Soil Microbial Communities Involved in Proteolysis and Sulfate-Ester Hydrolysis Are More Influenced by Interannual Variability than by Crop Sequence
Abstract
1. Introduction
2. Material and Methods
2.1. Experimental Site Description
2.2. Soil Sampling
2.3. DNA Extraction and Quantification of Microbial Genes
2.4. Quantification of Culturable Bacteria
2.5. Soil Protease and Arylsulfatase Activity
2.6. Net Sulfur and Nitrogen Mineralization
2.7. Data Analysis
3. Results
3.1. Effect of Crop Sequences on the Microbial Abundance, Enzymatic Activity and Net N and S Mineralization
3.2. Relative Influence of Crop Sequence, Month and Year of Sampling
3.3. Relationships among Microbial Abundance, Enzymatic Activity and Net Mineralization
4. Discussion
4.1. Microbial Abundances Control N and S Mineralization and Protease Activity but Not Arylsulfatase Activity
4.2. Cropping Oilseed Rape after Wheat Increased the Microbial Abundance and Activity
4.3. Yearly Meteorology More Strongly Impact Microbial Communities than Crops
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schimel, J.P.; Weintraub, M.N. The implications of exoenzyme activity on microbial carbon and nitrogen limitation in soil: A theoretical model. Soil Biol. 2003, 35, 549–563. [Google Scholar] [CrossRef]
- Wolters, V. Invertebrate control of soil organic matter stability. Biol. Fertil. Soils 2000, 31, 1–19. [Google Scholar] [CrossRef]
- Scherer, H.W.; Pacyna, S.; Manthey, N.; Schulz, M. Sulphur supply to peas (Pisum sativum L.) influences symbiotic N 2 fixation. Plant Soil Environ. 2006, 52, 72–77. [Google Scholar] [CrossRef]
- Jamal, A.; Moon, Y.S.; Abdin, M.Z. Sulphur—A general overview and interaction with nitrogen. Aust. J. Crop Sci. 2010, 4, 523–529. [Google Scholar]
- Haneklaus, S.; Bloem, E.; Schnug, E. Sulfur interactions in crop ecosystems. In Sulfur in Plants—An Ecological Perspective; Hawkesford, M.J., De Kok, L.J., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 17–58. ISBN 1-4020-5886-1. [Google Scholar]
- Zhao, F.J.; Withers, P.J.A.; Evans, E.J.; Monaghan, J.; Salmon, S.E.; Shewry, P.R.; McGrath, S.P. Sulphur nutrition: An important factor for the quality of wheat and rapeseed (Reprinted from Plant nutrition for sustainable food production and environment, 1997). Soil Sci. Plant Nutr. 1997, 43, 1137–1142. [Google Scholar] [CrossRef]
- Schulten, R.; Schnitzer, M. The chemistry of soil organic nitrogen: A review. Biol. Fertil. Soils 1998, 26, 1–15. [Google Scholar] [CrossRef]
- Jan, M.T.; Roberts, P.; Tonheim, S.K.; Jones, D.L. Protein breakdown represents a major bottleneck in nitrogen cycling in grassland soils. Soil Biol. Biochem. 2009, 41, 2272–2282. [Google Scholar] [CrossRef]
- Mishra, S.; Di, H.J.; Cameron, K.C.; Monaghan, R.; Carran, A. Gross nitrogen mineralisation rates in pastural soils and their relationships with organic nitrogen fractions, microbial biomass and protease activity under glasshouse conditions. Biol. Fertil. Soils 2005, 42, 45–53. [Google Scholar] [CrossRef]
- Yuan, L.; Bao, D.J.; Jin, Y.; Yang, Y.H.; Huang, J.G. Influence of fertilizers on nitrogen mineralization and utilization in the rhizosphere of wheat. Adv. Agron. 2011, 343, 187–193. [Google Scholar] [CrossRef]
- Zaman, M.; Di, H.J.; Cameron, K.C.; Frampton, C.M. Gross nitrogen mineralization and nitrification rates and their relationships to enzyme activities and the soil microbial biomass in soils treated with dairy shed effluent and ammonium fertilizer at different water potentials. Biol. Fertil. Soils 1999, 29, 178–186. [Google Scholar] [CrossRef]
- Hofmockel, K.S.; Fierer, N.; Colman, B.P.; Jackson, R.B. Amino acid abundance and proteolytic potential in North American soils. Oecologia 2010, 163, 1069–1078. [Google Scholar] [CrossRef]
- Sakurai, M.; Suzuki, K.; Onodera, M. Analysis of bacterial communities in soil by PCR–DGGE targeting protease genes. Soil Biol. Biochem. 2007, 39, 2777–2784. [Google Scholar] [CrossRef]
- Bach, H.-J.; Munch, J.C. Identification of bacterial sources of soil peptidases. Biol. Fertil. Soils 2000, 31, 219–224. [Google Scholar] [CrossRef]
- Bettany, J.R.; Stewart, J.W.B. Sulphur cycling in soils. In Proceedings of the 1982 International Sulphur Conference; British Sulphur Corp.: London, UK, 1983; Volume 2, pp. 767–786. [Google Scholar]
- Hawkesford, M.J. Sulfur and plant ecology: A central role of sulfate transporters in response to sulfur availability. In Sulfur in Plants—An Ecological Perspective; Hawkesford, M.J., De Kok, L.J., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 1–15. ISBN 1-4020-5886-1. [Google Scholar]
- Dedourge, O.; Vong, P.; Benizri, É.; Lasserre-joulin, F.; Guckert, A. Immobilization of sulphur-35, microbial biomass and arylsulphatase activity in soils from field-grown rape, barley and fallow. Biol. Fertil. Soils 2003, 38, 181–185. [Google Scholar] [CrossRef]
- Vong, P.; Dedourge, O.; Lasserre-Joulin, F.; Guckert, A. Immobilized-S, microbial biomass-S and soil arylsulfatase activity in the rhizosphere soil of rape and barley as affected by labile substrate C and N additions. Soil Biol. Biochem. 2003, 35, 1651–1661. [Google Scholar] [CrossRef]
- Vong, P.; Piutti, S.; Slezack-Deschaumes, S.; Benizri, E.; Guckert, A. Sulphur immobilization and arylsulphatase activity in two calcareous arable and fallow soils as affected by glucose additions. Geoderma 2008, 148, 79–84. [Google Scholar] [CrossRef]
- Legay, N.; Personeni, E.; Slezack-Deschaumes, S.; Piutti, S.; Cliquet, J.B. Grassland species show similar strategies for sulphur and nitrogen acquisition. Plant Soil 2014, 375, 113–126. [Google Scholar] [CrossRef]
- Vong, P.; Lasserre-Joulin, F.; Guckert, A. Mobilization of labelled organic sulfur in rhizosphere of rape and barley and in non-rhizosphere soil. J. Plant Nutr. 2002, 25, 2191–2204. [Google Scholar] [CrossRef]
- Cregut, M.; Piutti, S.; Vong, P.; Slezack-deschaumes, S.; Crovisier, I.; Benizri, E. Density, structure, and diversity of the cultivable arylsulfatase-producing bacterial community in the rhizosphere of field-grown rape and barley. Soil Biol. Biochem. 2009, 41, 704–710. [Google Scholar] [CrossRef]
- Slezack-Deschaumes, S.; Piutti, S.; Vong, P.; Benizri, E. Dynamics of cultivable arylsulfatase-producing bacterial and fungal communities along the phenology of field-grown rape. Eur. J. Soil Biol. 2012, 48, 66–72. [Google Scholar] [CrossRef]
- Bell, C.W.; Asao, S.; Calderon, F.; Wolk, B.; Wallenstein, M.D. Plant nitrogen uptake drives rhizosphere bacterial community assembly during plant growth. Soil Biol. Biochem. 2015, 85, 170–182. [Google Scholar] [CrossRef]
- Dinesh, R.; Suryanarayana, M.A.; Chaudhuri, S.G.; Sheeja, T.E. Long-term influence of leguminous cover crops on the biochemical properties of a sandy clay loam Fluventic Sulfaquent in a humid tropical region of India. Soil Tillage Res. 2004, 77, 69–77. [Google Scholar] [CrossRef]
- Gschwendtner, S.; Reichmann, M.; Müller, M.; Radl, V.; Munch, J.C. Abundance of bacterial genes encoding for proteases and chitinases in the rhizosphere of three different potato cultivars. Biol. Fertil. Soils 2010, 46, 649–652. [Google Scholar] [CrossRef]
- Hu, Z.; Yang, Z.; Xu, C.; Haneklaus, S.; Cao, Z.; Schnug, E. Effect of crop growth on the distribution and mineralization of soil sulfur fractions in the rhizosphere. J. Plant Nutr. Soil Sci. 2002, 165, 249–254. [Google Scholar] [CrossRef]
- Knauff, U.; Schulz, M.; Scherer, H.W. Arylsufatase activity in the rhizosphere and roots of different crop species. Eur. J. Agron. 2003, 19, 215–223. [Google Scholar] [CrossRef]
- Mougel, C.; Offre, P.; Ranjard, L.; Corberand, T.; Gamalero, E.; Robin, C.; Lemanceau, P. Dynamic of the genetic structure of bacterial and fungal communities at different developmental stages of Medicago truncatula Gaertn. cv. Jemalong line J5. New Phytol. 2006, 170, 165–175. [Google Scholar] [CrossRef]
- Xuan, D.T.; Guong, V.T.; Rosling, A.; Alström, S.; Chai, B.; Högberg, N. Different crop rotation systems as drivers of change in soil bacterial community structure and yield of rice, Oryza sativa. Biol. Fertil. Soils 2012, 48, 217–225. [Google Scholar] [CrossRef]
- Jensen, E.S. Rhizodeposition of N by pea and barley and its effect on soil N dynamics. Soil Biol. Biochem. 1996, 28, 65–71. [Google Scholar] [CrossRef]
- Dornbush, M.E. Grasses, litter, and their interaction affect microbial biomass and soil enzyme activity. Soil Biol. Biochem. 2007, 39, 2241–2249. [Google Scholar] [CrossRef]
- Lupwayi, N.Z.; Clayton, G.W.; O’Donovan, J.T.; Harker, K.N.; Turkington, T.K.; Soon, Y.K. Nitrogen release during decomposition of crop residues under conventional and zero tillage. Can. J. Soil Sci. 2006, 86, 11–19. [Google Scholar] [CrossRef]
- Marschner, P.; Kandeler, E.; Marschner, B. Structure and function of the soil microbial community in a long-term fertilizer experiment. Soil Biol. Biochem. 2003, 35, 453–461. [Google Scholar] [CrossRef]
- Sainju, U.M.; Lenssen, A.W.; Caesar-TonThat, T.; Jabro, J.D.; Lartey, R.T.; Evans, R.G.; Allen, B.L. Dryland soil nitrogen cycling influenced by tillage, crop rotation, and cultural practice. Nutr. Cycl. Agroecosyst. 2012, 93, 309–322. [Google Scholar] [CrossRef]
- Blagodatskaya, E.; Littschwager, J.; Lauerer, M.; Kuzyakov, Y. Plant traits regulating N capture define microbial competition in the rhizosphere. Eur. J. Soil Biol. 2014, 61, 41–48. [Google Scholar] [CrossRef]
- Knops, J.M.H.; Bradley, K.L.; Wedin, D.A. Mechanisms of plant species impacts on ecosystem nitrogen cycling. Ecol. Lett. 2002, 5, 454–466. [Google Scholar] [CrossRef]
- Moreau, D.; Pivato, B.; Bru, D.; Busset, H.; Deau, F.; Faivre, C.; Matejicek, A.; Strbik, F.; Philippot, L.; Mougel, C. Plant traits related to nitrogen uptake influence plant-microbe competition. Ecology 2015, 96, 2300–2310. [Google Scholar] [CrossRef]
- Paterson, E. Importance of rhizodeposition in the coupling of plant and microbial productivity. Eur. J. Soil Sci. 2003, 54, 741–750. [Google Scholar] [CrossRef]
- Arcand, M.M.; Knight, J.D.; Farrell, R.E. Estimating belowground nitrogen inputs of pea and canola and their contribution to soil inorganic N pools using 15N labeling. Plant Soil 2013, 371, 67–80. [Google Scholar] [CrossRef]
- Eriksen, J.; Mortensen, J.V.; Nielsen, J.D.; Nielsen, N.E. Sulphur mineralisation in five Danish soils as measured by plant uptake in a pot experiment. Agric. Ecosyst. Environ. 1995, 56, 43–51. [Google Scholar] [CrossRef]
- Gan, Y.T.; Campbell, C.A.; Janzen, H.H.; Lemke, R.; Liu, L.P.; Basnyat, P.; McDonald, C.L. Root mass for oilseed and pulse crops: Growth and distribution in the soil profile. Can. J. Plant Sci. 2009, 89, 883–893. [Google Scholar] [CrossRef]
- Haneklaus, S.; Bloem, E.; Schnug, E.; De Kok, L.; Stulen, I. Sulfur. In Handbook of Plant Nutrition; Barker, A., Pilbeam, D., Eds.; Taylor & Francis: Boca Raton, FL, USA, 2006; pp. 183–238. [Google Scholar]
- Soon, Y.K.; Arshad, M.A. Comparison of the decomposition and N and P mineralization of canola, pea and wheat residues. Biol. Fertil. Soils 2002, 36, 10–17. [Google Scholar] [CrossRef]
- Vong, P.; Piutti, S.; Slezack-Deschaumes, S.; Benizri, E.; Robin, C.; Guckert, A. Water-soluble carbon in roots of rape and barley: Impacts on labile soil organic carbon, arylsulphatase activity and sulphur mineralization. Plant Soil 2007, 294, 19–29. [Google Scholar] [CrossRef]
- Wichern, F.; Mayer, J.; Georg, R.; Müller, T. Rhizodeposition of C and N in peas and oats after 13C – 15 N double labelling under field conditions. Soil Biol. Biochem. 2007, 39, 2527–2537. [Google Scholar] [CrossRef]
- Wyszkowska, J.; Borowik, A.; Olszewski, J.; Kucharski, J. Soil Bacterial Community and Soil Enzyme Activity Depending on the Cultivation of Triticum aestivum, Brassica napus, and Pisum sativum ssp. arvense. Diversity 2019, 11, 246. [Google Scholar]
- Kwiatkowski, C.A.; Harasim, E.; Feledyn-Szewczyk, B.; Antonkiewicz, J. Enzymatic activity of loess soil in organic and conventional farming systems. Agriculture 2020, 10, 135. [Google Scholar] [CrossRef]
- Asmar, F.; Eiland, F.I.; Nielsen, N.E. Interrelationship between extracellular enzyme activity, ATP content, total counts of bacteria and CO2 evolution. Biol. Fertil. Soils 1992, 14, 288–292. [Google Scholar] [CrossRef]
- Geisseler, D.; Joergensen, R.G.; Ludwig, B. Potential soil enzyme activities are decoupled from microbial activity in dry residue-amended soil. Pedobiologia 2012, 55, 253–261. [Google Scholar] [CrossRef]
- Giacometti, C.; Scott, M.; Cavani, L.; Marzadori, C.; Ciavatta, C.; Kandeler, E. Chemical and microbiological soil quality indicators and their potential to differentiate fertilization regimes in temperate agroecosystems. Appl. Soil Ecol. 2013, 64, 32–48. [Google Scholar] [CrossRef]
- Gispert, M.; Emran, M.; Pardini, G.; Doni, S.; Ceccanti, B. The impact of land management and abandonment on soil enzymatic activity, glomalin content and aggregate stability. Geoderma 2013, 202–203, 51–61. [Google Scholar] [CrossRef]
- Tian, Y.; Cao, F.; Wang, G. Soil microbiological properties and enzyme activity in Ginkgo – tea agroforestry compared with monoculture. Agrofor. Syst. 2013, 87, 1201–1210. [Google Scholar] [CrossRef]
- Carr, P.M.; Cavigelli, M.A.; Darby, H.; Delate, K.; Eberly, J.O.; Gramig, G.G.; Heckman, J.R.; Mallory, E.B.; Reeve, J.R.; Silva, E.M.; et al. Nutrient cycling in organic field crops in canada and the united states. Agron. J. 2019, 111, 2769–2785. [Google Scholar] [CrossRef]
- Techen, A.K.; Helming, K.; Brüggemann, N.; Veldkamp, E.; Reinhold-Hurek, B.; Lorenz, M.; Bartke, S.; Heinrich, U.; Amelung, W.; Augustin, K.; et al. Soil research challenges in response to emerging agricultural soil management practices. Adv. Agron. 2020, 161, 179–240. [Google Scholar] [CrossRef]
- Burke, D.J. Effects of annual and interannual environmental variability on soil fungi associated with an old-growth, temperate hardwood forest. FEMS Microbiol. Ecol. 2015, 91, fiv053. [Google Scholar] [CrossRef]
- Gutknecht, J.L.M.; Field, C.B.; Balser, T.C. Microbial communities and their responses to simulated global change fluctuate greatly over multiple years. Glob. Chang. Biol. 2012, 18, 2256–2269. [Google Scholar] [CrossRef]
- Kaiser, C.; Koranda, M.; Kitzler, B.; Fuchslueger, L.; Schnecker, J.; Schweiger, P.; Rasche, F.; Zechmeister-Boltenstern, S.; Sessitsch, A.; Richter, A. Belowground carbon allocation by trees drives seasonal patterns of extracellular enzyme activities by altering microbial community composition in a beech forest soil. New Phytol. 2010, 187, 843–858. [Google Scholar] [CrossRef]
- Lipson, D.A.; Schmidt, S.K.; Monson, R.K. Links between microbial population dynamics and nitrogen availability in an alpine ecosystem. Ecology 1999, 80, 1623–1631. [Google Scholar] [CrossRef]
- Waldrop, M.P.; Firestone, M.K. Seasonal dynamics of microbial community composition and function in oak canopy and open grassland soils. Microb. Ecol. 2006, 52, 470–479. [Google Scholar] [CrossRef]
- Serna-Chavez, H.M.; Fierer, N.; van Bodegom, P.M. Global drivers and patterns of microbial abundance in soil. Glob. Ecol. Biogeogr. 2013, 22, 1162–1172. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Mancinelli, R.; Marinari, S.; di Felice, V.; Savin, M.C.; Campiglia, E. Soil property, CO2 emission and aridity index as agroecological indicators to assess the mineralization of cover crop green manure in a Mediterranean environment. Ecol. Indic. 2013, 34, 31–40. [Google Scholar] [CrossRef]
- Fraser, F.C.; Hallett, P.D.; Wookey, P.A.; Hartley, I.P.; Hopkins, D.W. How do enzymes catalysing soil nitrogen transformations respond to changing temperatures? Biol. Fertil. Soils 2013, 49, 99–103. [Google Scholar] [CrossRef]
- Vanegas, J.; Landazabal, G.; Melgarejo, L.M.; Beltran, M.; Uribe-Vélez, D. Structural and functional characterization of the microbial communities associated with the upland and irrigated rice rhizospheres in a neotropical Colombian savannah. Eur. J. Soil Biol. 2013, 55, 1–8. [Google Scholar] [CrossRef]
- Bell, T.H.; Henry, H.A.L. Fine scale variability in soil extracellular enzyme activity is insensitive to rain events and temperature in a mesic system. Pedobiologia 2011, 54, 141–146. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Lauber, L.; Weintraub, M.N.; Ahmed, B.; Allison, S.D.; Crenshaw, C.; Contosta, A.R.; Cusack, D.; Frey, S.; Gallo, M.E.; et al. Stoichiometry of soil enzyme activity at global scale. Ecol. Lett. 2008, 11, 1252–1264. [Google Scholar] [CrossRef]
- Allison, S.D.; Weintraub, M.N.; Gartner, T.B.; Waldrop, M.P. Evolutionary-Economic Principles as Regulators of Soil Enzyme Production and Ecosystem Function. In Soil Enzymology; Shukla, G., Varma, A., Eds.; Springer: Berlin, Germany, 2011; pp. 275–285. ISBN 978-3-642-14224-6. [Google Scholar]
- Nunan, N. The microbial habitat in soil: Scale, heterogeneity and functional consequences. J. Plant Nutr. Soil Sci. 2017, 180, 425–429. [Google Scholar] [CrossRef]
- Gray, S.B.; Classen, A.T.; Kardol, P.; Yermakov, Z.; Michael Mille, R. Multiple climate change factors interact to alter soil microbial community structure in an old-field ecosystem. Soil Sci. Soc. Am. J. 2011, 75, 2217. [Google Scholar] [CrossRef]
- Bassirirad, H. Kinetics of Nutrient Uptake by Roots: Responses to Global Change. New Phytol. 2000, 147, 155–169. [Google Scholar] [CrossRef]
- Canarini, A.; Dijkstra, F.A. Dry-rewetting cycles regulate wheat carbon rhizodeposition, stabilization and nitrogen cycling. Soil Biol. Biochem. 2015, 81, 195–203. [Google Scholar] [CrossRef]
- Mahieu, S.; Germon, F.; Aveline, A.; Hauggaard-Nielsen, H.; Ambus, P.; Jensen, E.S. The influence of water stress on biomass and N accumulation, N partitioning between above and below ground parts and on N rhizodeposition during reproductive growth of pea (Pisum sativum L.). Soil Biol. Biochem. 2009, 41, 380–387. [Google Scholar] [CrossRef]
- Lauber, C.L.; Ramirez, K.S.; Aanderud, Z.; Lennon, J.; Fierer, N. Temporal variability in soil microbial communities across land-use types. ISME J. 2013, 7, 1641–1650. [Google Scholar] [CrossRef]
- Classen, E.T.; Sundqvist, M.K.; Henning, J.A.; Newman, G.S.; Moore, J.A.M.; Cregger, M.A.; Moorhead, L.C.; Patterson, C.M. Direct and indirect effects of climate change on soil microbial and soil microbial-plant interactions: What lies ahead? Ecosphere 2015, 6, 1–21. [Google Scholar] [CrossRef]
- WRB. World Reference Base for Soil Resources 2014. International Soil Classification System for Naming Soils and Creating Legends for Soil Maps; World Soil Resources Reports No. 106; FAO: Rome, Italy, 2014; ISBN 978-92-5-108369-7. [Google Scholar]
- Yates, F. The analysis of experiments containing different crop rotations. Biometrics 1954, 10, 324–346. [Google Scholar] [CrossRef]
- Bach, H.; Hartmann, A.; Schloter, M.; Munch, J.C. PCR primers and functional probes for amplification and detection of bacterial genes for extracellular peptidases in single strains and in soil. J. Microbiol. Methods 2001, 44, 173–182. [Google Scholar] [CrossRef]
- López-Gutiérrez, J.C.; Henry, S.; Hallet, S.; Martin-Laurent, F.; Catroux, G.; Philippot, L. Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J. Microbiol. Methods 2004, 57, 399–407. [Google Scholar] [CrossRef]
- Prévost-Bourré, N.C.; Christen, R.; Dequiedt, S.; Mougel, C.; Lelivre, M.; Jolivet, C.; Shahbazkia, H.R.; Guillou, L.; Arrouays, D.; Ranjard, L. Validation and application of a PCR primer set to quantify fungal communities in the soil environment by Real-Time Quantitative PCR. PLoS ONE 2011, 6, e24166. [Google Scholar] [CrossRef]
- Bach, H.; Tomanova, J.; Schloter, M.; Munch, J.C. Enumeration of total bacteria and bacteria with genes for proteolytic activity in pure cultures and in environmental samples by quantitative PCR mediated amplification. J. Microbiol. Methods 2002, 49, 235–245. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Donachie, S.P.; Foster, J.S.; Brown, M.V. Culture clash: Challenging the dogma of microbial diversity. ISME J. 2007, 1, 97–99. [Google Scholar] [CrossRef]
- Wyss, C. Campylobacter-Wolinella group organisms are the only oral bacteria that form arylsulfatase-active colonies on a synthetic indicator medium. Infect. Immun. 1989, 57, 1380–1383. [Google Scholar] [CrossRef]
- Ladd, J.N.; Butler, J.H.A. Short-term assays of soil proteolytic enzyme activities using proteins and dipeptide derivatives as substrates. Soil Biol. Biochem. 1972, 4, 19–30. [Google Scholar] [CrossRef]
- Tabatabai, M.A.; Bremner, J.M. Arylsulfatase Activity of Soils. Soil Sci. Soc. Am. J. 1970, 34, 225–229. [Google Scholar] [CrossRef]
- Walker, D.R.; Doornenbal, G. Soil Sulfate II. As an index of the sulfur available to legumes. Can. J. Soil Sci. 1972, 52, 261–266. [Google Scholar] [CrossRef]
- Fox, J.; Weisberg, S. An {R} Companion to Applied Regression, 3rd ed.; Sage: Thousand Oaks, CA, USA, 2019. [Google Scholar]
- Ginot, V.; Gaba, S.; Beaudouin, R.; Aries, F.; Monod, H. Combined use of local and ANOVA-based global sensitivity analyses for the investigation of a stochastic dynamic model: Application to the case study of an individual-based model of a fish population. Ecol. Model. 2006, 193, 479–491. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Badalucco, L.; Kuikman, P.; Nannipieri, P. Protease and deaminase activities in wheat rhizosphere and their relation to bacterial and protozoan populations. Biol. Fertil. Soils 1996, 23, 99–104. [Google Scholar] [CrossRef]
- Fu, Q.; Liu, C.; Ding, N.; Lin, Y.; Guo, B.; Luo, J.; Wang, H. Soil microbial communities and enzyme activities in a reclaimed coastal soil chronosequence under rice – barley cropping. J. Soils Sediments 2012, 12, 1134–1144. [Google Scholar] [CrossRef]
- Cregut, M.; Piutti, S.; Slezack-deschaumes, S.; Benizri, E. Compartmentalization and regulation of arylsulfatase activities in Streptomyces sp., Microbacterium sp. and Rhodococcus sp. soil isolates in response to inorganic sulfate limitation. Microbiol. Res. 2013, 168, 12–21. [Google Scholar] [CrossRef]
- Dedourge, O.; Vong, P.C.; Lasserre-Joulin, F.; Benizri, E.; Guckert, A. Effects of glucose and rhizodeposits (with or without cysteine-S) on immobilized-35S, microbial biomass-35S and arylsulphatase activity in a calcareous and an acid brown soil. Eur. J. Soil Sci. 2004, 55, 649–656. [Google Scholar] [CrossRef]
- Piutti, S.; Slezack-Deschaumes, S.; Niknahad-Gharmakher, H.; Vong, P.-C.; Recous, S.; Benizri, E. Relationships between the density and activity of microbial communities possessing arylsulfatase activity and soil sulfate dynamics during the decomposition of plant residues in soil. Eur. J. Soil Biol. 2015, 70, 88–96. [Google Scholar] [CrossRef]
- Klose, S.; Moore, J.; Tabatabai, M. Arylsulfatase activity of microbial biomass in soils as affected by cropping systems. Biol. Fertil. Soils 1999, 29, 46–54. [Google Scholar] [CrossRef]
- Whalen, J.K.; Warman, P.R. Arylsulfatase activity in soil and soil extracts using natural and artificial substrates. Biol. Fertil. Soils 1996, 22, 373–378. [Google Scholar] [CrossRef]
- Shindo, H. Effect of continuous compost application on the activities of protease, beta-acetylglucosaminidase, and adenosine deaminase in soils of upland fields and relationships between the enzyme activities and the mineralization of organic nitrogen. Jpn. J. Soil Sci. Plant Nutr. 1992, 63, 190–195. [Google Scholar]
- Balota, E.L.; Yada, I.F.; Amaral, H.; Nakatani, A.S.; Dick, R.P.; Coyne, M.S. Long-term land use influences soil microbial biomass P and S, phosphatase and arylsulfatase activities, and S mineralization in a Brazilian oxisol L. Degrad. Dev. 2014, 25, 397–406. [Google Scholar] [CrossRef]
- Riffaldi, R.; Saviozzi, A.; Cardelli, R.; Cipolli, S.; Levi-Minzi, R. Sulphur mineralization kinetics as influenced by soil properties. Biol. Fertil. Soils 2006, 43, 209–214. [Google Scholar] [CrossRef]
- Aschi, A.; Aubert, M.; Riah-Anglet, W.; Nélieu, S.; Dubois, C.; Akpa-Vinceslas, M.; Trinsoutrot-Gattin, I. Introduction of Faba bean in crop rotation: Impacts on soil chemical and biological characteristics. Appl. Soil Ecol. 2017, 120, 219–228. [Google Scholar] [CrossRef]
- Garbeva, P.; van Elsas, J.D.; van Veen, J.A. Rhizosphere microbial community and its response to plant species and soil history. Plant Soil 2008, 302, 19–32. [Google Scholar] [CrossRef]
- Soman, C.; Li, D.; Wander, M.M.; Kent, A.D. Long-term fertilizer and crop-rotation treatments differentially affect soil bacterial community structure. Plant Soil 2017, 413, 145–159. [Google Scholar] [CrossRef]
- Meyer, A.; Focks, A.; Radl, V.; Keil, D.; Welzl, G.; Scho, I.; Boch, S.; Marhan, S.; Kandeler, E.; Schloter, M. Different land use intensities in grassland ecosystems drive ecology of microbial communities involved in nitrogen turnover in soil. PLoS ONE 2013, 8, e73536. [Google Scholar] [CrossRef]
- Thion, C.E.; Poirel, J.D.; Cornulier, T.; De Vries, F.T.; Bardgett, R.D.; Prosser, J.I. Plant nitrogen-use strategy as a driver of rhizosphere archaeal and bacterial ammonia oxidiser abundance. FEMS Microbiol. Ecol. 2016, 92, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Romillac, N.; Piutti, S.; Amiaud, B.; Slezack-Deschaumes, S. Influence of pea root traits modulating soil bioavailable C and N effects upon ammonification activity. Soil Biol. Biochem. 2015, 90, 148–151. [Google Scholar] [CrossRef]
- Kallenbach, C.; Grandy, A.S. Controls over soil microbial biomass responses to carbon amendments in agricultural systems: A meta-analysis. Agric. Ecosyst. Environ. 2011, 144, 241–252. [Google Scholar] [CrossRef]
- Gan, Y.T.A.; Liang, B.C.B.; Liu, L.P.C.; Wang, X.Y.A.; Mcdonald, C.L.A. C:N ratios and carbon distribution profile across rooting zones in oilseed and pulse crops. Crop Pasture Sci. 2011, 62, 496–503. [Google Scholar] [CrossRef]
- Jensen, B. Rhizodeposition by field-grown winter barley exposed to 14CO2 pulse labelling. Appl. Soil Ecol. 1994, 1, 65–74. [Google Scholar] [CrossRef]
- Swinnen, J. Rhizodeposition and turnover of root-derived organic material in barley and wheat under conventional and integrated management. Agric. Ecosyst. Environ. 1994, 51, 115–128. [Google Scholar] [CrossRef]
- Geisseler, D.; Horwath, W.R. Regulation of extracellular protease activity in soil in response to different sources and concentrations of nitrogen and carbon. Soil Biol. Biochem. 2008, 40, 3040–3048. [Google Scholar] [CrossRef]
- Romillac, N.; Piutti, S.; Amiaud, B.; Slezack-deschaumes, S. Effects of organic inputs derived from pea and wheat root functional traits on soil protease activities. Pedobiol. J. Soil Ecol. 2019, 77, 150576. [Google Scholar] [CrossRef]
- Sieling, K.; Christen, O. Crop rotation effects on yield of oilseed rape, wheat and barley and residual effects on the subsequent wheat. Arch. Agron. Soil Sci. 2015, 61, 1531–1549. [Google Scholar] [CrossRef]
- Brockett, B.F.T.; Prescott, C.E.; Grayston, S.J. Soil moisture is the major factor influencing microbial community structure and enzyme activities across seven biogeoclimatic zones in western Canada. Soil Biol. Biochem. 2012, 44, 9–20. [Google Scholar] [CrossRef]
- Drenovsky, R.; Steenwerth, K.; Jackson, L.; Scow, K.M. Land use and climatic factors structure regional patterns in soil microbial communities. Natl. Inst. Health 2010, 19, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Sandor, M.S.; Brad, T.; Maxim, A.; Toader, C. The influence of selected meteorological factors on microbial biomass and mineralization of two organic fertilizers. Not. Bot. Horti Agrobot. Cluj-Napoca 2011, 39, 107–113. [Google Scholar] [CrossRef]
- Zhang, N.; Liu, W.; Yang, H.; Yu, X.; Gutknecht, J.L.M.; Zhang, Z.; Wan, S.; Ma, K. Soil microbial responses to warming and increased precipitation and their implications for ecosystem C cycling. Oecologia 2013, 173, 1125–1142. [Google Scholar] [CrossRef] [PubMed]
- Reichardt, W.; Briones, A.; De Jesus, R.; Padre, B. Microbial population shifts in experimental rice systems. Appl. Soil Ecol. 2001, 17, 151–163. [Google Scholar] [CrossRef]
- Goux, X.; Amiaud, B.; Piutti, S.; Philippot, L.; Benizri, E. Spatial distribution of the abundance and activity of the sulfate ester-hydrolyzing microbial community in a rape field. J. Soils Sediments 2012, 12, 1360–1370. [Google Scholar] [CrossRef]
- Nadimi-Goki, M.; Bini, C.; Wahsha, M.; Kato, Y.; Fornasier, F. Enzyme dynamics in contaminated paddy soils under different cropping patterns (NE Italy). J. Soils Sediments 2018, 18, 2157–2171. [Google Scholar] [CrossRef]
- Chen, M.M.; Zhu, Y.G.; Su, Y.H.; Chen, B.D.; Fu, B.J.; Marschner, P. Effects of soil moisture and plant interactions on the soil microbial community structure. Eur. J. Soil Biol. 2007, 43, 31–38. [Google Scholar] [CrossRef]
- Quijano, J.C.; Kumar, P.; Drewry, D.T. Passive regulation of soil biogeochemical cycling by root water transport. Water Resour. Res. 2013, 49, 3729–3746. [Google Scholar] [CrossRef]
- Jones, D.L.; Hodge, A.; Kuzyakov, Y. Plant and mycorrhizal regulation of rhizodeposition. New Phytol. 2004, 163, 459–480. [Google Scholar] [CrossRef]
- Afzal, M.; Gagnon, A.S.; Mansell, M.G. Changes in the variability and periodicity of precipitation in Scotland. Theor. Appl. Climatol. 2015, 119, 135–159. [Google Scholar] [CrossRef]
- Vaughn, K.J.; Young, T.P. Contingent conclusions: Year of initiation influences ecological field experiments, but temporal replication is rare. Restor. Ecol. 2010, 18, 59–64. [Google Scholar] [CrossRef]
- Klemm, T.; Mcpherson, R.A. The Development of Seasonal Climate Forecasting for Agricultural Producers. Agric. For. Meteorol. 2017, 235, 384–399. [Google Scholar] [CrossRef]
- Rodriguez, D.; De Voil, P.; Hudson, D.; Brown, J.N.; Hayman, P.; Marrou, H.; Meinke, H. Predicting optimum crop designs using crop models and seasonal climate forecasts. Sci. Rep. 2018, 8, 2231. [Google Scholar] [CrossRef]
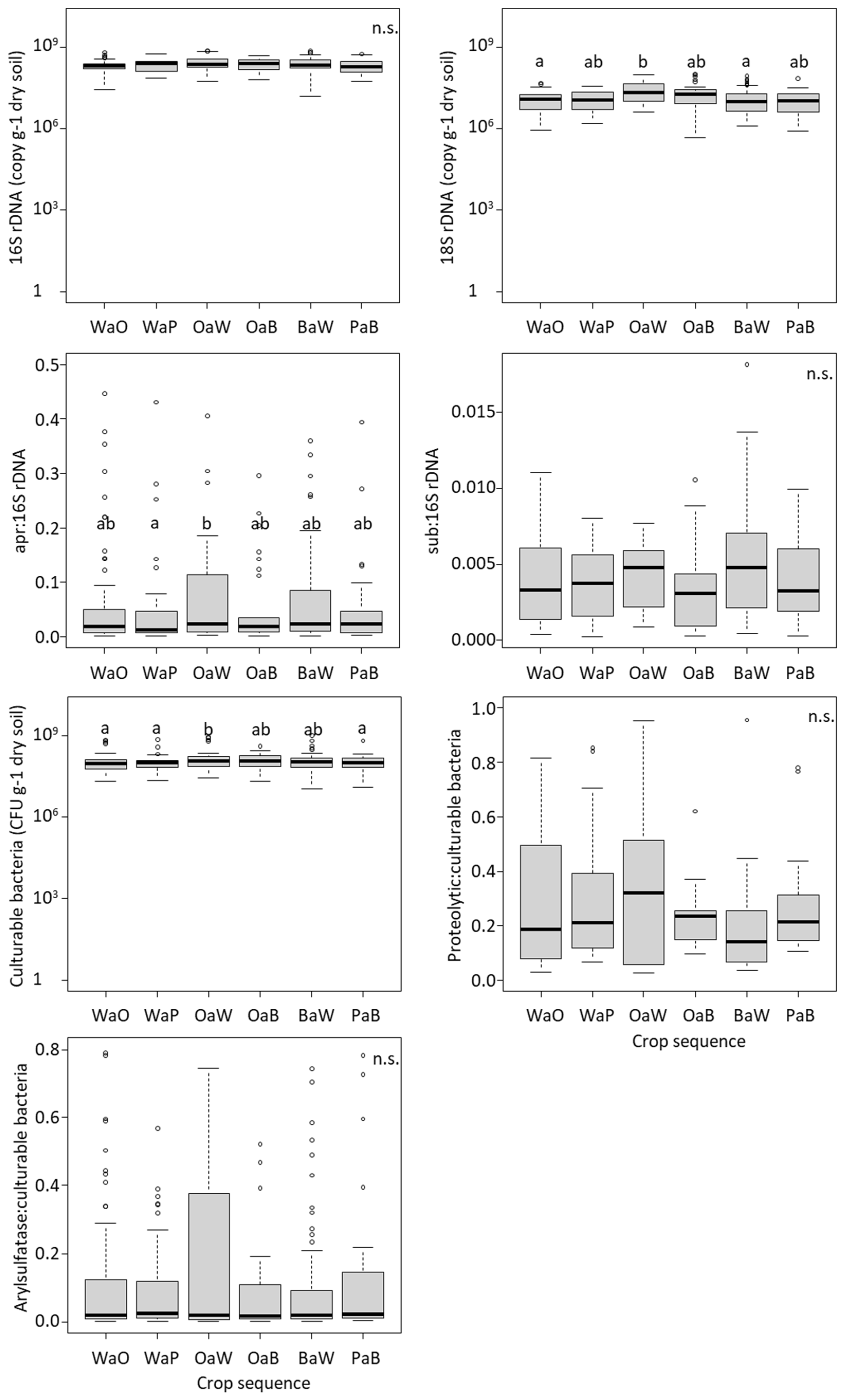


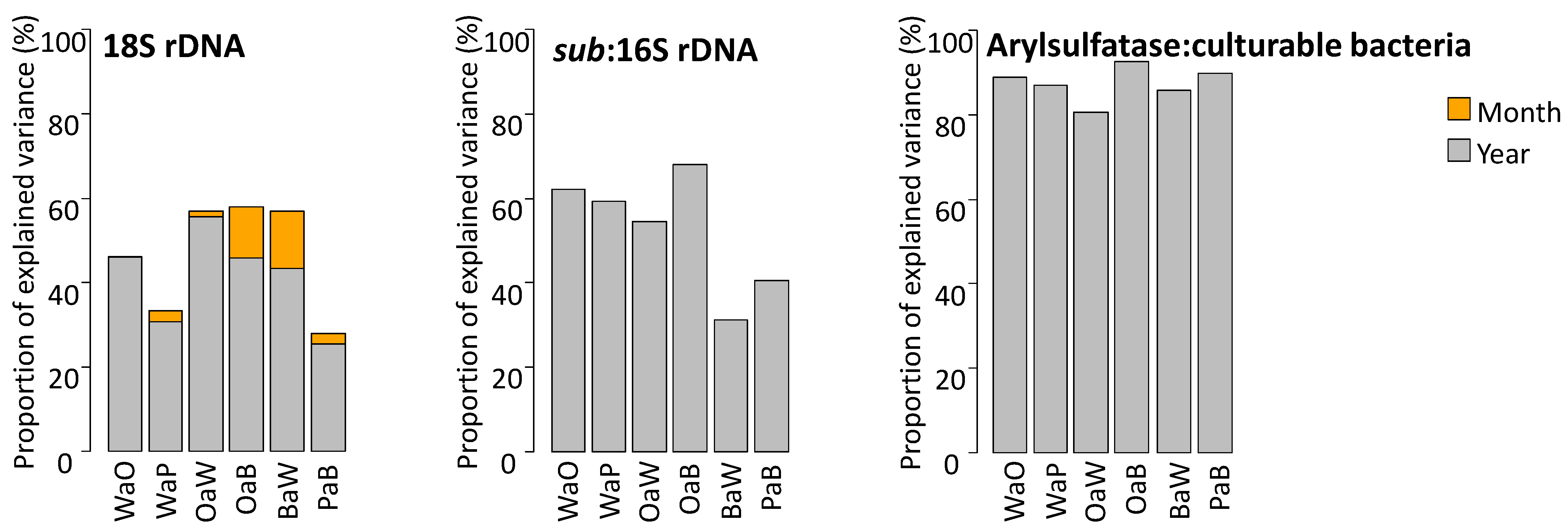
| Model (Transformation) | Crop Sequence | Month | Year | Crop Sequence X Month | Crop Sequence X Year | |
|---|---|---|---|---|---|---|
| Microbial abundance | ||||||
| 16S rDNA | GLM.nb | 1.77 | 23.15 *** | 26.17 *** | 1.10 | 0.92 |
| 18S rDNA | GLM.nb | 8.44 *** | 10.89 ** | 48.48 *** | 3.10 ** | 1.78 * |
| apr:16S rDNA | GLM.nb | 2.40 * | 5.65 * | 304.23 *** | 0.38 | 0.80 |
| sub:16S rDNA | GLM.nb | 2.20 | 0.11 | 37.45 *** | 0.22 | 2.23 ** |
| Culturable bacteria | GLM.nb | 2.92 * | 0.89 | 64.46 *** | 1.14 | 0.91 |
| Proteolytic: culturable bacteria | GLM.nb | 0.99 | 27.47 *** | 26.67 *** | 0.47 | 0.69 |
| Arylsulfatase: culturable bacteria | GLM.nb | 0.90 | 3.08 | 517.79 *** | 0.59 | 1.79 * |
| Enzymatic activity | ||||||
| Protease activity | LM (square root) | 2.53 * | 18.07 *** | 67.39 *** | 0.97 | 1.72 |
| Arylsulfatase activity | LM (square root) | 0.18 | 225.63 *** | 6.97 *** | 0.11 | 0.23 |
| Net mineralization | ||||||
| N mineralization | GLM.nb | 1.57 | -- | 20.69 *** | -- | 0.71 |
| S mineralization | LM | 0.31 | -- | 62.19 *** | -- | 0.34 |
| A | ||
| Protease Activity | Arylsulfatase Activity | |
| Transformation of the Response Variable | Square Root | Square Root |
| Log10(16S rDNA) | n.s. | n.s. |
| Log10(18S rDNA) | 0.298 ± 0.168 ** | n.s. |
| Log10(Culturable bacteria) | 0.336 ± 0.137 * | n.s. |
| apr:16S rDNA | n.s. | -- |
| sub:16S rDNA | n.s. | -- |
| Proteolytic: culturable bacteria | n.s. | -- |
| Arylsulfatase: culturable bacteria | -- | n.s. |
| F value | 8.20 | 2.50 |
| r2 | 0.29 *** | 0.02 n.s. |
| B | ||
| Net N Mineralization | Net S Mineralization | |
| Transformation of the Response Variable | Square Root | None |
| Log10(16S rDNA) | 0.417 ± 0.146 *** | n.s. |
| Log10(18S rDNA) | n.s. | n.s. |
| Log10(Culturable bacteria) | n.s. | n.s. |
| apr:16S rDNA | n.s. | -- |
| sub:16S rDNA | 0.265 ± 0.098 *** | -- |
| Proteolytic: culturable bacteria | −0.325 ± 0.115 ** | |
| Arylsulfatase: culturable bacteria | -- | 0.382 ± 0.100 *** |
| Protease activity | 0.366 ± 0.117 ** | -- |
| Arysulfatase activity | -- | −0.508 ± 0.065 *** |
| F value | 10.18 | 17.94 |
| r2 | 0.53 *** | 0.50 *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romillac, N.; Slezack-Deschaumes, S.; Amiaud, B.; Piutti, S. Soil Microbial Communities Involved in Proteolysis and Sulfate-Ester Hydrolysis Are More Influenced by Interannual Variability than by Crop Sequence. Agronomy 2023, 13, 180. https://doi.org/10.3390/agronomy13010180
Romillac N, Slezack-Deschaumes S, Amiaud B, Piutti S. Soil Microbial Communities Involved in Proteolysis and Sulfate-Ester Hydrolysis Are More Influenced by Interannual Variability than by Crop Sequence. Agronomy. 2023; 13(1):180. https://doi.org/10.3390/agronomy13010180
Chicago/Turabian StyleRomillac, Nicolas, Sophie Slezack-Deschaumes, Bernard Amiaud, and Séverine Piutti. 2023. "Soil Microbial Communities Involved in Proteolysis and Sulfate-Ester Hydrolysis Are More Influenced by Interannual Variability than by Crop Sequence" Agronomy 13, no. 1: 180. https://doi.org/10.3390/agronomy13010180
APA StyleRomillac, N., Slezack-Deschaumes, S., Amiaud, B., & Piutti, S. (2023). Soil Microbial Communities Involved in Proteolysis and Sulfate-Ester Hydrolysis Are More Influenced by Interannual Variability than by Crop Sequence. Agronomy, 13(1), 180. https://doi.org/10.3390/agronomy13010180






