Physiological and Metabolic Changes in ‘Xinyu Mandarin’ Following Natural Tetraploidization
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Sampling and Morphological Analysis
2.3. Determination of Physiological Indices
2.4. Non-Targeted Metabolic Profiling
2.5. Statistical Analysis
3. Results and Discussion
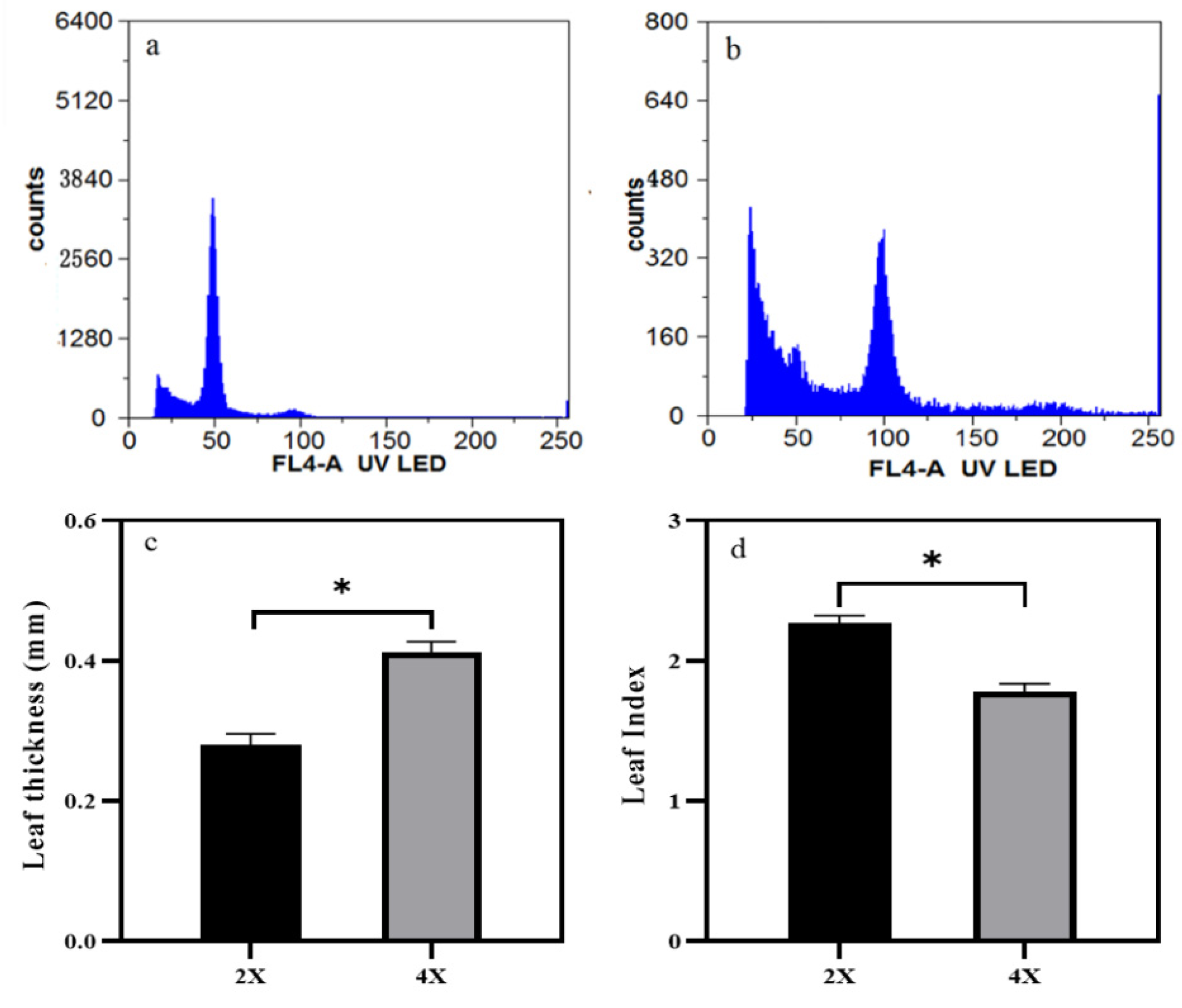
3.1. Changes of Morphology following Natural Tetraploidization
3.2. Changes of Physiological Indices following Natural Tetraploidization
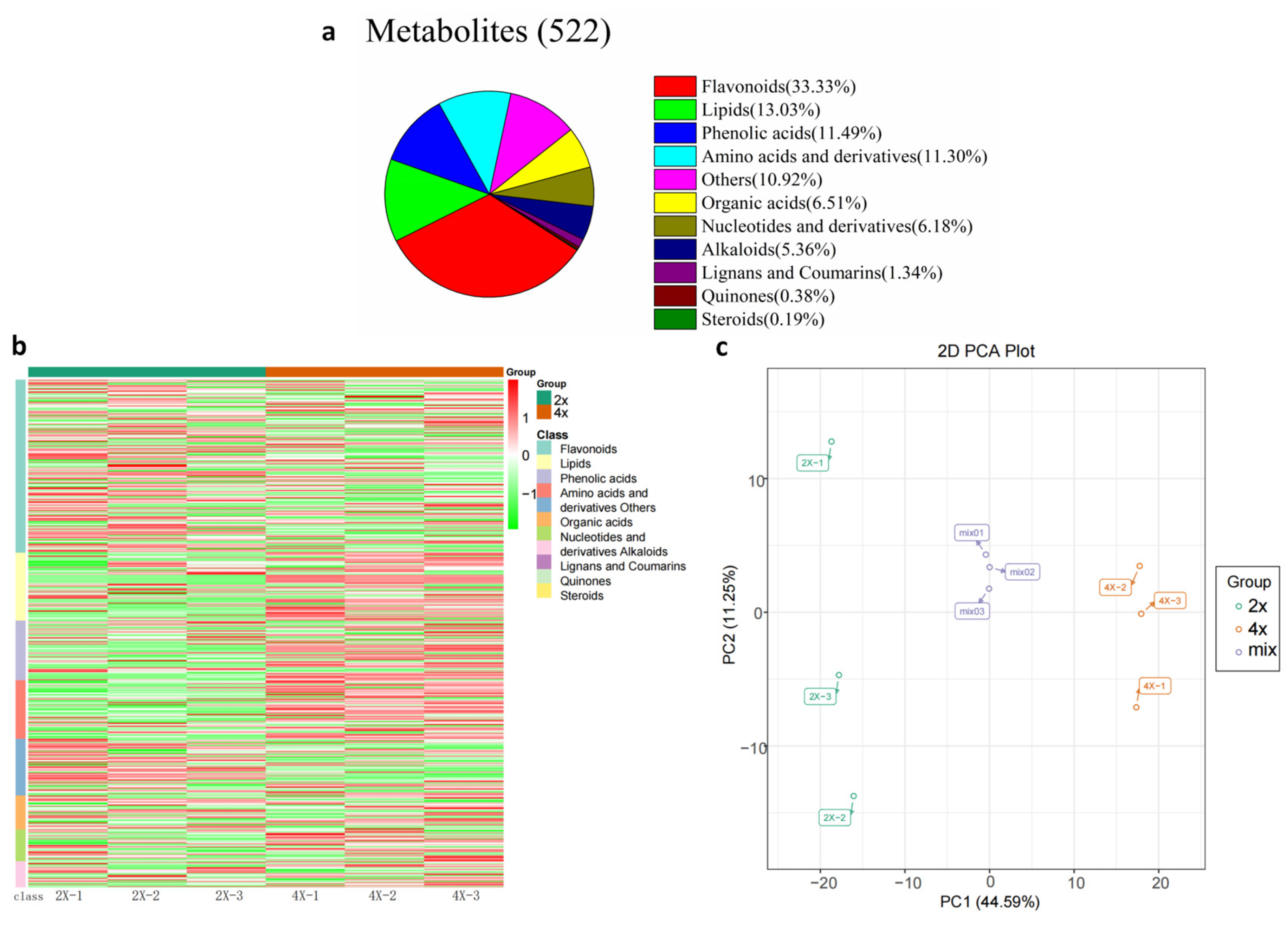
3.3. Non-Targeted Metabolic Analysis
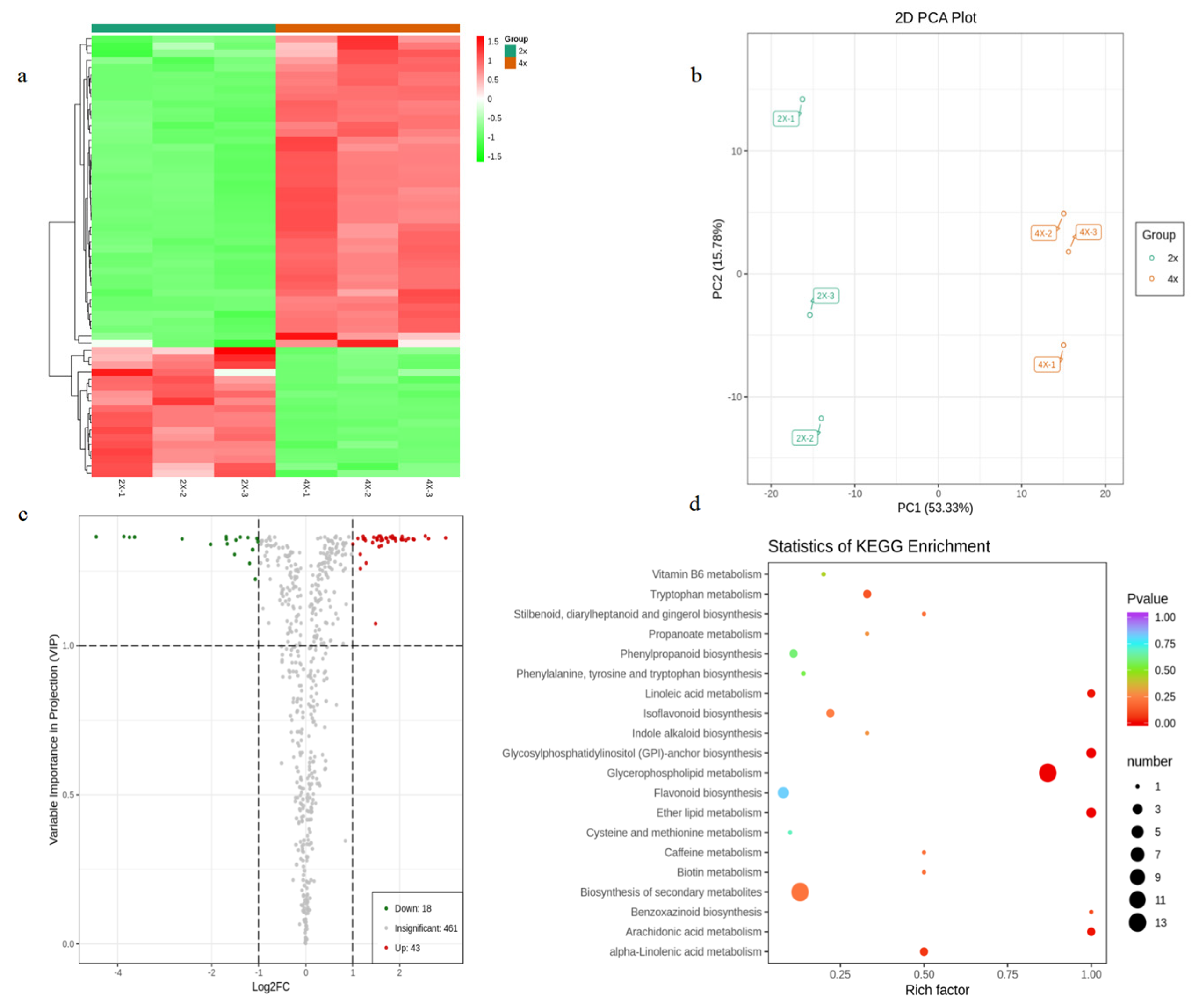
3.4. Changes of Metabolism following Natural Tetraploidization
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Van de Peer, Y.; Ashman, T.-L.; Soltis, P.S.; Soltis, D.E. Polyploidy: An evolutionary and ecological force in stressful times. Plant Cell 2021, 33, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Van De Peer, Y.; Mizrachi, Y.V.D.P.E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Bailey, E.C.; Kobielski, S.; Park, J.; Losick, V.P. Polyploidy in Tissue Repair and Regeneration. Cold Spring Harb. Perspect. Biol. 2021, 13, a040881. [Google Scholar] [CrossRef] [PubMed]
- Frawley, L.E.; Orr-Weaver, T.L. Polyploidy. Curr. Biol. 2015, 25, R353–R358. [Google Scholar] [CrossRef] [PubMed]
- Rothfels, C.J. Polyploid phylogenetics. New Phytol. 2021, 230, 66–72. [Google Scholar] [CrossRef]
- Chen, Z.J.; Sreedasyam, A.; Ando, A.; Song, Q.; De Santiago, L.M.; Hulse-Kemp, A.M.; Ding, M.; Ye, W.; Kirkbride, R.C.; Jenkins, J.; et al. Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat. Genet. 2020, 52, 525–533. [Google Scholar] [CrossRef]
- Huynh, S.; Broennimann, O.; Guisan, A.; Felber, F.; Parisod, C. Eco-genetic additivity of diploids in allopolyploid wild wheats. Ecol. Lett. 2020, 23, 663–673. [Google Scholar] [CrossRef]
- Ding, M.; Chen, Z.J. Epigenetic perspectives on the evolution and domestication of polyploid plant and crops. Curr. Opin. Plant Biol. 2018, 42, 37–48. [Google Scholar] [CrossRef]
- Liu, S.; Yang, Y.; Wei, F.; Duan, J.; Braynen, J.; Tian, B.; Cao, G.; Shi, G.; Yuan, J. Autopolyploidy leads to rapid genomic changes in Arabidopsis thaliana. Theory Biosci. 2017, 136, 199–206. [Google Scholar] [CrossRef]
- Scarrow, M.; Wang, Y.; Sun, G. Molecular regulatory mechanisms underlying the adaptability of polyploid plants. Biol. Rev. 2020, 96, 394–407. [Google Scholar] [CrossRef]
- Bao, Z.; Li, C.; Li, G.; Wang, P.; Peng, Z.; Cheng, L.; Li, H.; Zhang, Z.; Li, Y.; Huang, W.; et al. Genome architecture and tetrasomic inheritance of autotetraploid potato. Mol. Plant 2022, 15, 1211–1226. [Google Scholar] [CrossRef]
- Oliveira, I.D.B.; Amadeu, R.R.; Ferrão, L.F.V.; Muñoz, P.R. Optimizing whole-genomic prediction for autotetraploid blueberry breeding. Heredity 2020, 125, 437–448. [Google Scholar] [CrossRef]
- Garcia-Lozano, M.; Natarajan, P.; Levi, A.; Katam, R.; Lopez-Ortiz, C.; Nimmakayala, P.; Reddy, U.K. Altered chromatin conformation and transcriptional regulation in watermelon following genome doubling. Plant J. 2021, 106, 588–600. [Google Scholar] [CrossRef]
- Ollitrault, P.; Germana, M.A.; Froelicher, Y.; Cuenca, J.; Aleza, P.; Morillon, R.; Grosser, J.W.; Guo, W. Ploidy Manipulation for Citrus Breeding, Genetics, and Genomics. In The Citrus Genome; Gentile, A., la Malfa, S., Deng, Z., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 75–105. [Google Scholar]
- Zhao, N.; Grover, C.E.; Chen, Z.; Wendel, J.F.; Hua, J. Intergenomic gene transfer in diploid and allopolyploid Gossypium. BMC Plant Biol. 2019, 19, 492. [Google Scholar] [CrossRef]
- Coate, J.E.; Schreyer, W.M.; Kum, D.; Doyle, J.J. Robust Cytonuclear Coordination of Transcription in Nascent Arabidopsis thaliana Autopolyploids. Genes 2020, 11, 134. [Google Scholar] [CrossRef]
- Mattingly, K.Z.; Hovick, S.M. Autopolyploids of Arabidopsis thaliana are more phenotypically plastic than their diploid progenitors. Ann. Bot. 2021, 7, mcab081. [Google Scholar] [CrossRef]
- Dudits, D.; Török, K.; Cseri, A.; Paul, K.; Nagy, A.V.; Nagy, B.; Sass, L.; Ferenc, G.; Vankova, R.; Dobrev, P.; et al. Response of Organ Structure and Physiology to Autotetraploidization in Early Development of Energy Willow Salix viminalis. Plant Physiol. 2016, 170, 1504–1523. [Google Scholar] [CrossRef]
- Tan, F.-Q.; Tu, H.; Wang, R.; Wu, X.-M.; Xie, K.-D.; Chen, J.-J.; Zhang, H.-Y.; Xu, J.; Guo, W.-W. Metabolic adaptation following genome doubling in citrus doubled diploids revealed by non-targeted metabolomics. Metabolomics 2017, 13, 143. [Google Scholar] [CrossRef]
- Vergara, F.; Kikuchi, J.; Breuer, C. Artificial Autopolyploidization Modifies the Tricarboxylic Acid Cycle and GABA Shunt in Arabidopsis thaliana Col-0. Sci. Rep. 2016, 6, 26515. [Google Scholar] [CrossRef]
- Gantait, S.; Mukherjee, E. Induced autopolyploidy—A promising approach for enhanced biosynthesis of plant secondary metabolites: An insight. J. Genet. Eng. Biotechnol. 2021, 19, 4. [Google Scholar] [CrossRef]
- Chao, D.-Y.; Dilkes, B.; Luo, H.; Douglas, A.; Yakubova, E.; Lahner, B.; Salt, D.E. Polyploids Exhibit Higher Potassium Uptake and Salinity Tolerance in Arabidopsis. Science 2013, 341, 658–659. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xu, G.; Xia, X.; Wang, M.; Yin, X.; Zhang, B.; Zhang, X.; Cui, Y. Deciphering the physiological and molecular mechanisms for copper tolerance in autotetraploid Arabidopsis. Plant Cell Rep. 2017, 36, 1585–1597. [Google Scholar] [CrossRef] [PubMed]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2015, 243, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Coate, J.E. Autopolyploidy: An epigenetic macromutation. Am. J. Bot. 2020, 107, 1097–1100. [Google Scholar] [CrossRef] [PubMed]
- Cameron, J.W.; Frost, H.B. Genetic, breeding and nucellar embryony. In The Citrus Industry; Reuther, W., Batchelor, L.D., Webber, H.J., Eds.; University of California: Riverside, CA, USA, 1968; pp. 325–370. [Google Scholar]
- Gao, R.; Wang, H.; Dong, B.; Yang, X.; Chen, S.; Jiang, J.; Zhang, Z.; Liu, C.; Zhao, N.; Chen, F. Morphological, Genome and Gene Expression Changes in Newly Induced Autopolyploid Chrysanthemum lavandulifolium (Fisch. ex Trautv.) Makino. Int. J. Mol. Sci. 2016, 17, 1690. [Google Scholar] [CrossRef]
- Aleza, P.; Juárez, J.; Ollitrault, P.; Navarro, L. Production of tetraploid plants of non apomictic citrus genotypes. Plant Cell Rep. 2009, 28, 1837–1846. [Google Scholar] [CrossRef]
- Zeng, S.-H.; Chen, C.-W.; Hong, L.; Liu, J.-H.; Deng, X.-X. In vitro induction, regeneration and analysis of autotetraploids derived from protoplasts and callus treated with colchicine in Citrus. Plant Cell Tissue Organ Cult. PCTOC 2006, 87, 85–93. [Google Scholar] [CrossRef]
- Robinson, D.O.; Coate, J.E.; Singh, A.; Hong, L.; Bush, M.; Doyle, J.J.; Roeder, A.H. Ploidy and Size at Multiple Scales in the Arabidopsis Sepal. Plant Cell 2018, 30, 2308–2329. [Google Scholar] [CrossRef]
- Grosser, J.W.; Gmitter, F.G. Protoplast fusion for production of tetraploids and triploids: Applications for scion and rootstock breeding in citrus. Plant Cell Tissue Organ Cult. PCTOC 2010, 104, 343–357. [Google Scholar] [CrossRef]
- Guo, W.; Liang, W.; Xie, K.; Xia, Q.; Fu, J.; Guo, D.; Xie, Z.; Wu, X.; Xu, Q.; Yi, H.; et al. Exploitation of polyploids from 39 citrus seedling populations. Acta Hortic. 2016, 1135, 11–16. [Google Scholar] [CrossRef]
- Guo, W.-W.; Xiao, S.-X.; Deng, X.-X. Somatic cybrid production via protoplast fusion for citrus improvement. Sci. Hortic. 2013, 163, 20–26. [Google Scholar] [CrossRef]
- Li, L.-J.; Tan, W.-S.; Li, W.-J.; Zhu, Y.-B.; Cheng, Y.-S.; Ni, H. Citrus Taste Modification Potentials by Genetic Engineering. Int. J. Mol. Sci. 2019, 20, 6194. [Google Scholar] [CrossRef]
- Zheng, Z.; Chen, J.; Deng, X. Historical Perspectives, Management, and Current Research of Citrus HLB in Guangdong Province of China, Where the Disease has been Endemic for Over a Hundred Years. Phytopathology 2018, 108, 1224–1236. [Google Scholar] [CrossRef]
- Ye, J.; Liu, H.; Zhao, Z.; Xu, L.; Li, K.; Du, D. Fine mapping of the QTL cqSPDA2 for chlorophyll content in Brassica napus L. BMC Plant Biol. 2020, 20, 511. [Google Scholar] [CrossRef]
- Gujjar, R.S.; Banyen, P.; Chuekong, W.; Worakan, P.; Roytrakul, S.; Supaibulwatana, K. A Synthetic Cytokinin Improves Photosynthesis in Rice under Drought Stress by Modulating the Abundance of Proteins Related to Stomatal Conductance, Chlorophyll Contents, and Rubisco Activity. Plants 2020, 9, 1106. [Google Scholar] [CrossRef]
- Gabr, A.M.M.; Mabrok, H.B.; Abdel-Rahim, E.A.; El-Bahr, M.K.; Smetanska, I. Determination of lignans, phenolic acids and antioxidant capacity in transformed hairy root culture of Linum usitatissimum. Nat. Prod. Res. 2017, 32, 1867–1871. [Google Scholar] [CrossRef]
- Da Costa, M.V.J.; Ramegowda, V.; Sreeman, S.; Nataraja, K.N. Targeted Phytohormone Profiling Identifies Potential Regulators of Spikelet Sterility in Rice under Combined Drought and Heat Stress. Int. J. Mol. Sci. 2021, 22, 11690. [Google Scholar] [CrossRef]
- Beaulieu, J.M.; Leitch, I.J.; Patel, S.; Pendharkar, A.; Knight, C.A. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytol. 2008, 179, 975–986. [Google Scholar] [CrossRef]
- Tsukaya, H. Controlling Size in Multicellular Organs: Focus on the Leaf. PLOS Biol. 2008, 6, e174. [Google Scholar] [CrossRef]
- Ni, Z.; Kim, E.-D.; Ha, M.; Lackey, E.; Liu, J.; Zhang, Y.; Sun, Q.; Chen, Z.J. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 2008, 457, 327–331. [Google Scholar] [CrossRef]
- Hias, N.; Leus, L.; Davey, M.W.; Vanderzande, S.; Van Huylenbroeck, J.; Keulemans, J. Effect of polyploidization on morphology in two apple (Malus × domestica) genotypes. Hortic. Sci. 2017, 44, 55–63. [Google Scholar] [CrossRef]
- Allario, T.; Brumos, J.; Colmenero-Flores, J.M.; Iglesias, D.J.; Pina, J.A.; Navarro, L.; Talon, M.; Ollitrault, P.; Morillon, R. Tetraploid Rangpur lime rootstock increases drought tolerance via enhanced constitutive root abscisic acid production. Plant Cell Environ. 2012, 36, 856–868. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Haberer, G.; Matthes, M.C.; Rattei, T.; Mayer, K.F.X.; Gierl, A.; Torresruiz, R.A. Impact of natural genetic variation on the tran-scriptome of autotetraploid Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 17809–17814. [Google Scholar] [CrossRef] [PubMed]
- Samo, N.; Ebert, A.; Kopka, J.; Mozgová, I. Plant chromatin, metabolism and development—An intricate crosstalk. Curr. Opin. Plant Biol. 2021, 61, 102002. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, Y.; He, L.; Yang, J.; Fernie, A.R.; Luo, J. Natural variance at the interface of plant primary and specialized metabolism. Curr. Opin. Plant Biol. 2022, 67, 102201. [Google Scholar] [CrossRef]
- Rieseberg, T.P.; Dadras, A.; Fürst-Jansen, J.M.; Ashok, A.D.; Darienko, T.; de Vries, S.; Irisarri, I.; de Vries, J. Crossroads in the evolution of plant specialized metabolism. Semin. Cell Dev. Biol. 2022, 134, 37–58. [Google Scholar] [CrossRef]
- Liu, N.-J.; Hou, L.-P.; Bao, J.-J.; Wang, L.-J.; Chen, X.-Y. Sphingolipid metabolism, transport, and functions in plants: Recent progress and future perspectives. Plant Commun. 2021, 2, 100214. [Google Scholar] [CrossRef]
- Boutté, Y.; Jaillais, Y. Metabolic Cellular Communications: Feedback Mechanisms between Membrane Lipid Homeostasis and Plant Development. Dev. Cell 2020, 54, 171–182. [Google Scholar] [CrossRef]
- Igamberdiev, A.U.; Bykova, N.V. Role of organic acids in the integration of cellular redox metabolism and mediation of redox signalling in photosynthetic tissues of higher plants. Free Radic. Biol. Med. 2018, 122, 74–85. [Google Scholar] [CrossRef]
- Dehghanian, Z.; Habibi, K.; Dehghanian, M.; Aliyar, S.; Lajayer, B.A.; Astatkie, T.; Minkina, T.; Keswani, C. Reinforcing the bulwark: Unravelling the efficient applications of plant phenolics and tannins against environmental stresses. Heliyon 2022, 8, e09094. [Google Scholar] [CrossRef]
- Wen, W.; Alseekh, S.; Fernie, A.R. Conservation and diversification of flavonoid metabolism in the plant kingdom. Curr. Opin. Plant Biol. 2020, 55, 100–108. [Google Scholar] [CrossRef]
- Tan, F.-Q.; Zhang, M.; Xie, K.-D.; Fan, Y.-J.; Song, X.; Wang, R.; Wu, X.-M.; Zhang, H.-Y.; Guo, W.-W. Polyploidy remodels fruit metabolism by modifying carbon source utilization and metabolic flux in Ponkan mandarin (Citrus reticulata Blanco). Plant Sci. 2019, 289, 110276. [Google Scholar] [CrossRef]
- Watkins, J.L.; Facchini, P.J. Compartmentalization at the interface of primary and alkaloid metabolism. Curr. Opin. Plant Biol. 2022, 66, 102186. [Google Scholar] [CrossRef]

| Hormone Component | Group | |
|---|---|---|
| ‘Xinyu Mandarin’ 2X | ‘Xinyu Mandarin’ 4X | |
| Abscisic acid/ABA | 12.97 ± 1.16 | 20.5 ± 2.00 * |
| 3-Indoleacetic acid/IAA | N/A | 4.29 ± 0.26 |
| N6-(2-Isopentenyl) adenosine/IP | 0.0270 ± 0.0007 | 0.0381 ± 0.0014 * |
| BioReagent/tZ | 0.221 ± 0.012 | 0.154 ± 0.003 * |
| Compounds | Fold Change | Type | Compounds | Fold Change | Type |
|---|---|---|---|---|---|
| Amino acids and derivatives | |||||
| 4-Hydroxy–L–glutamic acid | 2.24 | up | L–Homocystine | 2.44 | up |
| organic acid compounds | |||||
| (S)–2–Hydroxybutanoicacid | 0.07 | down | 5–Hydroxyhexanoic acid | 0.31 | down |
| α–Hydroxyisobutyric acid | 0.08 | down | 2–Hydroxybutanoic acid | 0.07 | down |
| lipid compounds | |||||
| 1–Stearoyl–sn-glycero–3–phosphocholine | 4.55 | up | LysoPC 17:0 | 4.10 | up |
| LysoPE 18:1 | 4.20 | up | LysoPC 14:0(2n isomer) | 3.06 | up |
| LysoPC 18:3 | 2.35 | up | LysoPC 16:0(2n isomer) | 3.36 | up |
| LysoPC 16:0 | 3.29 | up | LysoPC 18:0 | 4.95 | up |
| LysoPE 18:1(2n isomer) | 4.10 | up | LysoPE 16:0(2n isomer) | 3.77 | up |
| LysoPC 16:2(2n isomer) | 2.87 | up | LysoPC (18:2) | 3.59 | up |
| LysoPE 14:0 | 3.06 | up | LysoPC (18:2) isomer | 3.75 | up |
| LysoPC 18:3(2n isomer) | 2.39 | up | LysoPC(16:1) | 2.86 | up |
| LysoPE 18:2(2n isomer) | 6.12 | up | LysoPC(18:2) | 3.68 | up |
| LysoPE 16:0 | 3.69 | up | LysoPC(16:0) | 3.11 | up |
| LysoPC 15:1 | 2.68 | up | LysoPC(18:1) | 3.03 | up |
| LysoPC 15:0 | 2.85 | up | LysoPC(18:0) | 4.87 | up |
| Compounds | Fold Change | Type | Compounds | Fold Change | Type |
|---|---|---|---|---|---|
| Phenolic acid compounds | |||||
| Di–p–Coumaroyltartaric acid | 0.44 | down | p–Coumaroylferuloyltartaric acid | 0.31 | down |
| Neochlorogenic acid (5–O–Caffeoylquinic acid) | 5.84 | up | 3–O–(E)–p–Coumaroyl quinic acid | 2.33 | up |
| Chlorogenic acid | 2.72 | up | 6’–trans–Cinnamoyl–8–epikingisidic acid | 4.14 | up |
| Chlorogenic acid methyl ester | 2.41 | up | Trans-3–O–p–coumaric quinic acid | 3.01 | up |
| Cryptochlorogenic acid | 7.88 | up | Cis–3–p–coumaric quinic acid | 3.59 | up |
| 3–O–Feruloyl quinic acid | 2.94 | up | Scopoletin(7-Hydroxy-5-methoxycoumarin) | 2.80 | up |
| Flavonoids | |||||
| Naringenin–7–O–glucoside | 2.24 | up | 5,6,7,8,3′,4′–Hexamethoxyflavanone | 0.43 | down |
| 6–C–Hexosy–apigenin O–feruloylhexoside | 0.25 | down | Apigenin–7–O–(6–O–Malonyl Glucoside) | 0.36 | down |
| Kaempferol 7–O–rhamnoside | 0.31 | down | Apigenin–7,4′–dimethylether | 0.38 | down |
| Apigenin 7–rutinoside (Isorhoifolin) | 0.49 | down | Linarin | 0.16 | down |
| Alkaloids | |||||
| Tryptamine | 2.95 | up | Indole | 2.16 | up |
| 3–{(2–Aminoethoxy) (hydroxy) phosphoryl] oxy}–2–12–octadecadienoate | 4.39 | up | 3–{(2–Aminoethoxy) (hydroxy) phosphoryl] oxy}–2–hydroxypropyl palmitate | 3.47 | up |
| Others | |||||
| Xanthosine | 2.00 | up | Propyl2–(trimethylammonio) ethyl phosphate | 3.27 | up |
| 1,1–Kestotetraose | 0.48 | down | Biotin | 0.35 | down |
| D–(+)–Melezitose | 0.46 | down | Limonin | 0.05 | down |
| Pyridoxine | 0.49 | down | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Wan, S.; Tang, Y.; Yang, H.; Xu, C.; Liu, X.; Hu, Z.; Hu, X. Physiological and Metabolic Changes in ‘Xinyu Mandarin’ Following Natural Tetraploidization. Agronomy 2023, 13, 29. https://doi.org/10.3390/agronomy13010029
Wang Y, Wan S, Tang Y, Yang H, Xu C, Liu X, Hu Z, Hu X. Physiological and Metabolic Changes in ‘Xinyu Mandarin’ Following Natural Tetraploidization. Agronomy. 2023; 13(1):29. https://doi.org/10.3390/agronomy13010029
Chicago/Turabian StyleWang, Yuting, Shuilin Wan, Yuqing Tang, Huidong Yang, Chao Xu, Xincheng Liu, Zhongdong Hu, and Xinlong Hu. 2023. "Physiological and Metabolic Changes in ‘Xinyu Mandarin’ Following Natural Tetraploidization" Agronomy 13, no. 1: 29. https://doi.org/10.3390/agronomy13010029
APA StyleWang, Y., Wan, S., Tang, Y., Yang, H., Xu, C., Liu, X., Hu, Z., & Hu, X. (2023). Physiological and Metabolic Changes in ‘Xinyu Mandarin’ Following Natural Tetraploidization. Agronomy, 13(1), 29. https://doi.org/10.3390/agronomy13010029





