Genome-Wide Identification and Expression Analysis of GATA Gene Family under Different Nitrogen Levels in Arachis hypogaea L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of GATA Genes in the Arachis hypogaea L. Genome
2.2. Phylogenetic Classification of GATA Genes in Arachis hypogaea L.
2.3. Analysis of GATA Gene Structures and Conserved Motifs
2.4. Chromosomal Distribution of GATAFamilyGenes in Arachis hypogaea L.
2.5. Determination of the Optimal Nitrogen Fertilizer Level for Peanut Growth
2.6. Expression Analysis of GATA Genes in Arachis hypogaea L. by Quantitative Real-Time PCR (qRT-PCR)
3. Results
3.1. Genome-Wide Identification and Sequence Characteristics of GATA Gene Family in Arachis hypogaea L.
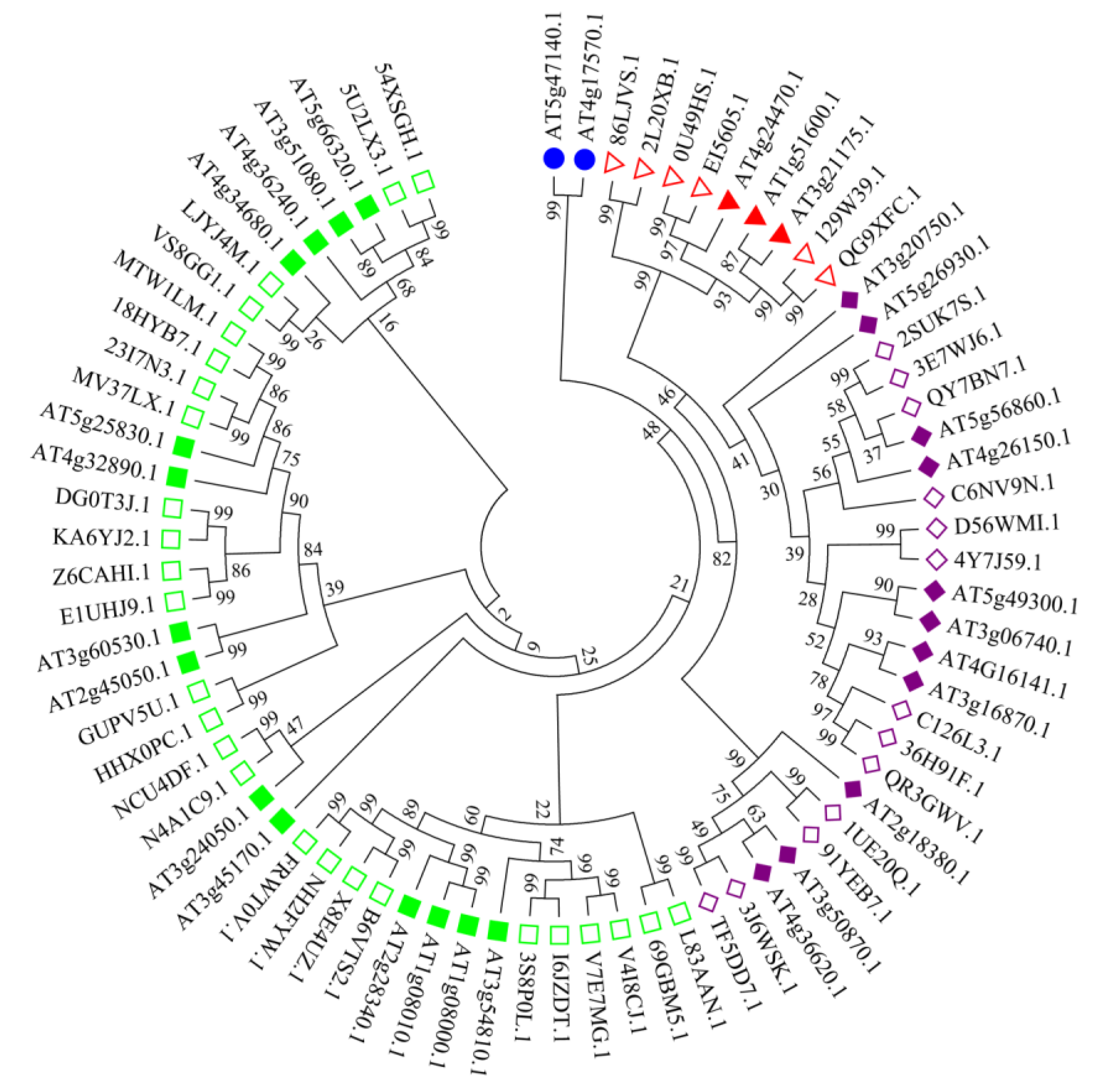
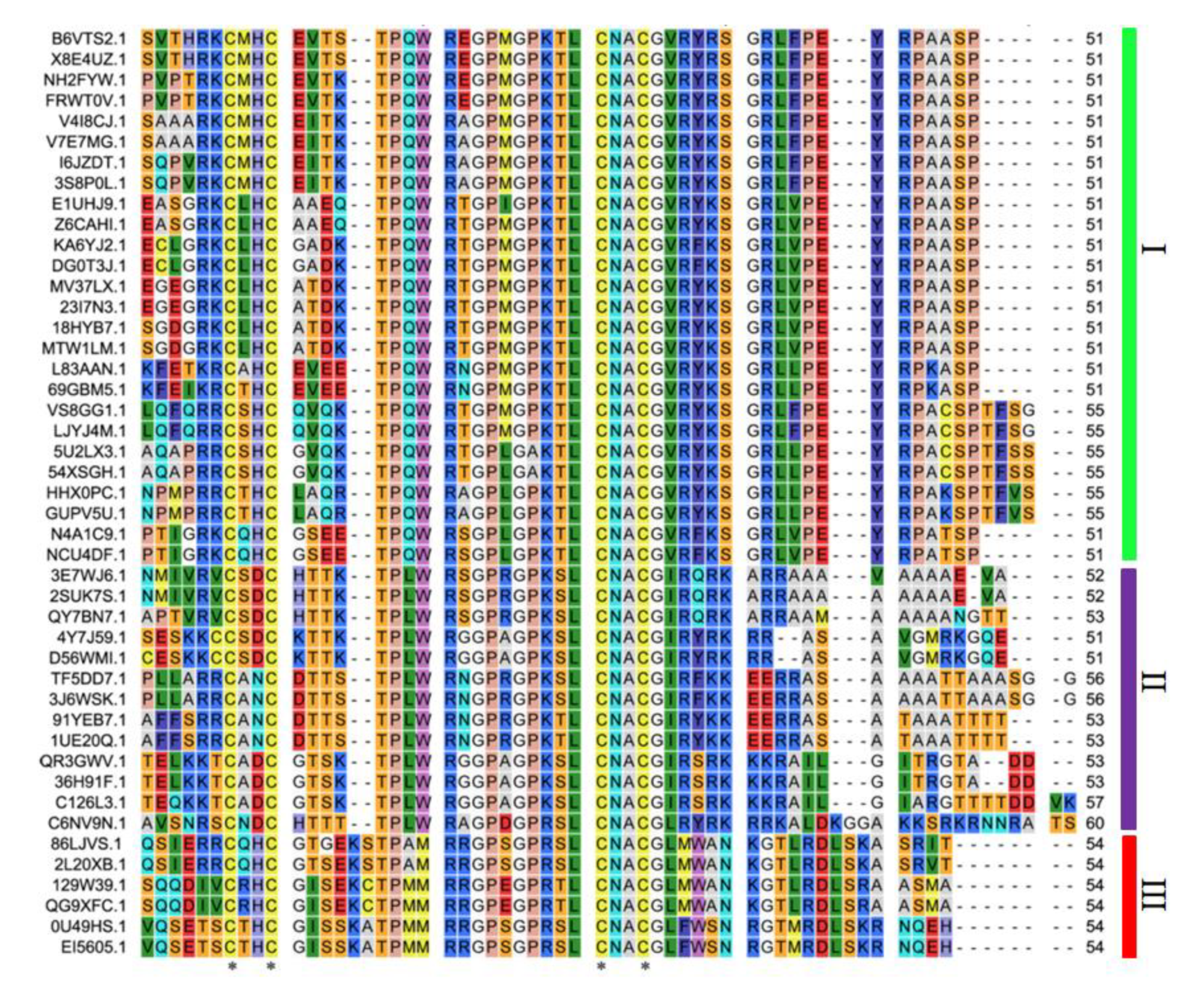
3.2. Sequence Alignment and Phylogenetic Analysis of Peanut GATA Genes
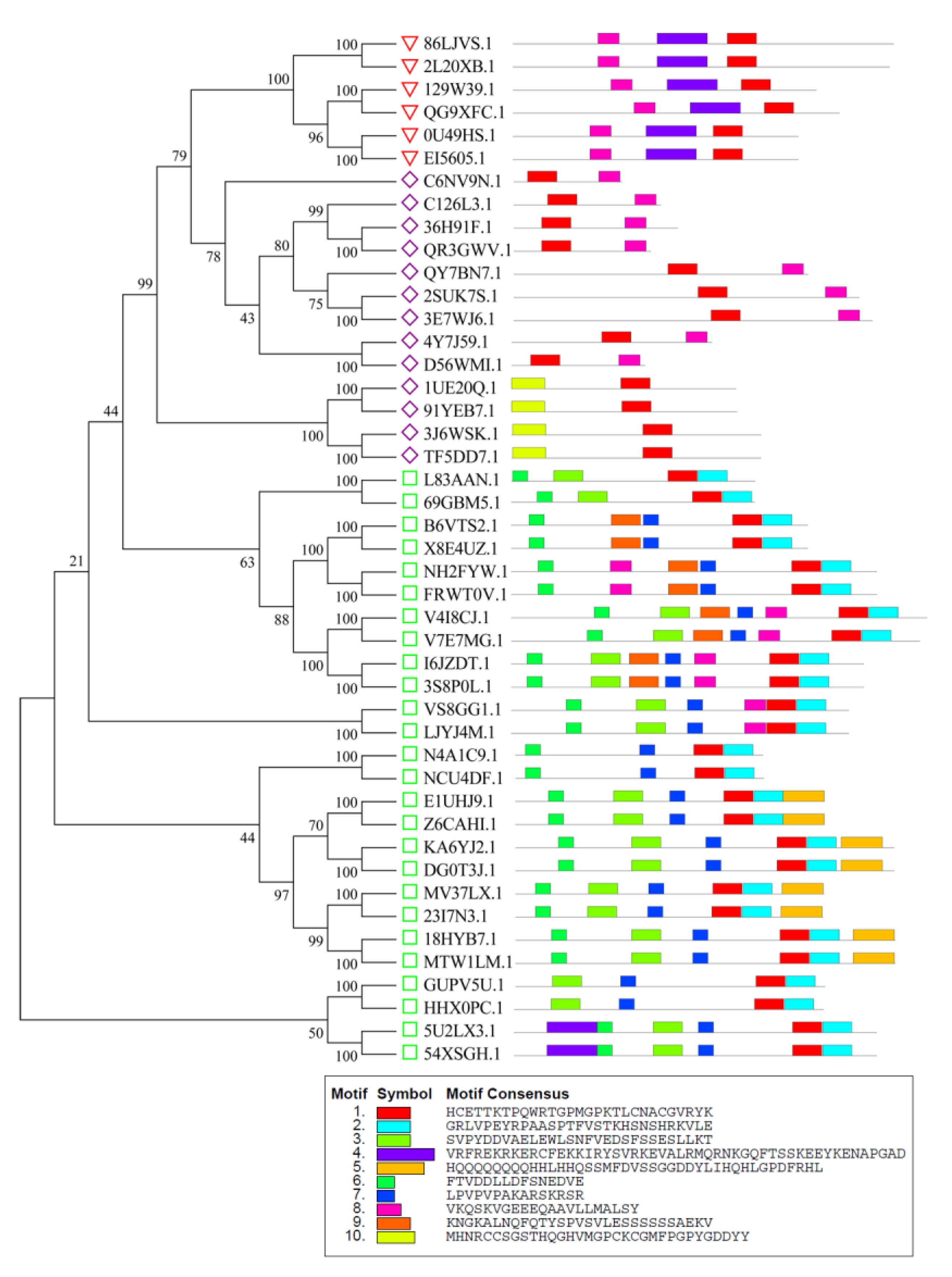
3.3. Analysis of Gene Structures and Conserved Motifs of Peanut GATA Family
3.4. Analysis of Chromosomal Distribution and Gene Duplication of Peanut GATA Genes
3.5. Optimal Level of Nitrogen Fertilizer on the Growth of Peanut Yuhua 23
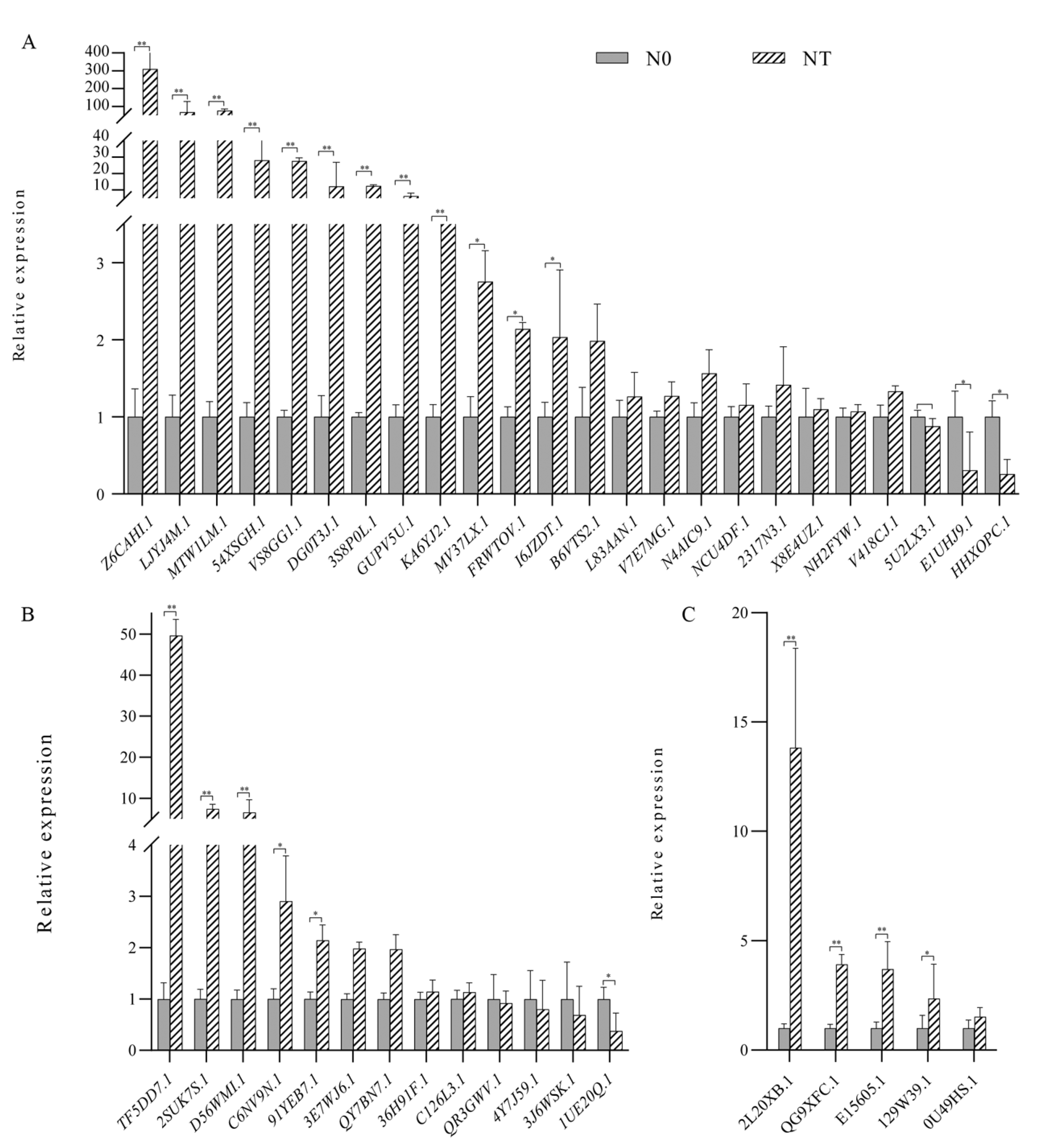
3.6. Expression Analysis of Peanut GATA Gene Family under Different Nitrogen Levels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schwechheimer, C.; Schroder, P.M.; Blaby-Haas, C.E. Plant GATA Factors: Their Biology, Phylogeny, and Phylogenomics. Annu. Rev. Plant Biol. 2022, 73, 123–148. [Google Scholar] [CrossRef] [PubMed]
- Lowry, J.A.; Atchley, W.R. Molecular evolution of the GATA family of transcription factors: Conservation within the DNA-binding domain. J. Mol. Evol. 2000, 50, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Patient, R.K.; McGhee, J.D. The GATA family (vertebrates and invertebrates). Curr. Opin. Genet. Dev. 2002, 12, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Teakle, G.R.; Gilmartin, P.M. Two forms of type IV zinc-finger motif and their kingdom-specific distribution between the flora, fauna and fungi. Trends Biochem. Sci. 1998, 23, 100–102. [Google Scholar] [CrossRef]
- Scazzocchio, C. The fungal GATA factors. Curr. Opin. Microbiol. 2000, 3, 126–131. [Google Scholar] [CrossRef]
- Reyes, J.C.; Muro-Pastor, M.I.; Florencio, F.J. The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 2004, 134, 1718–1732. [Google Scholar] [CrossRef]
- Pomraning, K.R.; Bredeweg, E.L.; Baker, S.E. Regulation of Nitrogen Metabolism by GATA Zinc Finger Transcription Factors in Yarrowia lipolytica. mSphere 2017, 2, e00038-17. [Google Scholar] [CrossRef]
- Daniel-Vedele, F.; Caboche, M. A tobacco cDNA clone encoding a GATA-1 zinc finger protein homologous to regulators of nitrogen metabolism in fungi. Mol. Gen. Genet. MGG 1993, 240, 365–373. [Google Scholar] [CrossRef]
- Hudson, D.; Guevara, D.R.; Hand, A.J.; Xu, Z.; Hao, L.; Chen, X.; Zhu, T.; Bi, Y.M.; Rothstein, S.J. Rice cytokinin GATA transcription Factor1 regulates chloroplast development and plant architecture. Plant Physiol 2013, 162, 132–144. [Google Scholar] [CrossRef]
- Shin, J.M.; Chung, K.; Sakamoto, S.; Kojima, S.; Yeh, C.M.; Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. The chimeric repressor for the GATA4 transcription factor improves tolerance to nitrogen deficiency in Arabidopsis. Plant Biotechnol. 2017, 34, 151–158. [Google Scholar] [CrossRef]
- Zhu, W.; Guo, Y.; Chen, Y.; Wu, D.; Jiang, L. Genome-wide identification, phylogenetic and expression pattern analysis of GATA family genes in Brassica napus. BMC Plant Biol. 2020, 20, 543. [Google Scholar] [CrossRef]
- Zhang, C.; Hou, Y.; Hao, Q.; Chen, H.; Chen, L.; Yuan, S.; Shan, Z.; Zhang, X.; Yang, Z.; Qiu, D.; et al. Genome-wide survey of the soybean GATA transcription factor gene family and expression analysis under low nitrogen stress. PLoS ONE 2015, 10, e0125174. [Google Scholar] [CrossRef]
- Luo, X.M.; Lin, W.H.; Zhu, S.; Zhu, J.Y.; Sun, Y.; Fan, X.Y.; Cheng, M.; Hao, Y.; Oh, E.; Tian, M.; et al. Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell 2010, 19, 872–883. [Google Scholar] [CrossRef]
- Jeong, M.J.; Shih, M.C. Interaction of a GATA factor with cis-acting elements involved in light regulation of nuclear genes encoding chloroplast glyceraldehyde-3-phosphate dehydrogenase in Arabidopsis. Biochem. Biophys. Res. Commun. 2003, 300, 555–562. [Google Scholar] [CrossRef]
- Zhao, Y.; Medrano, L.; Ohashi, K.; Fletcher, J.C.; Yu, H.; Sakai, H.; Meyerowitz, E.M. HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell 2004, 16, 2586–2600. [Google Scholar] [CrossRef]
- Nishii, A.; Takemura, M.; Fujita, H.; Shikata, M.; Yokota, A.; Kohchi, T. Characterization of a novel gene encoding a putative single zinc-finger protein, ZIM, expressed during the reproductive phase in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2000, 64, 1402–1409. [Google Scholar] [CrossRef]
- Liu, P.P.; Koizuka, N.; Martin, R.C.; Nonogaki, H. The BME3 (Blue Micropylar End 3) GATA zinc finger transcription factor is a positive regulator of Arabidopsis seed germination. Plant J. Cell Mol. Biol. 2005, 44, 960–971. [Google Scholar] [CrossRef]
- Wang, L.; Yin, H.; Qian, Q.; Yang, J.; Huang, C.; Hu, X.; Luo, D. NECK LEAF 1, a GATA type transcription factor, modulates organogenesis by regulating the expression of multiple regulatory genes during reproductive development in rice. Cell Res. 2009, 19, 598–611. [Google Scholar] [CrossRef]
- Liu, H.; Li, T.; Wang, Y.; Zheng, J.; Li, H.; Hao, C.; Zhang, X. TaZIM-A1 negatively regulates flowering time in common wheat (Triticum aestivum L.). J. Integr. Plant Biol. 2019, 61, 359–376. [Google Scholar] [CrossRef]
- Warning, H.O.; Hachtel, W. Functional analysis of a nitrite reductase promoter from birch in transgenic tobacco. Plant Sci. 2000, 155, 141–151. [Google Scholar] [CrossRef]
- Rastogi, R.; Bate, N.J.; Sivasankar, S.; Rothstein, S.J. Footprinting of the spinach nitrite reductase gene promoter reveals the preservation of nitrate regulatory elements between fungi and higher plants. Plant Mol. Biol. 1997, 34, 465–476. [Google Scholar] [CrossRef]
- Oliveira, I.C.; Coruzzi, G.M. Carbon and amino acids reciprocally modulate the expression of glutamine synthetase in Arabidopsis. Plant Physiol. 1999, 121, 301–310. [Google Scholar] [CrossRef]
- Krishna, G.; Singh, B.K.; Kim, E.K.; Morya, V.K.; Ramteke, P.W. Progress in genetic engineering of peanut (Arachis hypogaea L.)—A review. Plant Biotechnol. J. 2015, 13, 147–162. [Google Scholar] [CrossRef]
- Luo, H.; Ren, X.; Li, Z.; Xu, Z.; Li, X.; Huang, L.; Zhou, X.; Chen, Y.; Chen, W.; Lei, Y.; et al. Co-localization of major quantitative trait loci for pod size and weight to a 3.7 cM interval on chromosome A05 in cultivated peanut (Arachis hypogaea L.). BMC Genom. 2017, 18, 58. [Google Scholar] [CrossRef]
- Tegeder, M.; Masclaux-Daubresse, C. Source and sink mechanisms of nitrogen transport and use. New Phytol 2018, 217, 35–53. [Google Scholar] [CrossRef] [PubMed]
- Sarah, E.C. Nitrogen limitation of rhizoma peanut growth. J. Plant Nutr. 2013, 36, 311–328. [Google Scholar] [CrossRef]
- Hubick, K.T. Effects of Nitrogen Source and Water Limitation on Growth, Transpiration Efficiency and Carbon-Isotope Discrimination in Peanut Cultivars. Funct. Plant Biol. 1990, 17, 413–430. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2022, 51, D418–D427. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chow, C.N.; Lee, T.Y.; Hung, Y.C.; Li, G.Z.; Tseng, K.C.; Liu, Y.H.; Kuo, P.L.; Zheng, H.Q.; Chang, W.C. PlantPAN3.0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 2019, 47, D1155–D1163. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Abdalla, E.A.; Osman, A.K.; Maki, M.A.; Nur, F.M.; Ali, S.B.; Aune, J.B. The Response of Sorghum, Groundnut, Sesame, and Cowpea to Seed Priming and Fertilizer Micro-Dosing in South Kordofan State, Sudan. Agronomy 2015, 5, 476–490. [Google Scholar] [CrossRef]
- Jiang, C.; Li, X.; Zou, J.; Ren, J.; Jin, C.; Zhang, H.; Yu, H.; Jin, H. Comparative transcriptome analysis of genes involved in the drought stress response of two peanut (Arachis hypogaea L.) varieties. BMC Plant Biol. 2021, 21, 64. [Google Scholar] [CrossRef]
- Du, K.; Xia, Y.; Zhan, D.; Xu, T.; Lu, T.; Yang, J.; Kang, X. Genome-Wide Identification of the Eucalyptus urophylla GATA Gene Family and Its Diverse Roles in Chlorophyll Biosynthesis. Int. J. Mol. Sci. 2022, 23, 5251. [Google Scholar] [CrossRef]
- Zhang, K.; Jia, L.; Yang, D.; Hu, Y.; Njogu, M.K.; Wang, P.; Lu, X.; Yan, C. Genome-Wide Identification, Phylogenetic and Expression Pattern Analysis of GATA Family Genes in Cucumber (Cucumis sativus L.). Plants 2021, 10, 1626. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Lu, Y.; Sun, H.; Duan, W.; Hu, Y.; Yan, Y. Genome-Wide Analysis of Wheat GATA Transcription Factor Genes Reveals Their Molecular Evolutionary Characteristics and Involvement in Salt and Drought Tolerance. Int. J. Mol. Sci. 2023, 24, 27. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Yang, Y.; Lou, S.; Wei, W.; Zhao, Z.; Ren, Y.; Lin, C.; Ma, L. Genome-Wide Characterization and Gene Expression Analyses of GATA Transcription Factors in Moso Bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 2019, 21, 14. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, T.; Wang, B.; Li, H. Genome-Wide Characterization and Expression Analysis of GATA Transcription Factors in Response to Methyl Jasmonate in Salvia miltiorrhiza. Genes 2022, 13, 822. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Huang, Q.; Wang, Y.; Wang, C.; Zhu, R.; Zhang, S.; Kai, G. Genome-wide survey of the GATA gene family in camptothecin-producing plant Ophiorrhiza pumila. BMC Genom. 2022, 23, 256. [Google Scholar] [CrossRef]
- Yao, X.; Zhou, M.; Ruan, J.; He, A.; Ma, C.; Wu, W.; Lai, D.; Fan, Y.; Gao, A.; Weng, W.; et al. Genome-Wide Identification, Evolution, and Expression Pattern Analysis of the GATA Gene Family in Tartary Buckwheat (Fagopyrum tataricum). Int. J. Mol. Sci. 2022, 23, 12434. [Google Scholar] [CrossRef]
- Mondal, M.; Skalicky, M.; Garai, S.; Hossain, A.; Sarkar, S.; Banerjee, H.; Kundu, R.; Brestic, M.; Barutcular, C.; Erman, M.; et al. Supplementing Nitrogen in Combination with Rhizobium Inoculation and Soil Mulch in Peanut (Arachis hypogaea L.) Production System: Part II. Effect on Phenology, Growth, Yield Attributes, Pod Quality, Profitability and Nitrogen Use Efficiency. Agronomy 2020, 10, 1513. [Google Scholar] [CrossRef]
- Xin, C.; Qing-wei, Y.; Jia-lin, S.; Shuang, X.; Fu-chun, X.; Ya-jun, C. Research Progress on Nitrogen Use and Plant Growth. J. Northeast Agric. Univ. 2014, 21, 68–74. [Google Scholar] [CrossRef]
- Kihara, J.; Huising, J.; Nziguheba, G.; Waswa, B.S.; Njoroge, S.; Kabambe, V.; Iwuafor, E.; Kibunja, C.; Esilaba, A.O.; Coulibaly, A. Maize response to macronutrients and potential for profitability in sub-Saharan Africa. Nutr. Cycl. Agroecosyst. 2016, 105, 171–181. [Google Scholar] [CrossRef]
- Kiwia, A.; Kimani, D.; Harawa, R.; Jama, B.; Sileshi, G.W. Fertiliser use efficiency, production risks and profitability of maize on smallholder farms in East Africa. Exp. Agric. 2022, 58, e22. [Google Scholar] [CrossRef]
- Fu, Y.H.; Marzluf, G.A. nit-2, the major nitrogen regulatory gene of Neurospora crassa, encodes a protein with a putative zinc finger DNA-binding domain. Mol. Cell. Biol. 1990, 10, 1056–1065. [Google Scholar] [CrossRef]
- An, Y.; Han, X.; Tang, S.; Xia, X.; Yin, W. Poplar GATA transcription factor PdGNC is capable of regulating chloroplast ultrastructure, photosynthesis, and vegetative growth in Arabidopsis under varying nitrogen levels. Plant Cell Tissue Organ Cult. (PCTOC) 2014, 119, 313–327. [Google Scholar] [CrossRef]
- Garcia-Salcedo, R.; Casamayor, A.; Ruiz, A.; Gonzalez, A.; Prista, C.; Loureiro-Dias, M.C.; Ramos, J.; Arino, J. Heterologous expression implicates a GATA factor in regulation of nitrogen metabolic genes and ion homeostasis in the halotolerant yeast Debaryomyces hansenii. Eukaryot Cell 2006, 5, 1388–1398. [Google Scholar] [CrossRef]
- Bitter, G.A.; Egan, K.M. Expression of heterologous genes in Saccharomyces cerevisiae from vectors utilizing the glyceraldehyde-3-phosphate dehydrogenase gene promoter. Gene 1984, 32, 263–274. [Google Scholar] [CrossRef]


| IDs | Number of Amino Acid | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|
| 1UE20Q.1 | 224 | 25,348.52 | 6.82 | 41.79 | 29.69 | −1.135 |
| QY7BN7.1 | 294 | 33,214.55 | 9.06 | 56.06 | 65.78 | −0.937 |
| C126L3.1 | 147 | 15,730.66 | 10.02 | 55.42 | 47.28 | −0.849 |
| QR3GWV.1 | 137 | 14,504.58 | 10.21 | 43.32 | 59.93 | −0.669 |
| B6VTS2.1 | 296 | 32,708.73 | 8.74 | 56.5 | 64.46 | −0.606 |
| VS8GG1.1 | 337 | 37,006.44 | 5.62 | 58.49 | 62.2 | −0.603 |
| N4A1C9.1 | 247 | 27,876.6 | 8.63 | 57.37 | 66.68 | −0.619 |
| 3E7WJ6.1 | 358 | 40,639.2 | 8.96 | 45.52 | 58.1 | −1.146 |
| 3J6WSK.1 | 249 | 27,685.51 | 8.56 | 57.48 | 41.61 | −0.788 |
| 5U2LX3.1 | 362 | 39,790.7 | 6.02 | 69.26 | 57.38 | −0.602 |
| V4I8CJ.1 | 415 | 45,242.82 | 6.19 | 55.03 | 65.4 | −0.422 |
| 4Y7J59.1 | 200 | 22,493.01 | 9.19 | 56.66 | 71.15 | −0.292 |
| HHX0PC.1 | 309 | 34,119.05 | 6.95 | 51.22 | 63.07 | −0.771 |
| E1UHJ9.1 | 308 | 34,070.78 | 6.22 | 51.88 | 61.46 | −0.636 |
| 0U49HS.1 | 285 | 30,839.98 | 6.07 | 49.62 | 55.12 | −0.704 |
| NH2FYW.1 | 365 | 40,879.04 | 7.61 | 53.87 | 67.51 | −0.585 |
| L83AAN.1 | 243 | 28,861.03 | 6.26 | 45.46 | 50.91 | −1.19 |
| 129W39.1 | 303 | 33,350.89 | 6 | 42.61 | 56.63 | −0.846 |
| KA6YJ2.1 | 378 | 40,996.29 | 6.64 | 54.19 | 54.23 | −0.733 |
| I6JZDT.1 | 352 | 38,037.74 | 6.19 | 56.47 | 59.32 | −0.504 |
| 86LJVS.1 | 380 | 42,318.54 | 4.75 | 54.55 | 58.82 | −0.828 |
| MV37LX.1 | 308 | 34,852.79 | 6.6 | 56.81 | 59.58 | −0.836 |
| 91YEB7.1 | 225 | 25,302.44 | 6.78 | 41.82 | 28.71 | −1.091 |
| 36H91F.1 | 164 | 17,331.93 | 10.19 | 36.84 | 67.32 | −0.502 |
| NCU4DF.1 | 248 | 27,988.73 | 8.42 | 57.4 | 68.35 | −0.598 |
| X8E4UZ.1 | 296 | 32,718.72 | 8.53 | 58.05 | 65.14 | −0.622 |
| LJYJ4M.1 | 337 | 36,989.37 | 5.4 | 59.71 | 61.9 | −0.611 |
| 2SUK7S.1 | 345 | 38,773.25 | 8.98 | 47.31 | 60.58 | −1.069 |
| TF5DD7.1 | 249 | 27,663.46 | 8.36 | 57.18 | 41.61 | −0.789 |
| 54XSGH.1 | 362 | 39,784.73 | 6.02 | 70.16 | 59.53 | −0.581 |
| V7E7MG.1 | 408 | 45,052.88 | 7.19 | 54.66 | 67.94 | −0.425 |
| C6NV9N.1 | 108 | 12,119.71 | 9 | 54.77 | 56.11 | −0.849 |
| D56WMI.1 | 133 | 14,754.15 | 9.33 | 56.18 | 60.9 | −0.55 |
| GUPV5U.1 | 309 | 34,092.03 | 6.95 | 51.3 | 62.46 | −0.78 |
| 18HYB7.1 | 379 | 41,754.29 | 6.34 | 41.03 | 57.84 | −0.676 |
| MTW1LM.1 | 379 | 41,924.55 | 6.34 | 41.34 | 57.57 | −0.687 |
| EI5605.1 | 285 | 30,839.98 | 6.07 | 49.62 | 55.12 | −0.704 |
| Z6CAHI.1 | 308 | 34,178.92 | 6.37 | 51.35 | 60.19 | −0.679 |
| FRWT0V.1 | 365 | 40,728.87 | 7.59 | 52.79 | 66.44 | −0.556 |
| 69GBM5.1 | 243 | 29,025.26 | 5.95 | 54.14 | 47.28 | −1.137 |
| QG9XFC.1 | 326 | 36,002.16 | 6 | 41.96 | 59.51 | −0.703 |
| DG0T3J.1 | 378 | 41,109.54 | 6.61 | 53.65 | 52.94 | −0.731 |
| 3S8P0L.1 | 352 | 37,994.67 | 6.01 | 56.12 | 59.32 | −0.496 |
| 2L20XB.1 | 376 | 41,855.97 | 4.75 | 53.87 | 60.74 | −0.84 |
| 23I7N3.1 | 307 | 34,739.63 | 6.46 | 57.17 | 59.45 | −0.842 |
| Chromosome | Gene IDs | Start | End | Strand | Chromosome | Gene IDs | Start | End | Strand | Protein Identity (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Chr01 | C126L3.1 | 101,123,945 | 101,125,505 | + | Chr01 | QR3GWV.1 | 101,126,574 | 101,128,262 | − | 75.781 |
| Chr13 | 2SUK7S.1 | 41,897,732 | 41,900,296 | − | Chr03 | 3E7WJ6.1 | 39,900,001 | 39,903,171 | − | 92.265 |
| Chr06 | 4Y7J59.1 | 5,609,802 | 5,611,267 | − | Chr16 | D56WMI.1 | 16,199,511 | 16,200,847 | + | 93.431 |
| Chr08 | L83AAN.1 | 47,690,362 | 47,691,491 | − | Chr18 | 69GBM5.1 | 131,169,543 | 131,170,274 | − | 94.47 |
| Chr16 | 18HYB7.1 | 142,009,810 | 142,012,509 | − | Chr16 | MTW1LM.1 | 144,667,753 | 144,670,407 | − | 95.778 |
| Chr01 | 1UE20Q.1 | 1,145,984 | 1,148,294 | − | Chr11 | 91YEB7.1 | 15,059,031 | 15,060,930 | + | 96.444 |
| Chr05 | V4I8CJ.1 | 115,242,703 | 115,246,472 | + | Chr15 | V7E7MG.1 | 160,303,681 | 160,307,678 | + | 96.448 |
| Chr10 | 86LJVS.1 | 4,866,411 | 4,870,550 | − | Chr20 | 2L20XB.1 | 9,721,345 | 9,725,571 | − | 97.368 |
| Chr08 | NH2FYW.1 | 28,796,522 | 28,800,772 | − | Chr18 | FRWT0V.1 | 4,511,391 | 4,515,670 | − | 97.534 |
| Chr11 | 36H91F.1 | 142,387,755 | 142,389,602 | + | Chr01 | QR3GWV.1 | 101,126,574 | 101,128,262 | − | 97.81 |
| Chr03 | N4A1C9.1 | 14,791,317 | 14,793,255 | + | Chr12 | NCU4DF.1 | 70,664,814 | 70,666,699 | + | 97.984 |
| Chr09 | KA6YJ2.1 | 3,033,691 | 3,036,478 | − | Chr19 | DG0T3J.1 | 3,908,699 | 3,911,486 | − | 98.148 |
| Chr07 | E1UHJ9.1 | 52,962,210 | 52,963,745 | + | Chr18 | Z6CAHI.1 | 1,344,542 | 1,346,259 | − | 98.701 |
| Chr10 | MV37LX.1 | 99,237,013 | 99,238,881 | + | Chr20 | 23I7N3.1 | 122,307,538 | 122,309,406 | + | 98.701 |
| Chr16 | GUPV5U.1 | 125,861,212 | 125,862,943 | + | Chr06 | HHX0PC.1 | 94,867,241 | 94,868,959 | + | 98.706 |
| Chr03 | B6VTS2.1 | 1,591,608 | 1,595,718 | + | Chr13 | X8E4UZ.1 | 3,376,912 | 3,381,028 | + | 98.986 |
| Chr03 | VS8GG1.1 | 6,552,396 | 6,554,643 | − | Chr13 | LJYJ4M.1 | 7,985,629 | 7,987,892 | + | 99.11 |
| Chr09 | I6JZDT.1 | 118,856,896 | 118,860,690 | − | Chr19 | 3S8P0L.1 | 146,525,935 | 146,529,609 | + | 99.148 |
| Chr05 | 5U2LX3.1 | 11,738,072 | 11,740,438 | + | Chr15 | 54XSGH.1 | 12,325,129 | 12,327,494 | + | 99.448 |
| Chr05 | 3J6WSK.1 | 10,491,801 | 10,493,761 | + | Chr15 | TF5DD7.1 | 10,965,226 | 10,967,089 | + | 99.598 |
| Chr09 | 129W39.1 | 210,176 | 213,603 | − | Chr19 | QG9XFC.1 | 277,509 | 280,578 | + | 99.67 |
| Chr08 | 0U49HS.1 | 14,943,757 | 14,948,081 | + | Chr17 | EI5605.1 | 131,669,993 | 131,674,406 | + | 100 |
| Year | Nitrogen Levels (kg hm−2) | TPP | EPP | HPW (g) | Pod Yield (kg hm−2) | VCR |
|---|---|---|---|---|---|---|
| 2020 | 0 | 21.0 ± 1.3 d | 10.6 ± 0.3 c | 179.1 ± 3.5 c | 3595.8 ± 29.8 d | / |
| 75 | 26.8 ± 2.3 c | 12.2 ± 0.4 b | 184.6 ± 6.5 bc | 4265.7 ± 24.2 c | 15.0 ± 0.7 a | |
| 150 | 35.2 ± 1.6 b | 13.3 ± 0.3 a | 190.0 ± 3.4 ab | 4799.3 ± 25.2 b | 11.9 ± 0.8 a | |
| 225 | 41.8 ± 1.4 a | 13.6 ± 0.4 a | 192.0 ± 4.9 a | 4886.1 ± 20.2 a | 1.9 ± 0.5 b | |
| 300 | 43.7 ± 1.4 a | 13.3 ± 0.4 a | 194.7 ± 5.6 a | 4860.4 ± 22.8 a | −0.6 ± 0.7 b | |
| 2021 | 0 | 19.5 ± 1.4 d | 9.7 ± 0.3 b | 157.0 ± 5.6 b | 2884.0 ± 24.2 d | / |
| 75 | 25.6 ± 2.7 c | 11.7 ± 1.5 a | 158.9 ± 20.6 ab | 3489.8 ± 36.1 c | 13.2 ± 0.8 a | |
| 150 | 32.7 ± 2.1 b | 12.4 ± 0.1 a | 167.4 ± 1.1 ab | 3927.9 ± 16.1 b | 9.5 ± 0.5 ab | |
| 225 | 40.6 ± 1.4 a | 12.6 ± 0.3 a | 173.2 ± 3.4 a | 4144.0 ± 19.9 a | 4.7 ± 0.1 bc | |
| 300 | 42.1 ± 1.9 a | 12.6 ± 0.6 a | 172.5 ± 9.0 ab | 4137.8 ± 23.1 a | −0.1 ± 0.7 c | |
| 2022 | 0 | 23.4 ± 1.6 d | 10.7 ± 0.2 d | 185.1 ± 3.3 b | 3763.7 ± 9.6 d | / |
| 75 | 29.3 ± 0.8 c | 12.6 ± 0.1 c | 187.4 ± 2.1 b | 4478.1 ± 22.6 c | 11.8 ± 0.3 a | |
| 150 | 37.1 ± 4.2 b | 13.2 ± 0.5 b | 202.9 ± 7.3 a | 5064.3 ± 32.0 b | 9.7 ± 0.2 a | |
| 225 | 42.9 ± 3.0 a | 13.6 ± 0.3 a | 202.8 ± 4.2 a | 5178.6 ± 15.7 a | 1.9 ± 0.6 b | |
| 300 | 45.2 ± 2.5 a | 13.4 ± 0.4 ab | 206.1 ± 6.2 a | 5155.0 ± 26.0 a | −0.4 ± 0.3 b | |
| ANOVA | ||||||
| N | 558.16 * | 70.05 * | 13.56 * | 6681.25 * | 717.70 * | |
| Y | 4.69 * | 18.76 * | 96.46 * | 9627.63 * | 6.89 * | |
| N×Y | 0.28 | 0.27 | 0.43 | 18.10 * | 7.44 * | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Deng, X.; Han, S.; Zhang, X.; Dai, T. Genome-Wide Identification and Expression Analysis of GATA Gene Family under Different Nitrogen Levels in Arachis hypogaea L. Agronomy 2023, 13, 215. https://doi.org/10.3390/agronomy13010215
Li X, Deng X, Han S, Zhang X, Dai T. Genome-Wide Identification and Expression Analysis of GATA Gene Family under Different Nitrogen Levels in Arachis hypogaea L. Agronomy. 2023; 13(1):215. https://doi.org/10.3390/agronomy13010215
Chicago/Turabian StyleLi, Xiujie, Xiaoxu Deng, Suoyi Han, Xinyou Zhang, and Tingbo Dai. 2023. "Genome-Wide Identification and Expression Analysis of GATA Gene Family under Different Nitrogen Levels in Arachis hypogaea L." Agronomy 13, no. 1: 215. https://doi.org/10.3390/agronomy13010215
APA StyleLi, X., Deng, X., Han, S., Zhang, X., & Dai, T. (2023). Genome-Wide Identification and Expression Analysis of GATA Gene Family under Different Nitrogen Levels in Arachis hypogaea L. Agronomy, 13(1), 215. https://doi.org/10.3390/agronomy13010215





