Involvement of CYP51A and CYP51B in Growth, Reproduction, Pathogenicity, and Sensitivity to Fungicides in Colletotrichum siamense
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Colletotrichum spp.
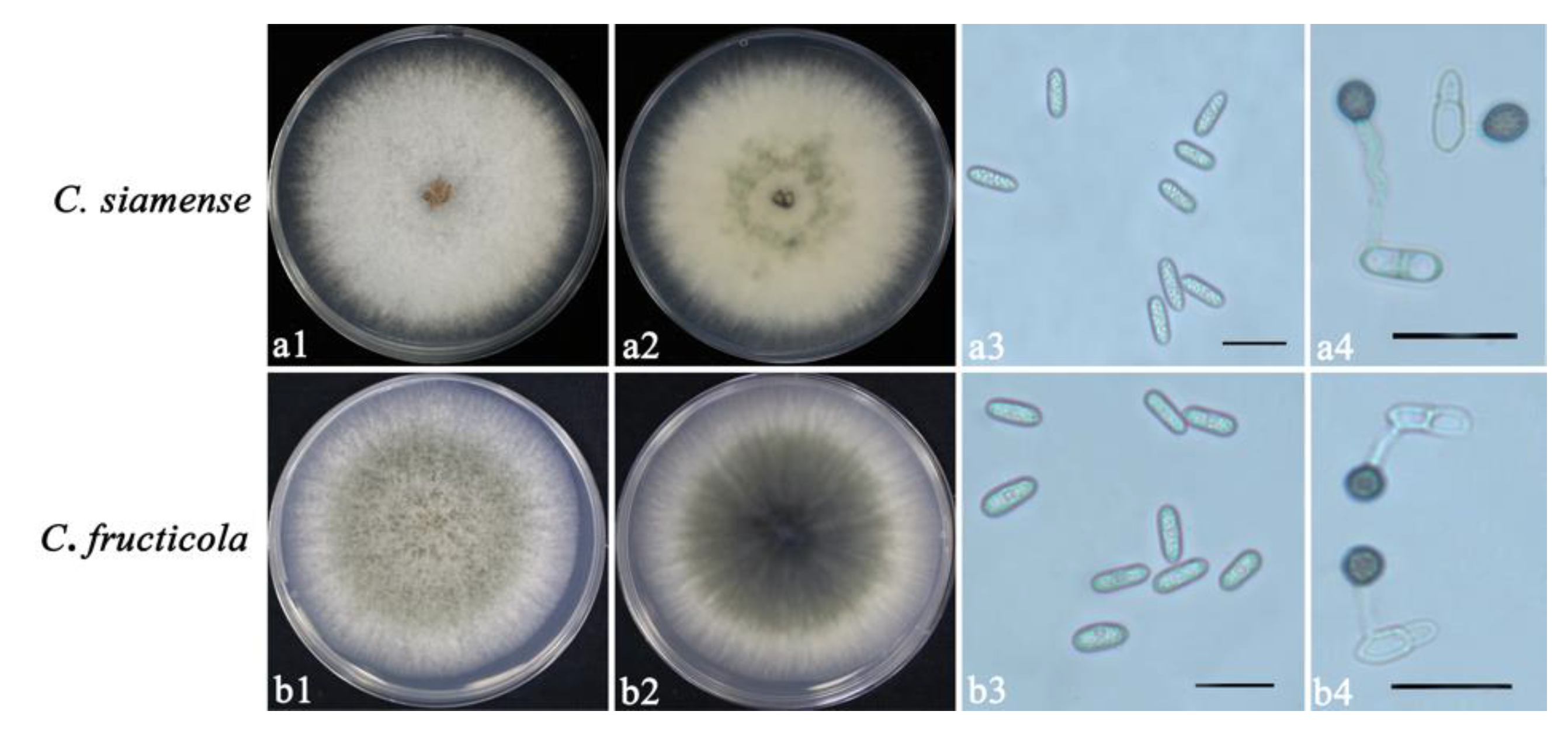
2.2. Morphological Characterization
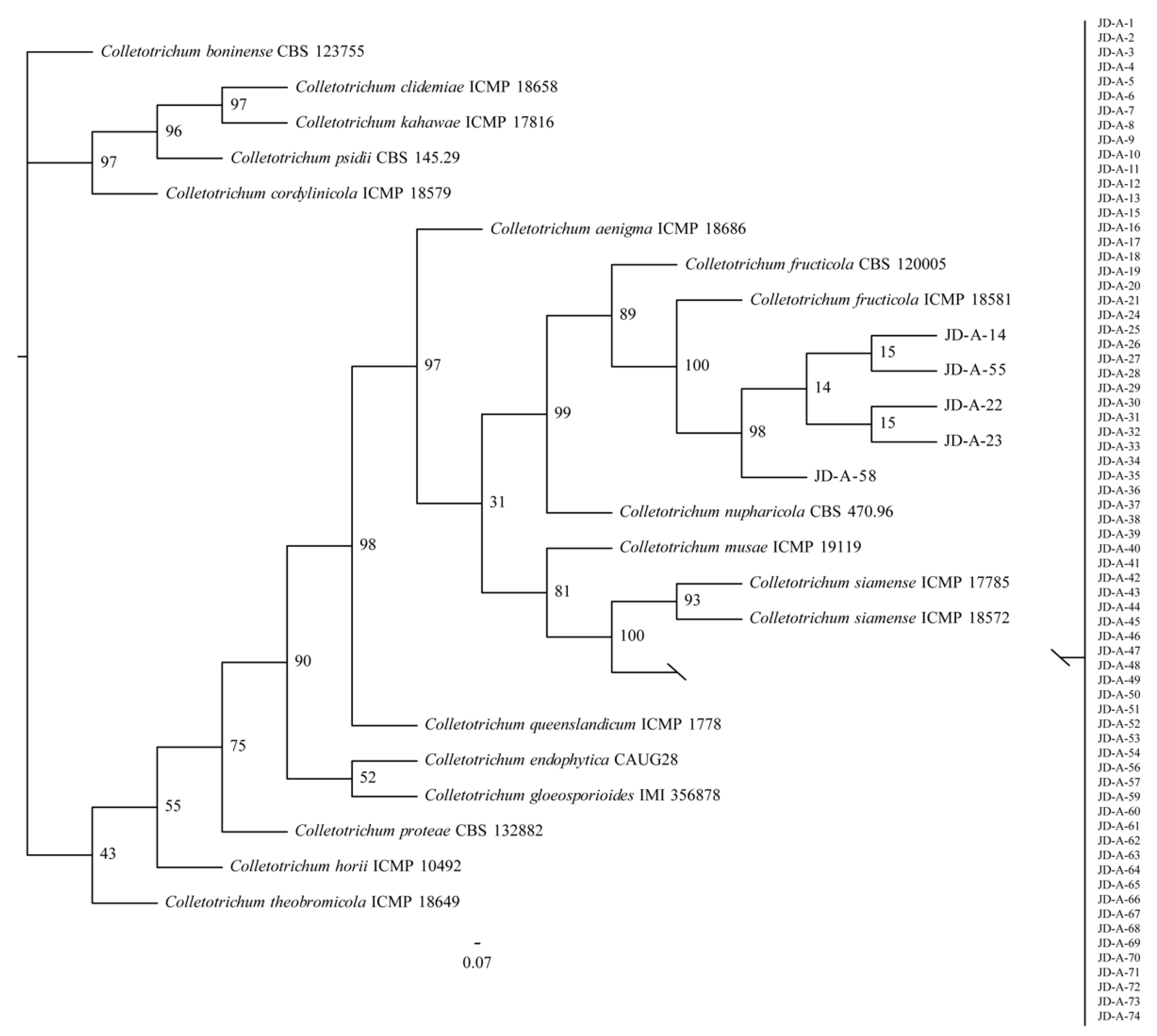
2.3. Molecular Identification and Phylogenetic Analysis
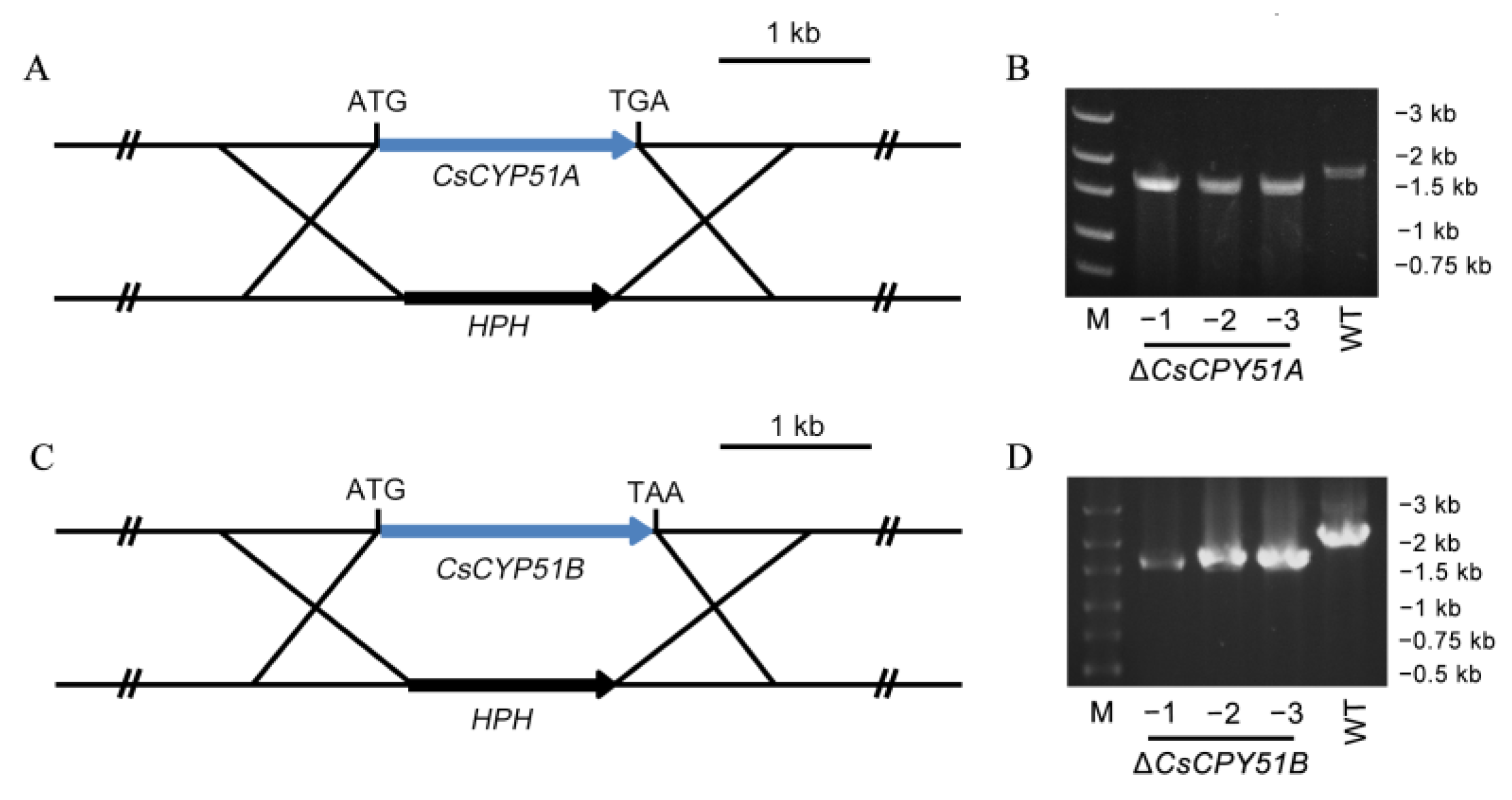
2.4. Construction of CsCYP51A and CsCYP51B Deletion Mutants
2.5. Transformation of C. siamense
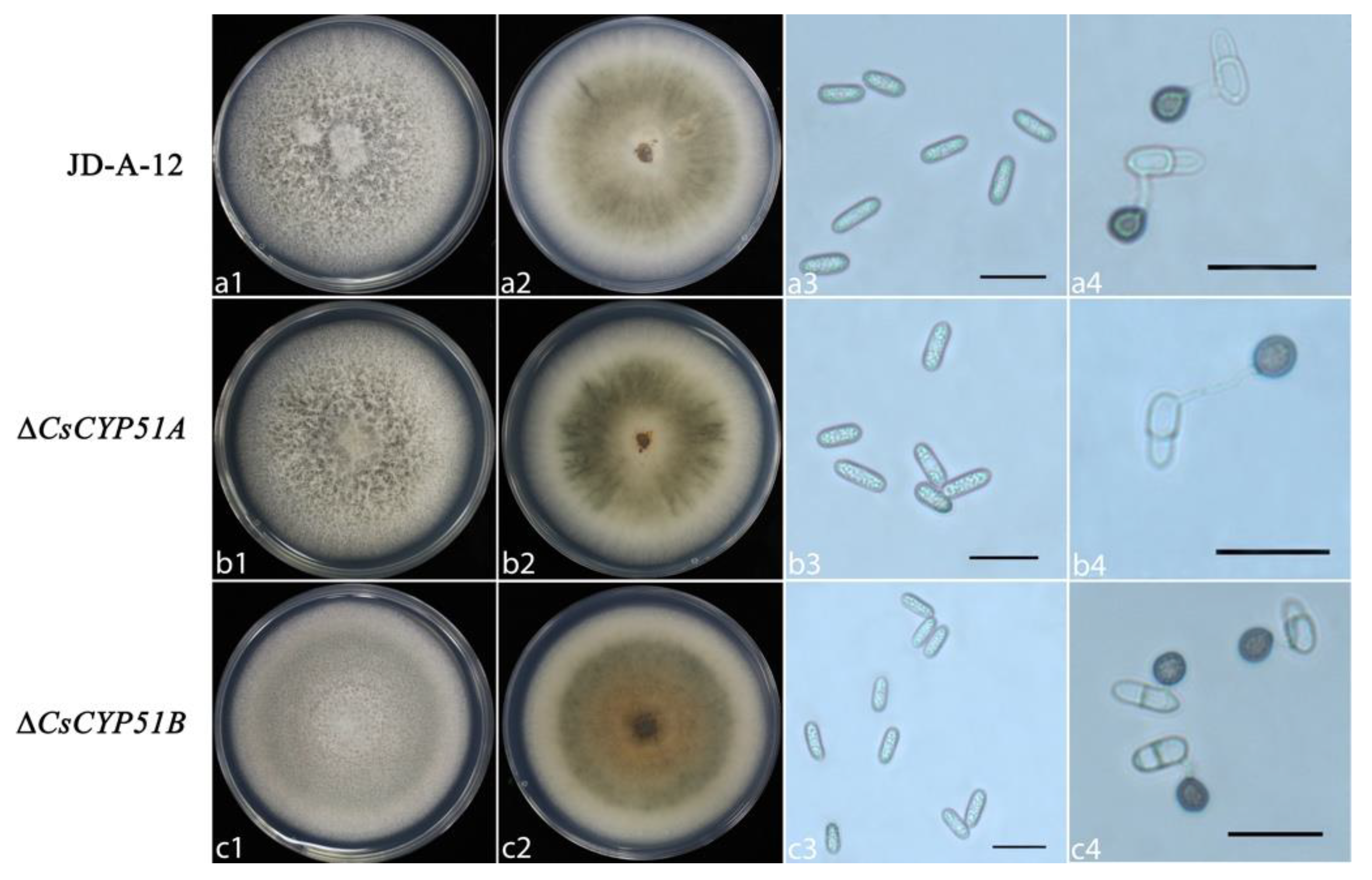
2.6. Phenotype Analysis
2.7. Pathogenicity Assays
2.8. Determination of Sensitivity to DMI Fungicides
3. Results
3.1. Isolation and Identification of Colletotrichum spp.
3.2. Deletion of CsCYP51 in C. siamense
3.3. Biological Characteristics of CsCYP51 Mutants
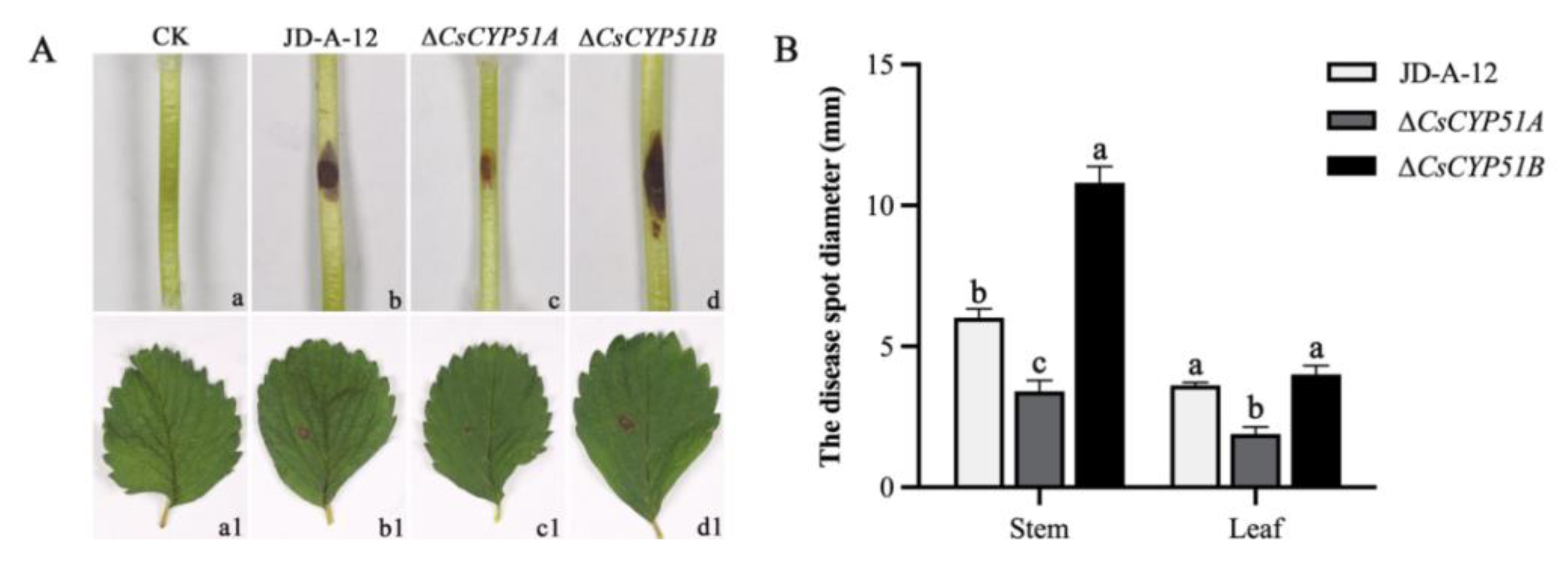
3.4. Effect of CYP51 on Pathogenicity
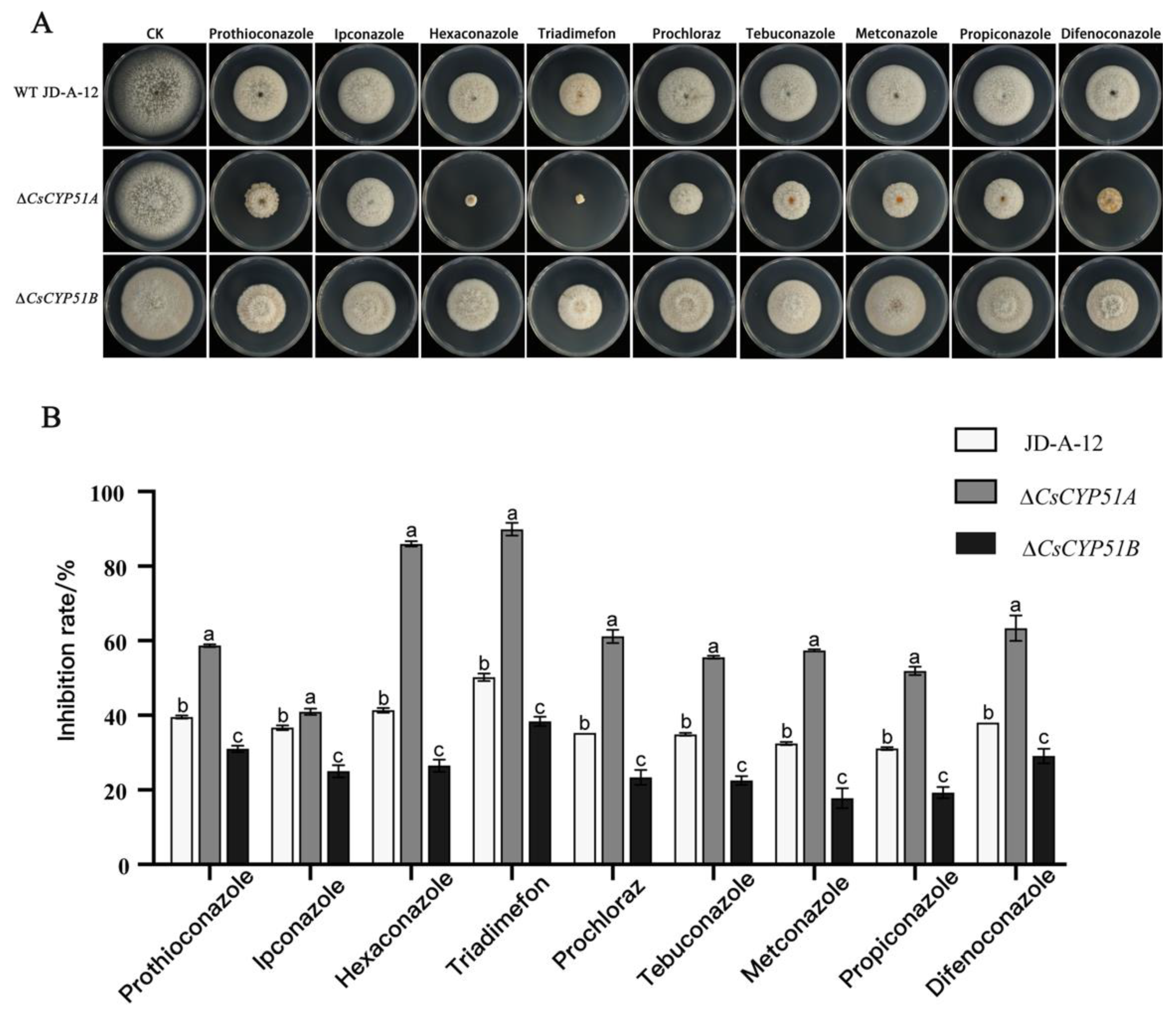
3.5. Effects of CYP51 Gene Deletion on the Sensitivity of C. siamense to DMIs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barbey, C.R.; Lee, S.; Verma, S.; Bird, K.A.; Yocca, A.E.; Edger, P.P.; Knapp, S.J.; Whitaker, V.M.; Folta, K.M. Disease resistance genetics and genomics in octoploid strawberry. G3 Genes Genom. Genet. 2019, 9, 3315–3332. [Google Scholar] [CrossRef] [PubMed]
- Paynter, M.; Gomez, A.; Ko, L.; Herringtonet, M.E. Research into crown rot and wilt diseases of strawberries in Queensland. Acta Hortic. 2016, 1117, 163–170. [Google Scholar] [CrossRef]
- Jacobs, R.L.; Adhikari, T.B.; Pattison, J.; Yencho, G.C.; Fernandez, G.E.; Louws, F.J. Assessing rate-reducing foliar resistance to anthracnose crown rot and fruit rot in strawberry. Plant Dis. 2020, 104, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Howard, C.M.; Maas, J.L.; Chandler, C.K.; Albregts, E.E. Anthracnose of strawberry caused by the Colletotrichum complex in Florida. Plant Dis. 1992, 76, 976–981. [Google Scholar] [CrossRef]
- Peres, N.; Timmer, L.; Adaskaveg Je Correll, J. Lifestyles of Colletotrichum acutatum. Plant Dis. 2005, 89, 784–796. [Google Scholar] [CrossRef]
- Zhang, Y.T.; Yu, H.; Hu, M.H.; Wu, J.Y.; Zhang, C.Q. Fungal pathogens associated with strawberry crown rot disease in China. J. Fungi 2022, 8, 1161. [Google Scholar] [CrossRef]
- Chen, X.Y.; Dai, D.J.; Zhao, S.F.; Shen, Y.; Wang, H.D.; Zhang, C.Q. Genetic diversity of Colletotrichum spp. causing strawberry anthracnose in Zhejiang, China. Plant Dis. 2020, 104, 1351–1357. [Google Scholar] [CrossRef]
- Baroncelli, R.; Zapparata, A.; Sarrocco, S.; Sukno, S.A.; Lane, C.R.; Thon, M.R.; Vannacci, G.; Holub, E.; Sreenivasaprasad, S. Molecular diversity of anthracnose pathogen populations associated with UK strawberry production suggests multiple introductions of three different Colletotrichum species. PLoS ONE 2015, 10, e0129140. [Google Scholar] [CrossRef]
- Fernández-Ortuño, D.; Loza-Reyes, E.; Atkins, S.L.; Fraaije, B.A. The CYP51C gene, a reliable marker to resolve interspecific phylogenetic relationships within the Fusarium species complex and a novel target for species-specific PCR. Int. J. Food Microbiol. 2010, 144, 301–309. [Google Scholar] [CrossRef]
- D’elye, C.; Bousset, L.; Corio-Costet, M.F. PCR cloning and detection of point mutations in the eburicol 14a-demethylase (CYP51) gene from Erysiphe graminis f. sp. hordei, a “recalcitrant” fungus. Curr. Genet. 1998, 34, 399–403. [Google Scholar]
- Dudakova, A.; Spiess, B.; Tangwattanachuleeporn, M.; Sasse, C.; Buchheidt, D.; Weig, M.; Groß, U.; Bader, O. Molecular tools for the detection and deduction of azole antifungal drug resistance phenotypes in Aspergillus species. Clin. Microbiol. Rev. 2017, 30, 1065–1091. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Proffer, T.J.; Jacobs, J.L.; Sundin, G.W. Overexpression of the 14a-demethylase target gene (CYP51) mediates fungicide resistance in Blumeriella jaapii. Appl. Environ. Microbiol. 2006, 72, 2581–2585. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, H.; Hasegawa, K.; Nakaune, R.; Lee, Y.J.; Makizumi, Y.; Akutsu, K.; Hibi, T. Tandem repeat of a transcriptional enhancer upstream of the sterol 14a-demethylase gene (CYP51) in Penicillium digitatum. Appl. Environ. Microbiol. 2000, 66, 3421–3426. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, M.; Leroch, M.; Mosbach, A.; Walker, A.S.; Fillinger, S.; Mernke, D.; Schoonbeek, H.J.; Pradier, J.M.; Leroux, P.; De Waard, M.A. Fungicide-driven evolution and molecular basis of multidrug resistance in field populations of the grey mould fungus Botrytis cinerea. PLoS Pathog. 2009, 5, e1000696. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.L.; Chen, W.C.; Zhao, W.C.; Wang, J.; Wang, B.R.; Li, F.J.; Wei, M.D.; Guo, J.; Chen, C.J.; Zhang, J.Q.; et al. Mutations and overexpression of CYP51 associated with DMI-resistance in Colletotrichum gloeosporioides from chili. Plant Dis. 2020, 104, 668–676. [Google Scholar] [CrossRef]
- Chen, F.; Liu, X.; Chen, S.; Schnabel, E.; Schnabel, G. Characterization of Monilinia fructicola strains resistant to both propiconazole and boscalid. Plant Dis. 2013, 97, 645–651. [Google Scholar] [CrossRef]
- Mellado, E.; Diaz-Guerra, T.M.; Cuenca-Estrella, M.; Rodriguez-Tudela, J.L. Identification of two different 14-α sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J. Clin. Microbiol. 2001, 39, 2431–2438. [Google Scholar] [CrossRef]
- Lu, C.; Zhang, H.; Wang, Y.; Zheng, X. Rapid diagnosis of Fusarium root rot in soybean caused by Fusarium equiseti or Fusarium graminearum using loop-mediated isothermal amplification (LAMP) assays. Australas. Plant Path. 2015, 44, 437–443. [Google Scholar] [CrossRef]
- Liu, X.; Yu, F.; Schnabel, G.; Wu, J.; Wang, Z.; Ma, Z. Paralogous cyp51 genes in Fusarium graminearum mediate differential sensitivity to sterol demethylation inhibitors. Fungal Genet. Biol. 2011, 48, 113–123. [Google Scholar] [CrossRef]
- Zheng, B.; Yan, L.; Liang, W.; Yang, Q. 2019. Paralogous Cyp51s mediate the differential sensitivity of Fusarium oxysporum to sterol demethylation inhibitors. Pest Manag. Sci. 2019, 75, 396–404. [Google Scholar] [CrossRef]
- Zhang, C.; Diao, Y.; Wang, W.; Hao, J.; Imran, M.; Duan, H.; Liu, X. Assessing the risk for resistance and elucidating the genetics of Colletotrichum truncatum that is only sensitive to some DMI fungicides. Front. Microbiol. 2017, 8, 1779. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shi, D.; Wei, L.; Chen, W.; Ma, W.; Chen, C.; Wang, K. Mutations at sterol 14α-demethylases (CYP51A &B) confer the DMI resistance in Colletotrichum gloeosporioides from grape. Pest Manag. Sci. 2020, 76, 4093–4103. [Google Scholar] [PubMed]
- Chen, S.; Wang, Y.; Schnabel, G.; Peng, C.A.; Lagishetty, S.; Smith, K.; Luo, C.; Yuan, H. Inherent Resistance to 14a-Demethylation Inhibitor Fungicides in Colletotrichum truncatum Is Likely Linked to CYP51A and/or CYP51B Gene Variants. Phytopathology 2018, 108, 1263–1275. [Google Scholar] [CrossRef]
- Phoulivong, S.; Cai, L.; Chen, H.; Mckenzie, E.H.C.; Abdelsalam, K.; Chukeatirote, E. Colletotrichum gloeosporioides is not a common pathogen on tropical fruits. Fungal Divers. 2020, 44, 33–43. [Google Scholar] [CrossRef]
- Noman, E.; Al-Gheethi, A.A.; Rahman, N.K.; Talip, B.; Mohamed, R.; Kadir, O.A. Single spore isolation as a simple and efficient technique to obtain fungal pure culture. Earth Environ. Sci. 2018, 140, 10255. [Google Scholar] [CrossRef]
- Cai, L.; Hyde, K.D.; Taylor, P.; Weir, B.S.; Waller, J.M.; Abang, M.M.; Zhang, J.Z.; Yang, Y.L.; Phoulivong, S.; Liu, Z.Y.; et al. A polyphasic approach for studying Colletotrichum. Fungal Divers. 2009, 39, 183–204. [Google Scholar]
- Weir, B.S.; Johnston, P.R.; Damm, U. The Colletotrichum gloeosporioides species complex. Stud. Mycol. 2012, 73, 115–180. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- O’Donnell, K.; Nirenberg, H.I.; Aoki, T.; Cigelnik, E. A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience 2000, 41, 61–78. [Google Scholar] [CrossRef]
- Damm, U.; Woudenberg, J.H.C.; Cannon, P.F.; Crous, P.W. Colletotrichum species with curved conidia from herbaceous hosts. Fungal Divers 2009, 39, 45. [Google Scholar]
- Templeton, M.D.; Rikkerink, E.H.; Solon, S.L.; Crowhurst, R.N. Cloning and molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase-encoding gene and cDNA from the plant pathogenic fungus Glomerella cingulata. Gene 1992, 122, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Damm, U.; Cannon, P.F.; Woudenberg, J.H.C.; Crous, P.W. The Colletotrichum acutatum species complex. Stud. Mycol. 2012, 73, 37–113. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA 5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. Sequence matrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Page, R.D. Tree view: An application to display phylogenetic trees on personal computers. Bioinformatics 1996, 12, 357–358. [Google Scholar] [CrossRef]
- Chen, S.; Yuan, N.; Schnabel, G.; Luo, C. Function of the genetic element ‘Mona’associated with fungicide resistance in Monilinia fructicola. Mol. Plant Pathol. 2017, 18, 90–97. [Google Scholar] [CrossRef]
- Lee, M.H.; Bostock, R.M. Agrobacterium T-DNA-mediated integration and gene replacement in the brown rot pathogen Monilinia fructicola. Curr. Genet. 2006, 49, 309–322. [Google Scholar] [CrossRef]
- Karimi, K.; Arzanlou, M.; Pertot, I. Weeds as potential inoculum reservoir for Colletotrichum nymphaeae causing strawberry anthracnose in Iran and Rep-PCR fingerprinting as useful marker to differentiate C. acutatum complex on strawberry. Front. Microbiol. 2019, 10, 129. [Google Scholar] [CrossRef]
- Zhang, Y.; Mao, C.X.; Zhai, X.Y.; Jamieson, P.A.; Zhang, C.Q. Mutation in cyp51b and overexpression of cyp51a and cyp51b confer multiple resistant to DMIs fungicide prochloraz in Fusarium fujikuroi. Pest. Manag. Sci. 2021, 77, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Talhinhas, P.; Baroncelli, R. Colletotrichum species and complexes: Geographic distribution, host range and conservation status. Fungal Divers. 2021, 110, 109–198. [Google Scholar]
- Oliveira, M.S.; Wang, N.Y.; Peres, N.A. Multilocus phylogenetic analyses of Colletotrichum gloeosporioides species complex causing crown rot on strawberry in Florida. Phytopathology 2022, 112, 898–906. [Google Scholar] [CrossRef]
- Nam, M.H.; Park, M.S.; Lee, H.D.; Yu, S.H. Taxonomic re-evaluation of Colletotrichum gloeosporioides isolated from strawberry in Korea. Plant Pathol. J. 2013, 29, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.D.; Zhang, Y.T.; Yu, H.; Zhou, J.Y.; Hu, M.H.; Liu, A.C.; Zhang, C.Q. Colletotrichum spp. diversity between leaf anthracnose and crown rot from the same strawberry plant. Front. Microbiol. 2022, 14, 860694. [Google Scholar] [CrossRef]
- Zhang, L.; Song, L.; Xu, X.; Zou, X.; Duan, K.; Gao, Q. Characterization and fungicide sensitivity of Colletotrichum species causing strawberry anthracnose in eastern China. Plant Dis. 2020, 104, 1960–1968. [Google Scholar] [CrossRef]
- Hu, W.; Sillaots, S.; Lemieux, S.; Davison, J.; Kauffman, S.; Breton, A. Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog. 2007, 3, e24. [Google Scholar] [CrossRef]
- Chen, S.; Hu, M.; Schnabel, G.; Yang, D.; Yan, X.; Yuan, H. Paralogous CYP51 genes of Colletotrichum spp. mediate differential sensitivity to sterol demethylation inhibitors. Phytopathology 2020, 110, 615–625. [Google Scholar] [CrossRef]
- Fang, Y.L.; Xia, L.M.; Wang, P.; Zhu, L.H.; Ye, J.R.; Huang, L. The MAPKKK CgMck1 is required for cell wall integrity, appressorium development, and pathogenicity in Colletotrichum gloeosporioides. Genes 2018, 9, 543. [Google Scholar] [CrossRef]
- Ishii, H.; Bryson, P.K.; Kayamori, M.; Miyamoto, T.; Yamaoka, Y.; Schnabel, G. Cross-resistance to the new fungicide mefentri fluconazole in DMI-resistant fungal pathogens. Pestic. Biochem. Phys. 2021, 171, 104737. [Google Scholar] [CrossRef]
- Ohno, S. The enormous diversity in genome sizes of fish as a reflection of nature’s extensive experiments with gene duplication. Trans. Am. Fish. Soc. 1970, 99, 120–130. [Google Scholar] [CrossRef]
| Primers | Direction | Length (bp) | Sequence (5′→3′) |
|---|---|---|---|
| CYP51a-UP-F | Forward | 987 | GGAGTCCTCGAATCTGAGTTC |
| CYP51a-UP-R | Reverse | AAAATAGGCATTGATGTGTTGACTCCCTCGG AAGTTCTATGCCTTC | |
| CYP51a-DOWN-F | Forward | 1017 | CTCGTCCGAGGGCAAAGGAATAGAGTAGCTGATGGCGACATGAACCGTG |
| CYP51a-DOWN-R | Reverse | CATGCTGGCAACGGAAGTG | |
| CYP51a-ID-F | Forward | 1649 | GGAAGCCATTATATGAGAAG |
| CYP51a-ID-R | Reverse | CATGCTGGCAACGGAAGTG | |
| CYP51a-Nest-F | Forward | 3188 | GGTGTCCATCTAAGGAATTGG |
| CYP51a-Nest-R | Reverse | CATGCTGGCAACGGAAGTG | |
| CYP51b-UP-F | Forward | 949 | GCAATTGCGAGCATGTGAGTG |
| CYP51b-UP-R | Reverse | AAAATAGGCATTGATGTGTTGACCTCCGCTGGTAGTGTGAAGGGAAG | |
| CYP51b-DOWN-F | Forward | 1016 | CTCGTCCGAGGGCAAAGGAATAGAGTAGGGGAATGTATATTGTAAGCC |
| CYP51b-DOWN-R | Reverse | CTTCTGCATCATGAGCTGGAC | |
| CYP51b-ID-F | Forward | 1541 | CTCTCTCGCGCCACTGCTG |
| CYP51b-ID-R | Reverse | GTGATGTCATAACGTCTTTTG | |
| CYP51b-Nest-F | Forward | 3096 | CTAGCGAATCGAAGACGGAG |
| CYP51b-Nest-R | Reverse | GCGCCGTCGACTCAGGGTAGG | |
| HPH-F | Forward | 1349 | GGAGGTCAACACATCAATGCCTATT |
| HPH-R | Reverse | CTACTCTATTCCTTTGCCCT |
| Species | Strain Number | Conidia y | Appresoria y | Growth Rate (mm/Day) y | Sporulation (×106) y | ||
|---|---|---|---|---|---|---|---|
| Length (μm) | Width (μm) | Length (μm) | Width (μm) | ||||
| C. siamense | JD-A-12 | 14.17 ± 0.63 b | 5.64 ± 0.46 b | 6.97 ± 0.45 b | 6.36 ± 0.70 ab | 13.54 ± 0.37 a | 21.06 ± 5.71 a |
| JD-A-26 | 15.57 ± 0.21 b | 6.87 ± 0.19 a | 7.10 ± 0.25 b | 5.69 ± 0.66 ab | 13.42 ± 0.08 a | 20.20 ± 2.44 ab | |
| C. fructicola | JD-A-14 | 17.14 ± 0.59 a | 6.21 ± 0.19 a | 8.19 ± 0.15 a | 6.83 ± 0.43 b | 13.48 ± 0.04 a | 16.36 ± 3.72 b |
| JD-A-22 | 16.68 ± 0.30 a | 6.63 ± 0.13 a | 8.90 ± 0.72 a | 7.03 ± 0.19 a | 13.51 ± 0.07 a | 14.53 ± 3.15 b | |
| Species | Growth Rate (mm/Day) y | Sporulation (×106) y | Appressorium Production Rate (%) y |
|---|---|---|---|
| JD-A-12 | 11.56 ± 0.07 a | 1.3 ± 0.09 b | 13.82 ± 1.74 b |
| ∆CsCYP51A | 10.39 ± 0.10 b | 1.7 ± 0.13 b | 14.47 ± 1.22 b |
| ∆CsCYP51B | 7.28 ± 0.09 c | 4.33 ± 0.30 a | 17.80 ± 1.25 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, S.; Wu, J.; Yang, X.; Xiao, W.; Yu, H.; Zhang, C. Involvement of CYP51A and CYP51B in Growth, Reproduction, Pathogenicity, and Sensitivity to Fungicides in Colletotrichum siamense. Agronomy 2023, 13, 239. https://doi.org/10.3390/agronomy13010239
Hu S, Wu J, Yang X, Xiao W, Yu H, Zhang C. Involvement of CYP51A and CYP51B in Growth, Reproduction, Pathogenicity, and Sensitivity to Fungicides in Colletotrichum siamense. Agronomy. 2023; 13(1):239. https://doi.org/10.3390/agronomy13010239
Chicago/Turabian StyleHu, Shuodan, Jianyan Wu, Xiaoqi Yang, Wenfei Xiao, Hong Yu, and Chuanqing Zhang. 2023. "Involvement of CYP51A and CYP51B in Growth, Reproduction, Pathogenicity, and Sensitivity to Fungicides in Colletotrichum siamense" Agronomy 13, no. 1: 239. https://doi.org/10.3390/agronomy13010239
APA StyleHu, S., Wu, J., Yang, X., Xiao, W., Yu, H., & Zhang, C. (2023). Involvement of CYP51A and CYP51B in Growth, Reproduction, Pathogenicity, and Sensitivity to Fungicides in Colletotrichum siamense. Agronomy, 13(1), 239. https://doi.org/10.3390/agronomy13010239






