BSA-Seq Approach Identified Candidate Region and Diagnostic Marker for Chilling Tolerance of High Oleic Acid Peanut at Germination Stage
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
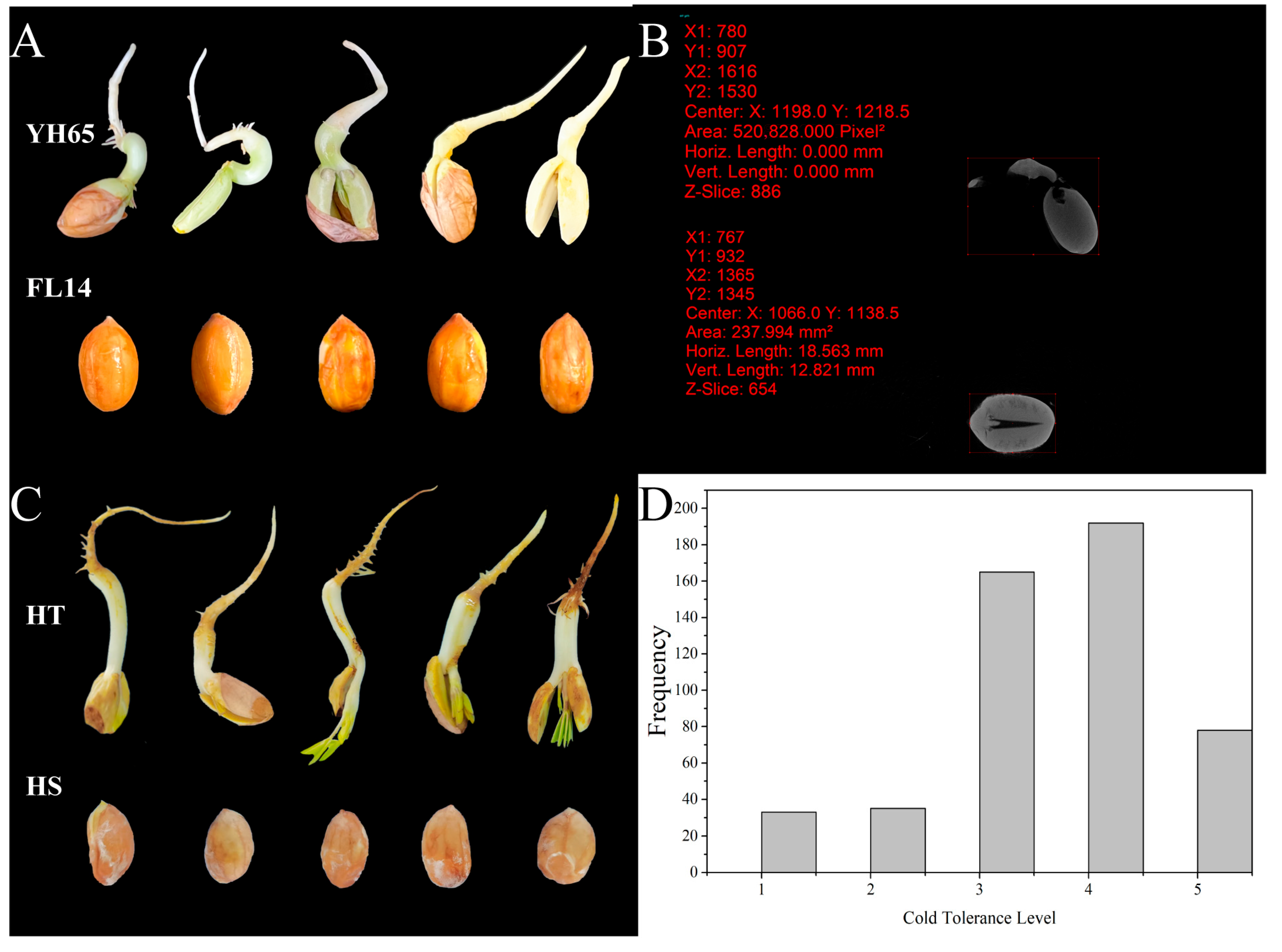
2.2. Evaluation of Chilling Tolerance at Germination Stage
2.3. Construction of Segregating Pools
2.4. Bulked Segregant Analysis (BSA)
2.5. Candidate Region Gene Function Annotation
2.6. KASP Marker Development and Validation
2.7. Statistical Analysis of Experimental Data
3. Results
3.1. Phenotypic Analysis of the Parents and F2 Population
3.2. Whole-Genome Resequencing and Identification of SNPs
3.3. Identification of Candidate Genomic Regions for LTG Tolerance
3.4. Functional Enrichment Analysis of Genes in Candidate Region
3.5. Validation of SNPs in Candidate Region with Different Germplasm Resources
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Varshney, R.; Mohan, S.; Gaur, P.; Gangarao, N.; Pandey, M.; Bohra, A.; Sawargaonkar, S.; Chitikineni, A.; Kimurto, P.; Janila, P.; et al. Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics. Biotechnol. Adv. 2013, 31, 1120–1134. [Google Scholar] [CrossRef]
- Huang, B.; Qi, F.; Sun, Z.; Miao, L.; Zhang, Z.; Liu, H.; Fang, Y.; Dong, W.; Tang, F.; Zheng, Z.; et al. Marker-assisted backcrossing to improve seed oleic acid content in four elite and popular peanut (Arachis hypogaea L.) cultivars with high oil content. Breed Sci. 2019, 69, 234–243. [Google Scholar] [CrossRef]
- Barkley, N.; Isleib, T.; Wang, M.; Pittman, R. Genotypic effect of ahFAD2 on fatty acid profiles in six segregating peanut (Arachis hypogaea L.) populations. BMC Genet. 2013, 14, 62. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yu, S.; Zhu, L. High Oleic Acid Peanuts in China, 1st ed.; Shanghai Scientific and Technical Publishers: Shanghai, China, 2021; pp. 19–27. [Google Scholar]
- Liu, H.; Li, H.; Gu, J.; Deng, L.; Ren, L.; Hong, Y.; Lu, Q.; Chen, X.; Liang, X. Identification of the candidate proteins related to oleic acid accumulation during peanut (Arachis hypogaea L.) seed development through comparative proteome analysis. Int. J. Mol. Sci. 2018, 19, 1235. [Google Scholar] [CrossRef] [PubMed]
- Norden, A.; Corbet, D.; Knauft, D.; Young, C. Variability in oil quality among peanut genotypes in the Florida breeding program. Peanut Sci. 1987, 14, 7–11. [Google Scholar] [CrossRef]
- Gorbet, D.; Knauft, D. Registration of ‘SunOleic 95R’ peanut. Crop Sci. 1997, 37, 1392. [Google Scholar] [CrossRef]
- Chu, Y.; Ramos, L.; Holbrook, C. Ozias-Akins P: Frequency of a loss-of-function mutation in oleoyl-PC desaturase (ahFAD2A) in the mini-core of the US peanut germplasm collection. Crop Sci. 2007, 47, 2372–2378. [Google Scholar] [CrossRef]
- Huang, B.; Qi, F.; Sun, Z.; Miao, L.; Fang, Y.; Zheng, Z.; Shi, L.; Zhang, Z.; Liu, H.; Dong, W.; et al. Improvement of oleic acid content in peanut (Arachis hypogaea L.) by marker assisted successive backcross and agronomic evaluation of derived lines [EB/OL]. Acta Agron. Sin. 2019, 45, 546–555. [Google Scholar] [CrossRef]
- Wang, X.; Tang, Y.; Wu, Q.; Sun, X.; Wang, Y.; Hu, D.; Wang, C. Characterization of high-oleic peanut natural mutants derived from an intersectional cross. Grasas Aceites 2015, 66, e091. [Google Scholar] [CrossRef]
- Zhang, H.; Dong, J.; Zhao, X.; Zhang, Y.; Ren, J.; Xing, L.; Jiang, C.; Wang, X.; Wang, J.; Zhao, S.; et al. Research progress in membrane lipid metabolism and molecular mechanism in peanut cold tolerance. Front. Plant Sci. 2019, 10, 838. [Google Scholar] [CrossRef]
- Jiang, C.; Zhang, H.; Ren, J.; Dong, J.; Zhao, X.; Wang, X.; Wang, J.; Zhong, C.; Zhao, S.; Liu, X.; et al. Comparative Transcriptome-Based Mining and Expression Profiling of Transcription Factors Related to Cold Tolerance in Peanut. Int. J. Mol. Sci. 2020, 21, 1921. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Y.; Sun, Q.; Chen, S. Negative effects of heat stress on maize yield were compensated by increasing thermal time and declining cold stress in northeast China. Int. J. Biometeorol. 2022, 66, 2395–2403. [Google Scholar] [CrossRef]
- Bera, S.; Kamdar, J.; Kasundra, S.; Patel, S.; Jasani, M.; Maurya, A.; Dash, P.; Chandrashekar, A.; Rani, K.; Manivannan, N.; et al. Steady expression of high oleic acid in peanut bred by marker-assisted backcrossing for fatty acid desaturase mutant alleles and its effect on seed germination along with other seedling traits. PLoS ONE 2019, 14, 12. [Google Scholar] [CrossRef]
- Jungman, B.; Schubert, A. The effect of fatty acid profiles on peanut seed germination at low soil temperatures. Proc. Am. Peanut Res. Educ. Soc. 2000, 32, 36. [Google Scholar]
- Sun, H.; Yu, S.; Ren, L.; Zhang, J.; Yu, B. Field Identification of Cold Tolerance of High Oleic Acid Peanut at Germination Stage and Verification of Physiological Indices. In Proceedings of the Fourth International Conference on Biological Information and Biomedical Engineering, Chengdu, China, 21–23 July 2020. [Google Scholar]
- Sun, D.; Bian, N.; Wang, X.; Xing, X.; Shen, Y.; Xu, Z.; Qi, Y.; Wang, X. Comprehensive evaluation of cold tolerance and germplasm screening of high oleic acid peanut at germination stage. Acta Agric. Nucl. Sin. 2021, 35, 1263–1272. [Google Scholar]
- Wang, X.; Zou, B.; Shao, Q.; Cui, Y.; Lu, S.; Zhang, Y.; Huang, Q.; Huang, J.; Hua, J. Natural variation reveals that OsSAP16 controls low-temperature germination in rice. J. Exp. Bot. 2018, 69, 413–421. [Google Scholar] [CrossRef]
- Ding, Y.L.; Shi, Y.; Yang, S. Molecular regulation of plant responses to environmental temperatures. Mol. Plant 2020, 13, 544–564. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, S. Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 2022, 64, 301–341. [Google Scholar] [CrossRef]
- Ntare, B.; Williams, J.; Dougbedji, F. Evaluation of groundnut genotypes for tolerance under fifield conditions in a sahelian environment using a simple physiological model for yield. J. Agric. Sci. 2001, 136, 81–88. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; Han, Z.; Chen, Y.; Huai, D.; Kang, Y.; Wang, Z.; Yan, L.; Jiang, H.; Lei, Y.; et al. Integrated transcriptomics and metabolomics analysis reveal Key metabolism pathways contributing to cold tolerance in peanut. Front. Plant Sci. 2021, 12, 752474. [Google Scholar] [CrossRef]
- Zhang, X.; Ren, C.; Xue, Y.; Tian, Y.; Zhang, H.; Li, N.; Sheng, C.; Jiang, H.; Bai, D. Small RNA and Degradome deep sequencing reveals the roles of microRNAs in peanut (Arachis hypogaea L.) cold response. Front. Plant Sci. 2022, 13, 920195. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jiang, C.; Lei, J.; Dong, J.; Ren, J.; Shi, X.; Zhong, C.; Wang, X.; Zhao, X.; Yu, H. Comparative physiological and transcriptomic analyses reveal key regulatory networks and potential hub genes controlling peanut chilling tolerance. Genomics 2022, 114, 110285. [Google Scholar] [CrossRef]
- Raza, A.; Sharif, Y.; Chen, K.; Wang, L.; Fu, H.; Zhuang, Y.; Chitikineni, A.; Chen, H.; Zhang, C.; Varshney, R.; et al. Genome-wide characterization of ascorbate peroxidase gene family in peanut (Arachis hypogea L.) revealed their crucial role in growth and multiple stress tolerance. Front. Plant Sci. 2022, 13, 962182. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, J.; Wing, R.; Ganal, M.; Tanksley, S. Isolation of molecular markers from specific chromosome intervals using DNA pools from existing mapping populations. Nucleic Acids Res. 1991, 19, 6553–6558. [Google Scholar] [CrossRef]
- Michelmore, R.; Paran, I.; Kesseli, R. Identification of markers linked to disease resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.; Terauchi, R.; McCouch, S. Harvesting the promising fruits of genomics: Applying genome sequencing technologies to crop breeding. PLoS Biol. 2014, 12, e1001883. [Google Scholar] [CrossRef]
- Li, Z.; Xu, Y. Bulk segregation analysis in the NGS era: A review of its teenage years. Plant J. 2022, 109, 1355–1374. [Google Scholar] [CrossRef]
- Guo, Z.; Cai, L.; Chen, Z.; Wang, R.; Zhang, L.; Guan, S.; Zhang, S.; Ma, W.; Liu, C.; Pan, G. Identification of candidate genes controlling chilling tolerance of rice in the cold region at the booting stage by BSA-Seq and RNA-Seq. R. Soc. Open Sci. 2020, 7, 201081. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, X.; Zhu, W.; Qin, X.; Xu, J.; Cheng, C.; Lou, Q.; Li, J.; Chen, J. Complete resistance to powdery mildew and partial resistance to downy mildew in a Cucumis hystrix introgression line of cucumber were controlled by a co-localized locus. Theor. Appl. Genet. 2018, 131, 2229–2243. [Google Scholar] [CrossRef]
- Zhong, C.; Sun, S.L.; Li, Y.P.; Duan, C.X.; Zhu, Z. Next-generation sequencing to identify candidate genes and develop diagnostic markers for a novel phytophthora resistance gene, RpsHC18, in soybean. Theor. Appl. Genet. 2018, 131, 525–538. [Google Scholar] [CrossRef]
- Luo, H.; Pandey, M.; Khan, A.; Guo, J.; Wu, B.; Cai, Y.; Huang, L.; Zhou, X.; Chen, Y.; Chen, W.; et al. Discovery of genomic regions and candidate genes controlling shelling percentage using QTL-seq approach in cultivated peanut (Arachis hypogaea L.). Plant Biotechnol. J. 2019, 7, 1248–1260. [Google Scholar] [CrossRef]
- Bertioli, D.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Bertioli, S.; Ren, L.; Farmer, A.; Pandey, M.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.; Zhang, C.; Chang, W.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Luo, H.; Pandey, M.; Zhi, Y.; Zhang, H.; Xu, S.; Guo, J.; Wu, B.; Chen, H.; Ren, X.; Zhou, X.; et al. Discovery of two novel and adjacent QTLs on chromosome B02 controlling resistance against bacterial wilt in peanut variety Zhonghua 6. Theor. Appl. Genet. 2020, 133, 1133–1148. [Google Scholar] [CrossRef]
- Zhao, Y.; Ma, J.; Li, M.; Deng, L.; Li, G.; Xia, H.; Zhao, S.; Hou, L.; Li, P.; Ma, C.; et al. Whole-genome resequencing-based QTL-seq identifified AhTc1 gene encoding a R2R3-MYB transcription factor controlling peanut purple testa colour. Plant Biotechnol. J. 2020, 18, 96–105. [Google Scholar] [CrossRef]
- Peng, Z.; Chen, H.; Tan, L.; Shu, H.; Varshney, R.; Zhou, Z.; Zhao, Z.; Luo, Z.; Chitikineni, A.; Wang, L.; et al. Natural polymorphisms in a pair of NSP2 homoeologs can cause loss of nodulation in peanut. J. Exp. Bot. 2021, 72, 1104–1118. [Google Scholar] [CrossRef]
- Zheng, Z.; Sun, Z.; Fang, Y.; Qi, F.; Liu, H.; Miao, L.; Du, P.; Shi, L.; Gao, W.; Han, S.; et al. Genetic Diversity, Population Structure, and Botanical Variety of 320 Global Peanut Accessions Revealed Through Tunable Genotyping-by-Sequencing. Sci. Rep. 2018, 8, 14500. [Google Scholar] [CrossRef]
- Yang, L.; Lei, L.; Li, P.; Wang, J.; Wang, C.; Yang, F.; Chen, J.; Liu, H.; Zheng, H.; Xin, W.; et al. Identification of Candidate Genes Conferring Cold Tolerance to Rice (Oryza sativa L.) at the Bud-Bursting Stage Using Bulk Segregant Analysis Sequencing and Linkage Mapping. Front. Plant Sci. 2021, 12, 647239. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J. Isolation of plant DNA from fresh tissue. Focus 1900, 12, 13–15. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, W.; Zhao, D.; Liu, B.; Shi, Y.; Chen, B.; Yang, H.; Guo, P.; Geng, X.; Shang, Z.; et al. Caenorhabditis elegans transthyretin-like protein TTR-52 mediates recognition of apoptotic cells by the CED-1 phagocyte receptor. Nat. Cell Biol. 2010, 12, 655–664. [Google Scholar] [CrossRef]
- Takagi, H.; Abe, A.; Yoshida, K.; Kosugi, S.; Natsume, S.; Mitsuoka, C.; Uemura, A.; Utsushi, H.; Tamiru, M.; Takuno, S.; et al. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013, 74, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.; Demarest, B.; Bisgrove, B.; Gorsi, B.; Su, Y.; Yost, H. MMAPPR: Mutation mapping analysis pipeline for pooled RNA-seq. Genome Res. 2013, 23, 687–697. [Google Scholar] [CrossRef]
- Altschul, S.; Madden, T.; Schäffer, A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs; Oxford University Press: Oxford, UK, 1997; Volume 25. [Google Scholar]
- Rasheed, A.; He, Z.; Gao, F.; Zhai, S.; Jin, H.; Liu, J.; Guo, Q.; Zhang, Y.; Dreisigacker, S.; Xia, X.; et al. Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor. Appl. Genet. 2016, 129, 1843–1860. [Google Scholar] [CrossRef]
- Semagn, K.; Babu, R.; Hearne, S. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): Overview of the technology and its application in crop improvement. Mol. Breed. 2014, 33, 1–14. [Google Scholar] [CrossRef]
- Pan, Y.; Liang, H.; Gao, L.; Dai, G.; Chen, W.; Yang, X.; Qing, D.; Gao, J.; Wu, H.; Huang, J.; et al. Transcriptomic profiling of germinating seeds under cold stress and characterization of the cold-tolerant gene LTG5 in rice. BMC Plant Biol. 2020, 20, 371. [Google Scholar] [CrossRef]
- Gao, C.; Mumtaz, M.; Zhou, Y.; Yang, Z.; Shu, H.; Zhu, J.; Bao, W.; Cheng, S.; Yin, L.; Huang, J.; et al. Integrated transcriptomic and metabolomic analyses of cold-tolerant and cold-sensitive pepper species reveal key genes and essential metabolic pathways involved in response to cold stress. Int. J. Mol. Sci. 2022, 23, 6683. [Google Scholar] [CrossRef]
- Raza, A.; Su, W.; Jia, Z.; Luo, D.; Zhang, Y.; Gao, A.; Hussain, M.; Mehmood, S.; Cheng, Y.; Lv, Y.; et al. Mechanistic insights into trehalose-mediated cold stress tolerance in rapeseed (Brassica napus L.) seedlings. Front. Plant Sci. 2022, 13, 857980. [Google Scholar] [CrossRef]
- Yousefi, S.; Marchese, A.; Salami, S.; Benny, J.; Giovino, A.; Perrone, A.; Caruso, T.; Gholami, M.; Sarikhani, H.; Buti, M.; et al. Identifying conserved genes involved in crop tolerance to cold stress. Funct. Plant Biol. 2022, 49, 861–873. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Woo, O.; Jang, H.; Lee, J. Characterization and comparative expression analysis of CUL1 genes in rice. Genes Genom. 2018, 40, 233–241. [Google Scholar] [CrossRef]
- Pan, W.; Tao, J.; Cheng, T.; Shen, M.; Ma, J.; Zhang, W.; Lin, Q.; Ma, B.; Chen, S.; Zhang, J. Soybean NIMA-related kinase1 promotes plant growth and improves salt and cold tolerance. Plant Cell Physiol. 2017, 58, 1268–1278. [Google Scholar] [CrossRef]
- Zhang, G.; Yu, S.; Wang, H.; Wang, X. Transcriptome profiling of high oleic peanut under low temperatureduring germination. Yi Chuan 2019, 41, 1050–1059. [Google Scholar]
- He, X.; Ni, X.; Xie, P.; Liu, W.; Yao, M.; Kang, Y.; Qin, L.; Hua, W. Comparative transcriptome analyses revealed conserved and novel responses to cold and freezing stress in Brassica napus L. G3 Genes Genomes Genet. 2019, 9, 2723–2737. [Google Scholar] [CrossRef]
- Feng, Q.; Yang, S.; Wang, Y.; Lu, L.; Sun, M.; He, C.; Wang, J.; Li, Y.; Yu, X.; Li, Q.; et al. Physiological and molecular mechanisms of ABA and CaCl2 regulating chilling tolerance of cucumber seedlings. Plants 2021, 10, 2746. [Google Scholar] [CrossRef]
- Dubrovina, A.; Kiselev, K.; Khristenko, V.; Aleynova, O. VaCPK20, a calcium-dependent protein kinase gene of wild grapevine vitis amurensis Rupr.; mediates cold and drought stress tolerance. J. Plant Physiol. 2015, 185, 1–12. [Google Scholar] [CrossRef]
- Chen, Q.; Xu, L.; Tan, W.; Chen, L.; Qi, H.; Xie, L.; Chen, M.; Liu, B.; Yu, L.; Yao, N.; et al. Disruption of the arabidopsis defense regulator genes SAG101, EDS1, and PAD4 confers enhanced freezing tolerance. Mol. Plant 2015, 8, 1536–1549. [Google Scholar] [CrossRef]
- Shen, X.; Liu, B.; Xue, Z.; Jiang, M.; Lu, X.; Zhang, Q. Spatiotemporal variation in vegetation spring phenology and its response to climate change in freshwater marshes of Northeast China. Sci. Total Environ. 2019, 666, 1169–1177. [Google Scholar] [CrossRef]
- Upadhyaya, H.; Reddy, L.; Dwivedi, S.; Gowda, C.; Singh, S. Phenotypic diversity in cold-tolerant peanut (Arachis hypogaea L.) germplasm. Euphytica 2009, 2, 279–291. [Google Scholar] [CrossRef]
- Sun, M.; Spears, J.; Isleib, T.; Jordan, D.; Penny, B.; Johnson, D.; Copeland, S. Effect of production environment on seed quality of normal and high-oleate large seeded Virginia-type peanut (Arachis hypogaea L.). Peanut Sci. 2014, 41, 90–99. [Google Scholar] [CrossRef]
- Tang, Y.; Wang, C.; Gao, H.; Feng, T.; Zhang, S.; Wang, X.; Zhang, J.; Yu, S. Low temperature tolerance during seed imbibition and its relationship to main quality traits in peanut. J. Nucl. Agric. Sci. 2011, 25, 436–442. [Google Scholar]
- Chang, B.; Zhong, P.; Liu, J.; Tang, Z.; Gao, Y.; Yu, H.; Guo, W. Effect of low-temperature stress and gibberellin on seed germination and seed-ling physiological responses in peanut. Acta Agron. Sin. 2019, 45, 118–130. [Google Scholar] [CrossRef]
- Varshney, R.; Pandey, M.; Bohra, A.; Singh, V.; Thudi, M. Toward the sequence-based breeding in legumes in the post-genome sequencing era. Theor. Appl. Genet. 2018, 17, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.; Thudi, M.; Pandey, M.; Tardieu, F.; Ojiewo, C.; Vadez, V.; Whitbread, A.; Siddique, K.; Nguyen, H.; Carberry, P.; et al. Accelerating genetic gains in legumes for the development of prosperous smallholder agriculture: Integrating genomics, phenotyping, systems modelling and agronomy. J. Exp. Bot. 2018, 69, 3293–3312. [Google Scholar] [CrossRef]
- Nawade, B.; Bosamia, T.; Thankappan, R.; Rathnakumar, A.; Kumar, A.; Dobaria, J.; Kundu, R.; Mishra, G. Insights into the indian peanut genotypes for ahFAD2 gene polymorphism regulating its oleic and linoleic acid fluxes. Front. Plant Sci. 2016, 7, 1271. [Google Scholar] [CrossRef]
- Zhang, M.; Zeng, Q.; Liu, H.; Qi, F.; Sun, Z.; Miao, L.; Li, X.; Li, C.; Liu, D.; Guo, J.; et al. Identification of a stable major QTL for fresh-seed germination on chromosome Arahy.04 in cultivated peanut (Arachis hypogaea L.). Crop J. 2022; in press. [Google Scholar] [CrossRef]
- Kumar, R.; Janila, P.; Vishwakarma, M.; Khan, A.; Manohar, S.; Gangurde, S.; Variath, M.; Shasidhar, Y.; Pandey, M.; Varshney, R. Whole-genome resequencing-based QTL-seq identified candidate genes and molecular markers for fresh seed dormancy in groundnut. Plant Biotechnol. J. 2020, 18, 992–1003. [Google Scholar] [CrossRef]
- Steuernagel, B.; Periyannan, S.; Hernandez-Pinzon, I.; Witek, K.; Rouse, M.; Yu, G.; Hatta, A.; Ayliffe, M.; Bariana, H.; Jones, J.; et al. Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat. Biol. 2016, 34, 652–655. [Google Scholar] [CrossRef]
- Das, S.; Upadhyaya, H.; Bajaj, D.; Kujur, A.; Badoni, S.; Kumar, V.; Tripathi, S.; Gowda, C.; Sharma, S.; Singh, S.; et al. Deploying QTL-seq for rapid delineation of a potential candidate gene underlying major trait-associated QTL in chickpea. DNA Res. 2015, 22, 193–203. [Google Scholar] [CrossRef]
- Singh, V.; Khan, A.; Saxena, R.; Kumar, V.; Kale, S.; Sinha, P.; Chitikineni, A.; Pazhamala, L.; Garg, V.; Sharma, M.; et al. Next-generation sequencing for identification of candidate genes for Fusarium wilt and sterility mosaic disease in pigeonpea (Cajanus cajan). Plant Biotechnol. J. 2016, 14, 1183–1194. [Google Scholar] [CrossRef]
- Huang, G.; Ma, S.; Bai, L.; Zhang, L.; Ma, H.; Jia, P.; Liu, J.; Zhong, M.; Guo, Z. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 2012, 39, 969–987. [Google Scholar] [CrossRef] [PubMed]
- Iordachescu, M.; Imai, R. Trehalose biosynthesis in response to abiotic stresses. J. Integr. Plant Biol. 2008, 50, 1223–1229. [Google Scholar] [CrossRef]
- He, W.; Gao, J.; Dou, T.; Shao, X.; Bi, F.; Sheng, O.; Deng, G.; Li, C.; Hu, C.; Liu, J.; et al. Early cold-induced peroxidases and aquaporins are associated with high cold tolerance in Dajiao (Musaspp. ‘Dajiao’). Front. Plant Sci. 2018, 9, 282. [Google Scholar] [CrossRef]
- Su, C.; Wang, Y.; Hsieh, T.; Lu, C.A.; Tseng, T.; Yu, S. A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol. 2010, 153, 145–158. [Google Scholar] [CrossRef]



| Item | Parents | F2 Population | ||||||
|---|---|---|---|---|---|---|---|---|
| YH65 | FL14 | Mean | Range | SD | Skewness | Kurtosis | CV (%) | |
| LTG scale | 1 | 5 | 3.49 | 1–5 | 1.05 | 0.109 | 0.217 | 0.300 |
| Bud length (cm) | 5.10 | 0.00 | 0.34 | 0.00–8.30 | 0.923 | 0.109 | 0.217 | 2.681 |
| Sample | Total Data (bp) | High-Quality Reads | Mapping (%) | GC (%) | Q30 (%) | Average Depth (×) | Genome Coverage (%) |
|---|---|---|---|---|---|---|---|
| YU65 | 55,039,185,600 | 332,684,456 | 99.96 | 39.64 | 88.22 | 15.23 | 98.87 |
| FuL14 | 53,770,119,000 | 344,017,168 | 99.88 | 40.69 | 92.46 | 16.26 | 98.24 |
| HT | 79,259,289,000 | 508,218,616 | 99.94 | 37.70 | 92.58 | 23.84 | 99.35 |
| HS | 80,996,380,200 | 494,885,654 | 99.97 | 37.58 | 88.66 | 22.2 | 99.35 |
| Chromosome | Overlapping Region (bp) | Size (Mb) | HQ_SNP | No. of Gene |
|---|---|---|---|---|
| 05 | 107,340,001~109,630,000 | 2.29 | 9 | 122 |
| 12 | 109,310,001~111,950,000 | 2.64 | 6 | 135 |
| 13 | 21,530,001~24,620,000 | 3.09 | 5 | 116 |
| 14 | 118,310,001~120,570,000 | 2.26 | 2 | 84 |
| 16 | 54,260,001~56,260,000 | 2.00 | 1 | 7 |
| 16 | 85,540,001~88,220,000 | 2.68 | 2 | 13 |
| 17 | 122,360,001~124,460,000 | 2.1 | 0 | 6 |
| ID | BSA-Seq Data | Primer Type | Primer Sequence | Allele | Parent | |
|---|---|---|---|---|---|---|
| Arahy.A05_107485885 | ΔSNP-index | 0.3272 | Allele-specific (HEX) | GAAGGTGACCAAGTTCATGCTGGTCAGTAATCAAGCTAGAATAAAAATTGG | G | YH65 |
| ED | 0.2141 | Allele-specific (FAM) | GAAGGTCGGAGTCAACGGATTGTCAGTAATCAAGCTAGAATAAAAATTGA | A | FL14 | |
| Common | ATTATCCTGGCAATATAATTCTTTGTAGTATTTC | |||||
| Arahy.A05_108044810 | ΔSNP-index | 0.5926 | Allele-specific (HEX) | GAAGGTGACCAAGTTCATGCTGATGGATTGATTTTTATTTTTTGTC | C | YH65 |
| ED | 0.7023 | Allele-specific (FAM) | GAAGGTCGGAGTCAACGGATTGATGGATTGATTTTTATTTTTTGTT | T | FL14 | |
| Common | TTAAGCCAATTTTTTTTGCTAATGATATCAT | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, H.; Ren, L.; Qi, F.; Wang, H.; Yu, S.; Sun, Z.; Huang, B.; Han, S.; Shi, P.; Wang, Y.; et al. BSA-Seq Approach Identified Candidate Region and Diagnostic Marker for Chilling Tolerance of High Oleic Acid Peanut at Germination Stage. Agronomy 2023, 13, 18. https://doi.org/10.3390/agronomy13010018
Sun H, Ren L, Qi F, Wang H, Yu S, Sun Z, Huang B, Han S, Shi P, Wang Y, et al. BSA-Seq Approach Identified Candidate Region and Diagnostic Marker for Chilling Tolerance of High Oleic Acid Peanut at Germination Stage. Agronomy. 2023; 13(1):18. https://doi.org/10.3390/agronomy13010018
Chicago/Turabian StyleSun, Hongxi, Liang Ren, Feiyan Qi, Haixin Wang, Shutao Yu, Ziqi Sun, Bingyan Huang, Suoyi Han, Puxiang Shi, Yibo Wang, and et al. 2023. "BSA-Seq Approach Identified Candidate Region and Diagnostic Marker for Chilling Tolerance of High Oleic Acid Peanut at Germination Stage" Agronomy 13, no. 1: 18. https://doi.org/10.3390/agronomy13010018
APA StyleSun, H., Ren, L., Qi, F., Wang, H., Yu, S., Sun, Z., Huang, B., Han, S., Shi, P., Wang, Y., Zhang, X., & Yu, G. (2023). BSA-Seq Approach Identified Candidate Region and Diagnostic Marker for Chilling Tolerance of High Oleic Acid Peanut at Germination Stage. Agronomy, 13(1), 18. https://doi.org/10.3390/agronomy13010018






