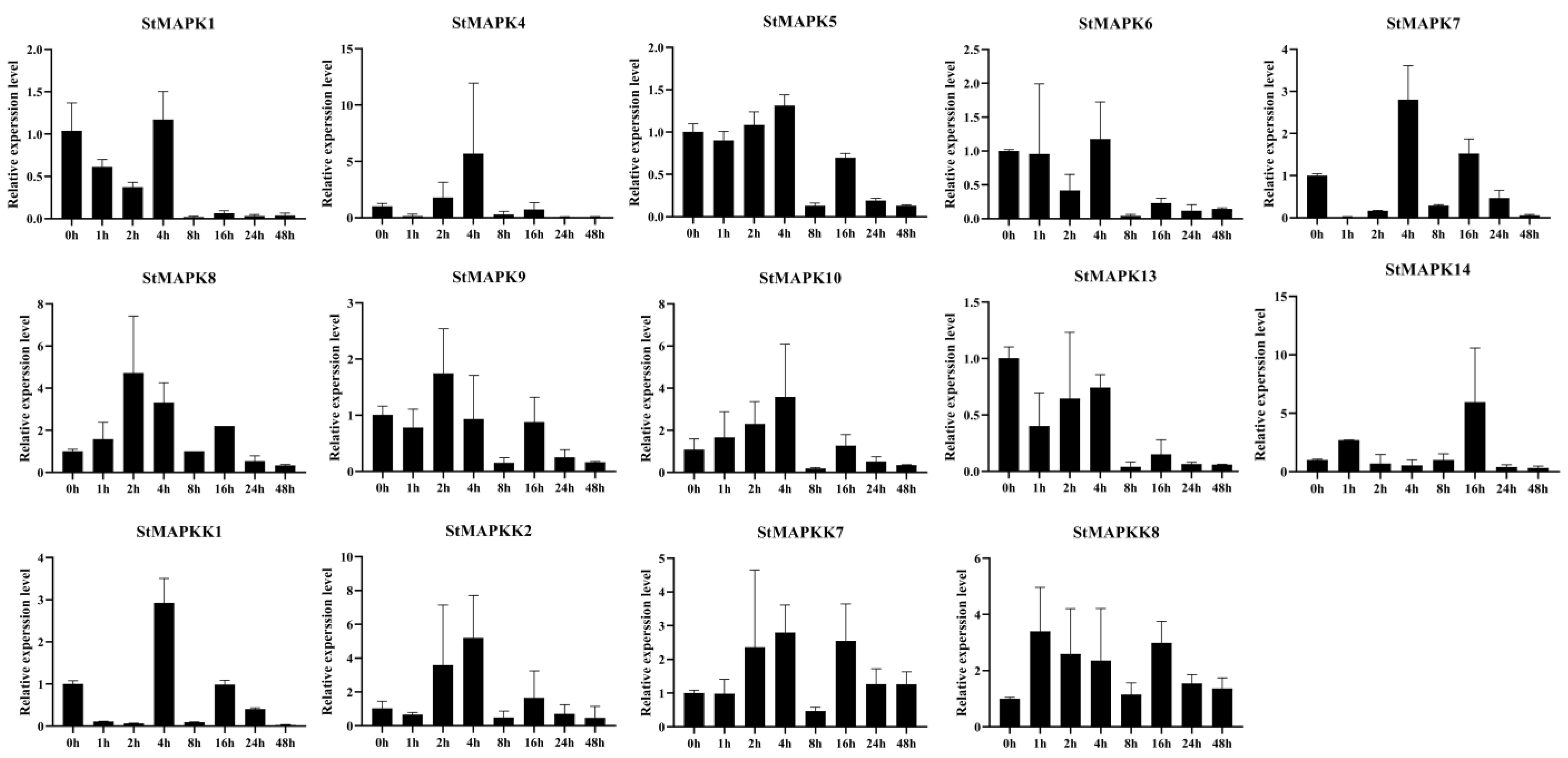
2.6. Expression Analysis of StMAPK and StMAPKK Family Members
3. Results
3.1. Identification of MAPK and MAPKK Genes and Multiple Alignment Analysis in Potato
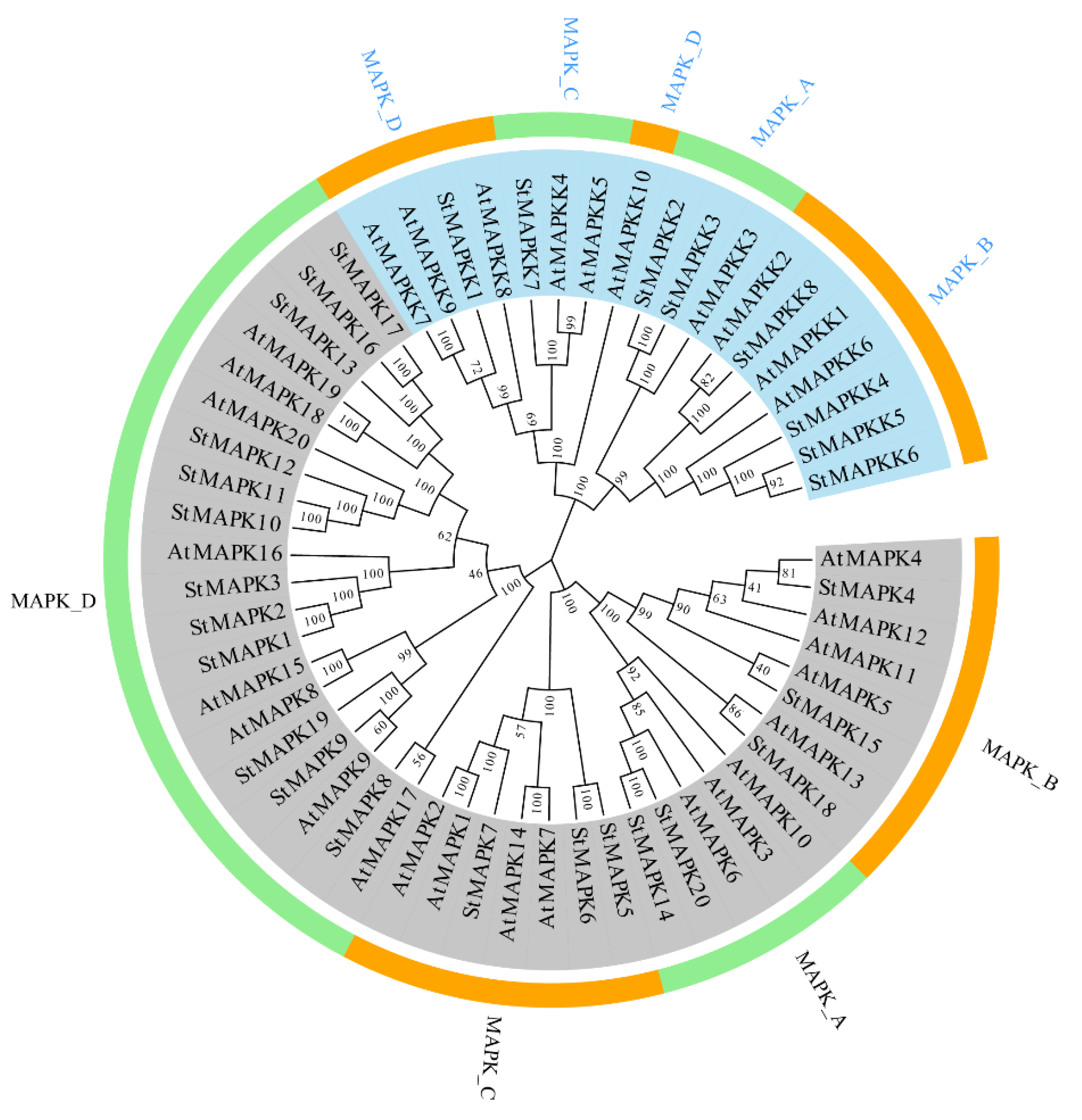
3.2. Phylogenetic Analysis of the MAPK and MAPKK Proteins
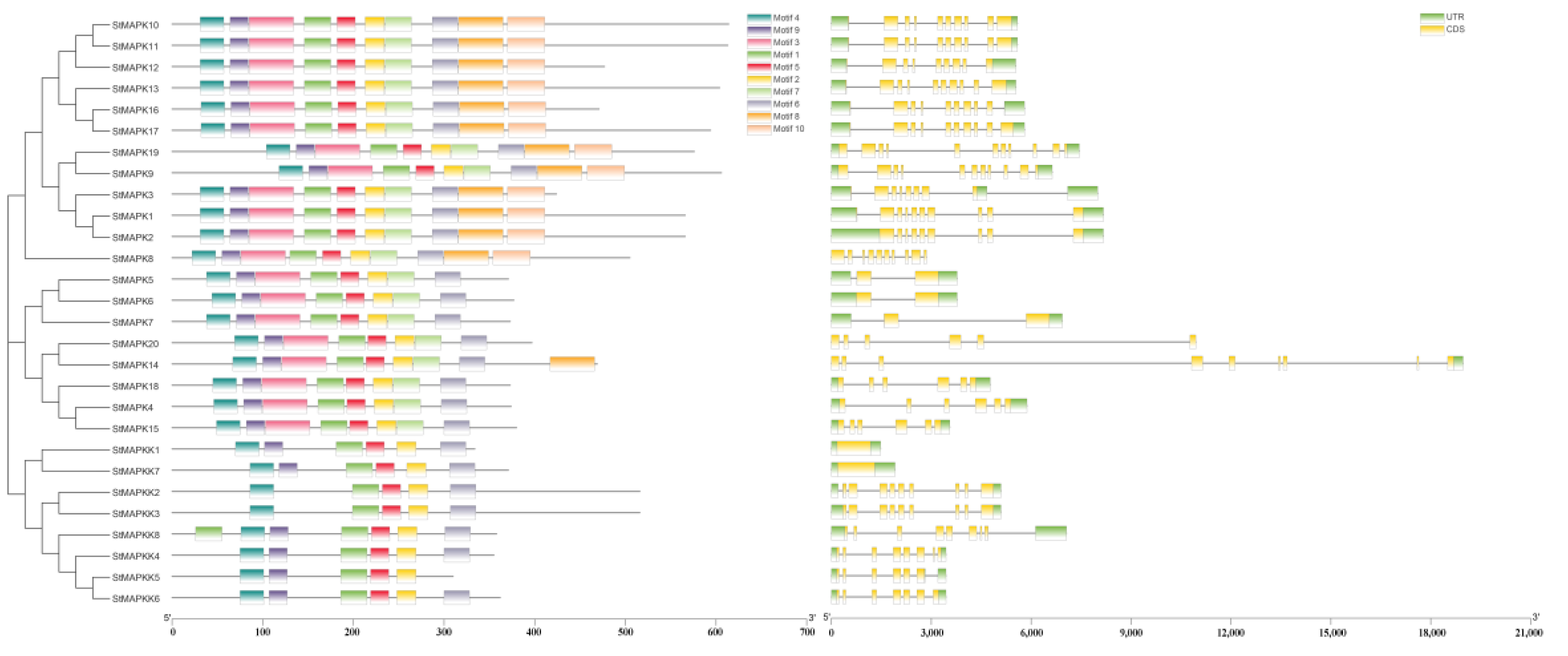
3.3. MAPK and MAPKK Gene Structure and Conserved Motif Analysis
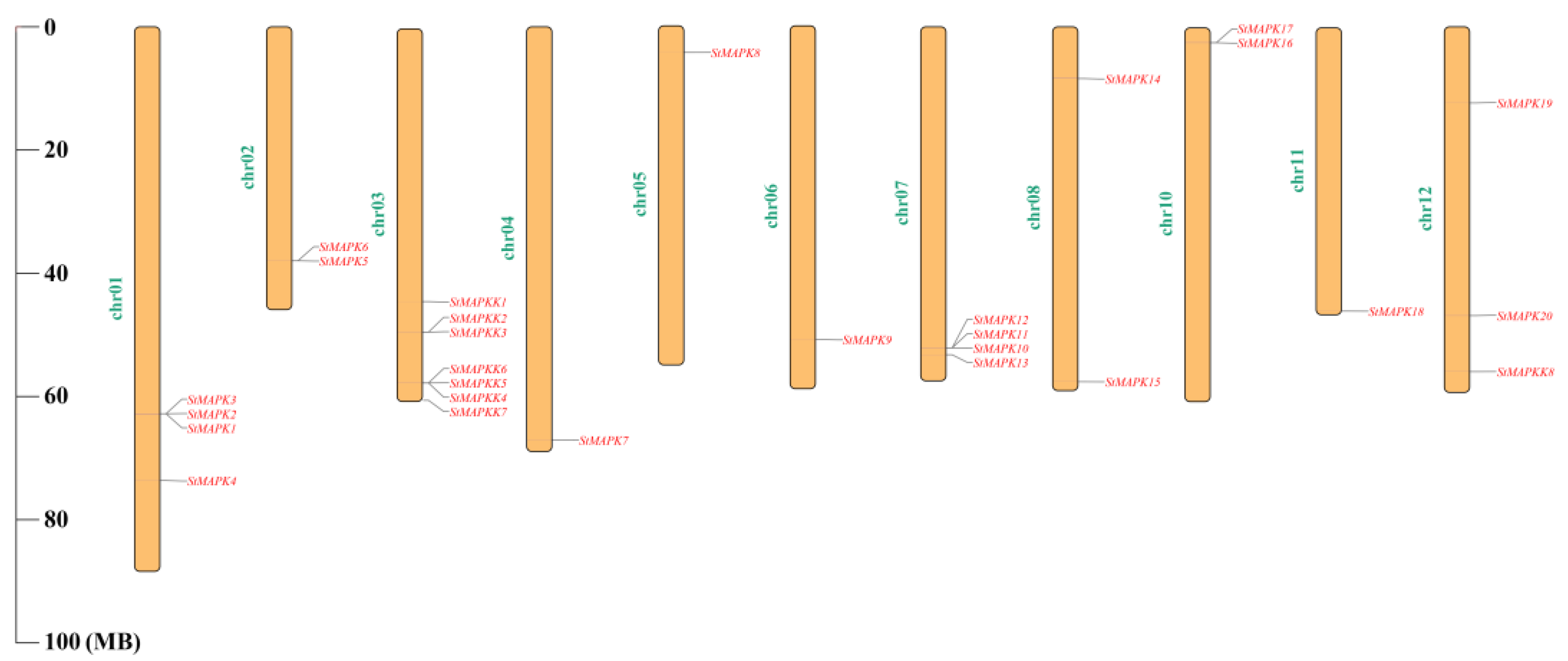
3.4. Chromosome Location of StMAPK and StMAPKK Genes
3.5. Promoter Sequence Analysis
3.6. Prediction of StMAPK and StMAPKK Protein Sequence Features
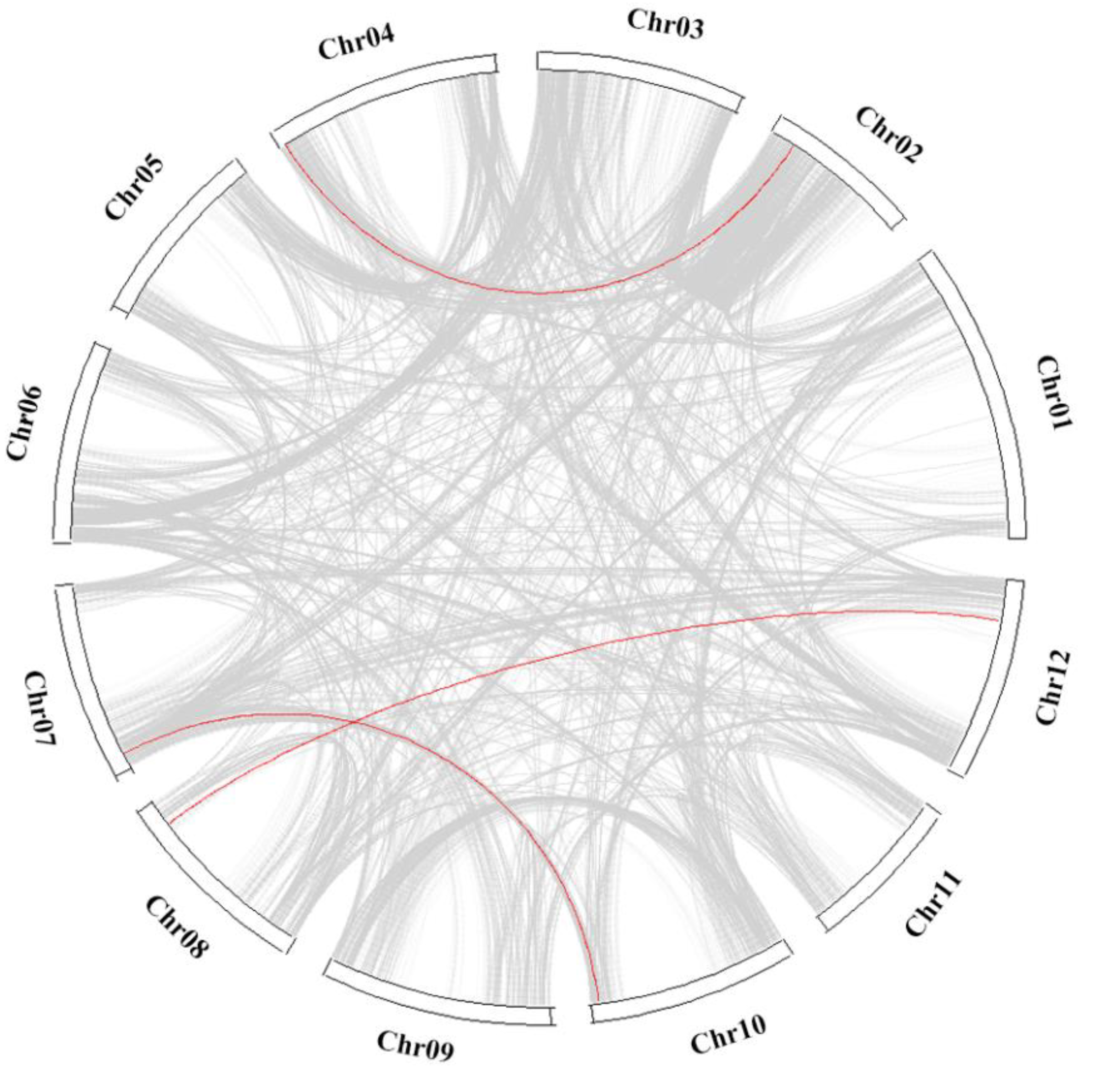
3.7. Collinearity of StMAPK and StMAPKK with Solanaceae Species
3.8. Expression Analysis of StMAPK and StMAPKK under Low-Temperature Stress
4. Discussion
4.1. Characterization and Evolution of StMAPKs and StMAPKKs in Potato
4.2. Expression Profile Analysis of Cold Resistance-Related StMAPK and StMAPKK Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mccully, M.; Canny, M.; Huang, C.X. The Management of Extracellular Ice by Petioles of Frost-resistant Herbaceous Plants. Ann. Bot. 2004, 94, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.T.; Ding, Y.; Yang, S.H. Molecular Regulation of CBF Signaling in Cold Acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ding, Y.L.; Shi, Y.T.; Zhang, X.Y.; Zhang, S.Q.; Gong, Z.Z.; Yang, S.H. MPK3 and MPK6 Mediated ICE1 Phosphorylation Negatively Regulates ICE1 Stability and Freezing Tolerance in Arabidopsis. Dev. Cell 2017, 43, 630–642.e4. [Google Scholar] [CrossRef] [PubMed]
- Danquah, A.; Zelicourt, A.D.; Colcombet, J.; Hirt, H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 2014, 32, 40–52. [Google Scholar] [CrossRef]
- Zhang, M.M.; Zhang, S.Q. Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 2022, 64, 301–341. [Google Scholar] [CrossRef]
- Tena, G.; Asai, T.; Chiu, W.L.; Sheen, J. Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 2001, 4, 392–400. [Google Scholar] [CrossRef]
- Zhang, M.M.; Su, J.B.; Zhang, Y.; Xu, J.; Zhang, S.Q. Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr. Opin. Plant Biol. 2018, 45, 1–10. [Google Scholar] [CrossRef]
- Boudsocq, M.; Danquah, A.; Zélicourt, A.; Hirt, H.; Colcombet, J. Plant MAPK cascades: Just rapid signaling modules? Plant Signal. Behav. 2015, 10, e1061197. [Google Scholar] [CrossRef][Green Version]
- Chen, Y.; Chen, L.; Sun, X.M.; Kou, S.; Liu, T.T.; Dong, J.K.; Tu, W.; Zhang, Y.L.; Song, B.T. The mitogen-activated protein kinase kinase MKK2 positively regulates constitutive cold resistance in the potato. Environ. Exp. Bot. 2022, 194, 104702. [Google Scholar] [CrossRef]
- Zhang, A.Y.; Jiang, M.Y.; Zhang, J.H.; Tan, M.P.; Hu, X.L. Mitogen-activated protein kinase is involved in abscisic acid-induced antioxidant defense and acts downstream of reactive oxygen species production in leaves of maize plants. Plant Physiol. 2006, 141, 475–487. [Google Scholar] [CrossRef]
- Kim, T.W.; Michniewicz, M.; Bergmann, D.C.; Wang, Z.Y. Brassinosteroid regulates stomatal development by gsk3-mediated inhibition of a MAPK pathway. Nature 2012, 482, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Adachi, H.; Nakano, T.; Miyagawa, N.; Ishihama, N.; Yoshioka, M.; Katou, Y.; Yaeno, T.; Shirasu, K.; Yoshioka, H. Wrky transcription factors phosphorylated by MAPK regulate a plant immune NADPH Oxidase in nicotiana benthamiana. Plant Cell. 2015, 27, 2645–2663. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lu, W.J.; He, X.W.; Wang, F.; Zhou, Y.L.; Guo, X.L.; Guo, X.Q. The cotton mitogen-activated protein kinase kinase 3 functions in drought tolerance by regulating stomatal responses and root growth. Plant Cell Physiol. 2016, 57, 1629–1642. [Google Scholar] [CrossRef] [PubMed]
- Smékalová, V.; Luptovčiak, I.; Komis, G.; Šamajová, O.; Ovečka, M.; Doskočilová, A.; Takáč, T.; Vadovič, P.; Novák, O.; Pechan, T.; et al. Involvement of YODA and mitogen activated protein kinase 6 in Arabidopsis post-embryogenic root development through auxin up-regulation and cell division plane orientation. New Phytol. 2014, 203, 1175–1193. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Boller, T. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–982. [Google Scholar] [CrossRef]
- Ding, Y.L.; Li, H.; Zhang, X.Y.; Xie, Q.; Gong, Z.Z.; Yang, S.H. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 2015, 32, 278–289. [Google Scholar] [CrossRef]
- Li, F.; Bian, C.S.; Xu, J.F.; Pang, W.F.; Liu, J.; Duan, S.G.; Lei, Z.G.; Jiwan, P.; Jin, L.P. Cloning and functional characterization of SAD genes in potato. PLoS ONE 2015, 10, e0122036. [Google Scholar] [CrossRef]
- Kou, S.; Chen, L.; Tu, W.; Scossa, F.; Wang, Y.; Liu, J.; Fernie, A.R.; Song, B.; Xie, C.H. The arginine decarboxylase gene ADC1, associated to the putrescine pathway, plays an important role in potato coldacclimated freezing tolerance as revealed by transcriptome and metabolome analyses. Plant J. 2018, 96, 1283–1298. [Google Scholar] [CrossRef]
- Chen, L.; Zhan, H.B.; Chen, Y.; Jiang, F.J.; Zhou, F.Y.; Liu, Q.; Fan, Y.Q.; Liu, T.T.; Tu, W.; Walther, D.; et al. Comparative transcriptomics analysis reveals a calcineurin B-like gene to positively regulate constitutive and acclimated freezing tolerance in potato. Plant Cell Environ. 2022, 45, 3305–3321. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef]
- Lozano, R.; Hamblin, M.T.; Prochnik, S.; Jannink, J. Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genom. 2015, 16, 360. [Google Scholar] [CrossRef] [PubMed]
- Hall, B. Building Phylogenetic Trees from Molecular Data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Hereditas 2007, 29, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Firth, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME Suite:tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Li, Q.H.; Yin, H.; Qi, K.J.; Li, L.T.; Wang, R.Z.; Zhang, S.L.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; Debarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Zhan, H.S.; Xian, H.Y.; Wang, Z.M.; Song, W.N.; Nie, X.J. Genome-Wide Identification and Analysis of MAPK and MAPKK Gene Families in Bread Wheat (Triticum aestivum L.). Genes 2017, 8, 284. [Google Scholar] [CrossRef]
- Hamel, L.P.; Nicole, M.C.; Sritubtim, S.; Morency, M.J.; Ellis, M.; Ehlting, J.; Beaudoin, N.; Barbazuk, B.; Klessig, D.; Lee, J.; et al. Ancient signals: Comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 2006, 11, 192–198. [Google Scholar] [CrossRef]
- Ning, J.; Li, X.; Hicks, L.M.; Xiong, L.Z. A Raf-Like MAPKKK Gene DSMT1 Mediates Drought Resistance through Reactive Oxygen Species Scavenging in Rice. Plant Physiol. 2010, 152, 876–890. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.K.; Shao, Y.M.; Ge, S.; Zhang, M.M.; Zhang, T.S.; Hu, X.T.; Liu, Y.D.; Walker, J.; Zhang, S.Q.; Xu, J. AMAPK cascade downstream of IDA-HAE/HSL2 ligand-receptor pair in lateral rootemergence. Nat. Plants 2019, 5, 414–423. [Google Scholar] [CrossRef]
- Chen, X.B.; Wang, J.; Zhu, M.; Jia, H.H.; Liu, D.D.; Hao, L.L.; Guo, X.Q. A cotton Raf-like MAP3K gene, GhMAP3K40, mediates reduced tolerance to biotic and abiotic stress in Nicotianabenthamiana by negatively regulating growth and development. Plant Sci. 2015, 240, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; He, X.W.; Li, Y.Z.; Wang, L.J.; Guo, X.L.; Guo, X.Q. The Cotton MAPK kinase GhMPK20 negatively regulates resistance to fusarium oxysporum by mediating the mkk4-mpk20-wrky40 cascade. Mol. Plant Pathol. 2017, 19, 1624–1638. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.D.; Zhang, X.H.; Xu, S.C.; Sun, L.L.; Jiang, M.Y.; Zhang, A.Y.; Jin, Y. Induction of protection against paraquat-induced oxidative damage by abscisic acid in maize leaves is mediated through mitogen-activated protein kinase. J. Integr. Plant Biol. 2009, 51, 961–972. [Google Scholar] [CrossRef]
- Reyna, N.S.; Yang, Y. Molecular analysis of the Rice MAP kinase gene family in relation to Magnaporthe grisea infection. Mol. Mol. Plant-Microbe Interact. 2006, 19, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Nicole, M.C.; Hamel, L.P.; Morency, M.J.; Beaudion, N.; Ellis, B.; Séguin, A. MAP-ping genomic organization and organ-specific expression profiles of poplar MAP kinases and MAP kinase kinases. BMC Genom. 2006, 7, 223. [Google Scholar] [CrossRef]
- Zhang, X.L.; Li, Y.P.; Xing, Q.J.; Qi, H.Y. Genome-wide identification of mitogen-activated protein kinase (MAPK) cascade and expression profiling of CmMAPKs in melon (Cucumis melo L.). PLoS ONE 2020, 15, e0232756. [Google Scholar] [CrossRef]
- Johnson, G.L.; Lapadat, R. Mitogen-activated protein kinase path ways mediated by ERK, JNK, and p38 protein kinases. Science 2002, 298, 1911–1912. [Google Scholar] [CrossRef]
- Krens, S.F.G.; Spaink, H.P.; Snaar-Jagalska, B.E. Functions of the MAPK family invertebrate-development. FEBS Lett. 2006, 580, 4984–4990. [Google Scholar] [CrossRef]
- Nakagami, H.; Pitzschke, A.; Hirt, H. Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 2005, 10, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.H.; Meng, X.Z.; Liu, Y.D.; Zheng, Z.Y.; Chen, Z.X.; Zhang, S.Q. Phosphorylation of a WRKY transcription factor by two pathogen responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell. 2012, 3, 1639–1653. [Google Scholar] [CrossRef]
- Teige, M.; Scheikl, E.; Eulgem, T.; Do’czi, R.; Ichimura, K.; Shinozak, K.; Dang, J.L.; Hirt, H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 2004, 15, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Xu, X.Y.; Yu, Y.J.; Chen, C.; Wang, J.; Cai, C.P.; Guo, W.Z. Integration analysis of MKK and MAPK family members highlights potential MAPK signaling modules incotton. Sci. Rep. 2016, 6, 29781. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, J.; Pan, C.T.; Guan, X.Y.; Wang, Y.; Liu, S.Y.; He, Y.J.; Chen, L.F.; Lu, G. Genome-wide identification of MAPKK and MAPKKK gene families in tomato andtranscriptional profiling analysis during development and stress response. PLoS ONE 2014, 9, e10303. [Google Scholar] [CrossRef]
- Chen, L.H.; Hu, W.; Tan, S.L.; Wang, M.; Ma, Z.B.; Zhou, S.Y.; Deng, X.M.; Zhang, Y.; Huang, C.; Yang, G.X. Genome-wide identification and analysis of MAPK and MAPKKgene families in Brachypodium distachyon. PLoS ONE 2012, 7, e46744. [Google Scholar] [CrossRef]
- Cui, L.C.; Yang, G.; Yan, J.L.; Pan, Y.; Nie, X.J. Genome-wide identification, expression profiles and regulatory network of MAPK cascade gene family in barley. BMC Genom. 2019, 20, 750. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.P.; Richa, T.; Kumar, K.; Raghuram, B.; Sinha, A.K. In silico analysis reveals 75 members of mitogen-activated protein kinase kinase kinase gene family in Rice. DNA Res. 2010, 17, 139–153. [Google Scholar] [CrossRef]
- Liang, W.; Yang, B.; Yu, B.-J.; Zhou, Z.; Li, C.; Jia, M.; Sun, Y.; Zhang, Y.; Wu, F.; Zhang, H.; et al. Identification and analysis of MKK and MPK gene families incanola (Brassica napus L.). BMC Genom. 2013, 14, 392. [Google Scholar] [CrossRef]
- Xie, G.S.; Kato, H.; Lmai, R. Biochemical identification of the OsMKK6-OsMPK3 signalling pathway for chilling stress tolerance in Rice. Biochem. J. 2012, 443, 95–102. [Google Scholar] [CrossRef]
- Zhao, C.Z.; Wang, P.C.; Si, T.; Hsu, C.C.; Wang, L.; Zayed, O.; Yu, Z.P.; Zhu, Y.F.; Dong, J.; Tao, W.; et al. Map kinase cascades regulate the cold response by modulating ice1 protein stability. Dev. Cell 2017, 43, 618–629.e5. [Google Scholar] [CrossRef]
- Agrawal, G.K.; Rakwal, R.; Iwahashi, H. Isolation of novel rice (Oryza sativa L.) multiple stress responsive MAP kinase gene, OsMSRMK2, whose mRNA accumulates rapidly in responseto environmental cues. Biochem. Biophys. Res. Commun. 2002, 295, 1009–1016. [Google Scholar] [CrossRef]
- Wang, J.X.; Ding, H.D.; Zhang, A.Y.; Ma, F.F.; Cao, J.M.; Jiang, M.Y. A novel mitogen-activated protein kinase gene in maize (Zea mays), ZmMPK3, is involved in response to diverse environmental cues. J. Integr. Plant Biol. 2010, 52, 442–452. [Google Scholar] [CrossRef]
- Berberich, T.; Sano, H.; Kusano, T. Involvement of a MAP kinase, ZmMPK5, in senescence and recovery from low-temperature stress in maize. Mol. Gen. Genet. 1999, 262, 534–542. [Google Scholar] [CrossRef]
- Cai, G.H.; Wang, G.D.; Wang, L.; Pan, J.W.; Liu, Y.; Li, D. ZmMKK1, a novel group a mitogen-activated protein kinase kinase gene in maize, conferred chilling stress tolerance and was involved in pathogen defense in transgenic tobacco. Plant Sci. 2014, 214, 57–73. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, R.R.; Zheng, Y.Y.; Chen, L.; Li, R.; Ma, J.F.; Hong, X.F.; Ma, P.; Sheng, J.P.; Shen, L. SlMAPK1/2/3 and antioxidant enzymes are associated with H2O2 induced chilling tolerance in tomato plants. J. Agric. Food Chem. 2017, 65, 6812–6820. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shang, Y.; Luo, X.; Zhang, H.; Chen, M.; Yin, W.; Cao, Z.; Deng, R.; Li, Y.; Li, F. Genome-Wide Identification and Analysis of the MAPK and MAPKK Gene Families in Potato (Solanum tuberosum L.). Agronomy 2023, 13, 93. https://doi.org/10.3390/agronomy13010093
Shang Y, Luo X, Zhang H, Chen M, Yin W, Cao Z, Deng R, Li Y, Li F. Genome-Wide Identification and Analysis of the MAPK and MAPKK Gene Families in Potato (Solanum tuberosum L.). Agronomy. 2023; 13(1):93. https://doi.org/10.3390/agronomy13010093
Chicago/Turabian StyleShang, Yutong, Xiaobo Luo, Heng Zhang, Mingjun Chen, Wang Yin, Zhenju Cao, Renju Deng, Yan Li, and Fei Li. 2023. "Genome-Wide Identification and Analysis of the MAPK and MAPKK Gene Families in Potato (Solanum tuberosum L.)" Agronomy 13, no. 1: 93. https://doi.org/10.3390/agronomy13010093
APA StyleShang, Y., Luo, X., Zhang, H., Chen, M., Yin, W., Cao, Z., Deng, R., Li, Y., & Li, F. (2023). Genome-Wide Identification and Analysis of the MAPK and MAPKK Gene Families in Potato (Solanum tuberosum L.). Agronomy, 13(1), 93. https://doi.org/10.3390/agronomy13010093





