Comprehensive Analysis of GASA Family Members in the Peanut Genome: Identification, Characterization, and Their Expressions in Response to Pod Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Screening and Identification of the GASA Family in the Peanut
2.2. Phylogenetic Analysis of GASA Proteins
2.3. Conserved Motif and Gene Structure Analyses
2.4. Chromosome Localization, Duplications, and Evolutionary Analysis of AhGASAs
2.5. Promoter Analysis of AhGASAs
2.6. Plant Materials
2.7. Gene Expression Analysis and Functional Annotation of AhGASAs
2.8. Validation of Gene Expression
3. Results
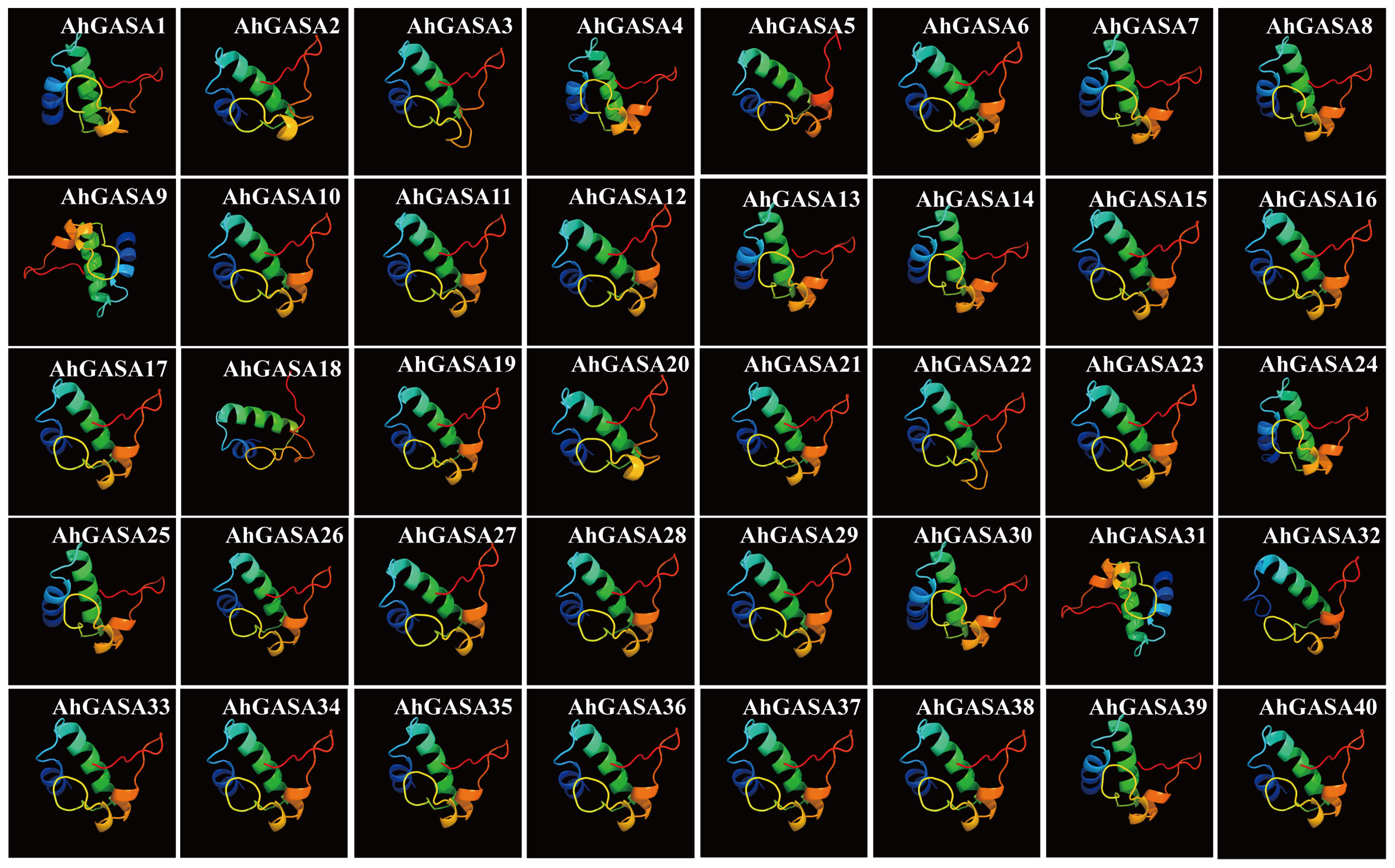
3.1. Composition and Protein Characteristics of the Peanut GASA-Gene Family
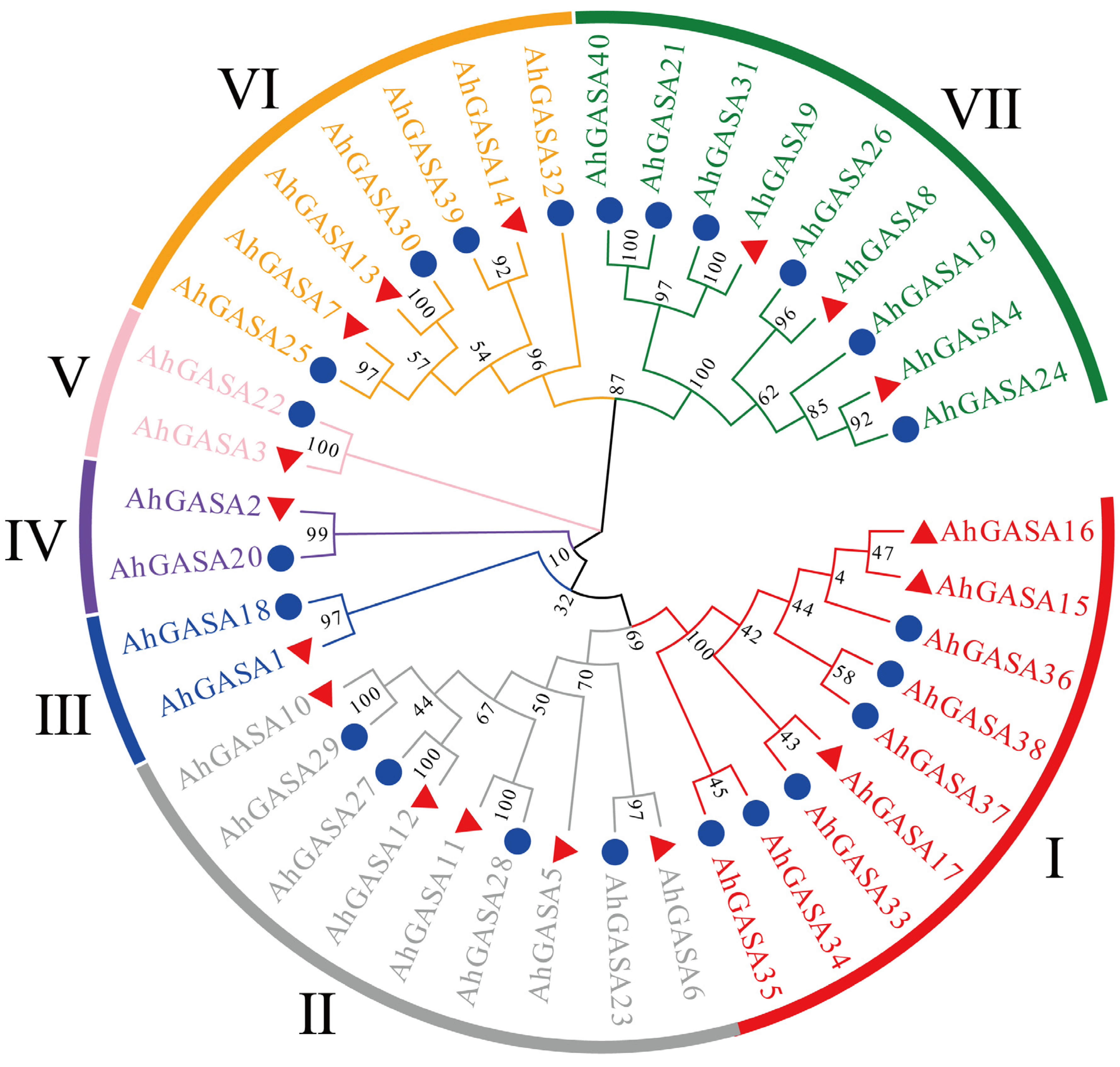
3.2. Phylogenetic Analysis of GASAs
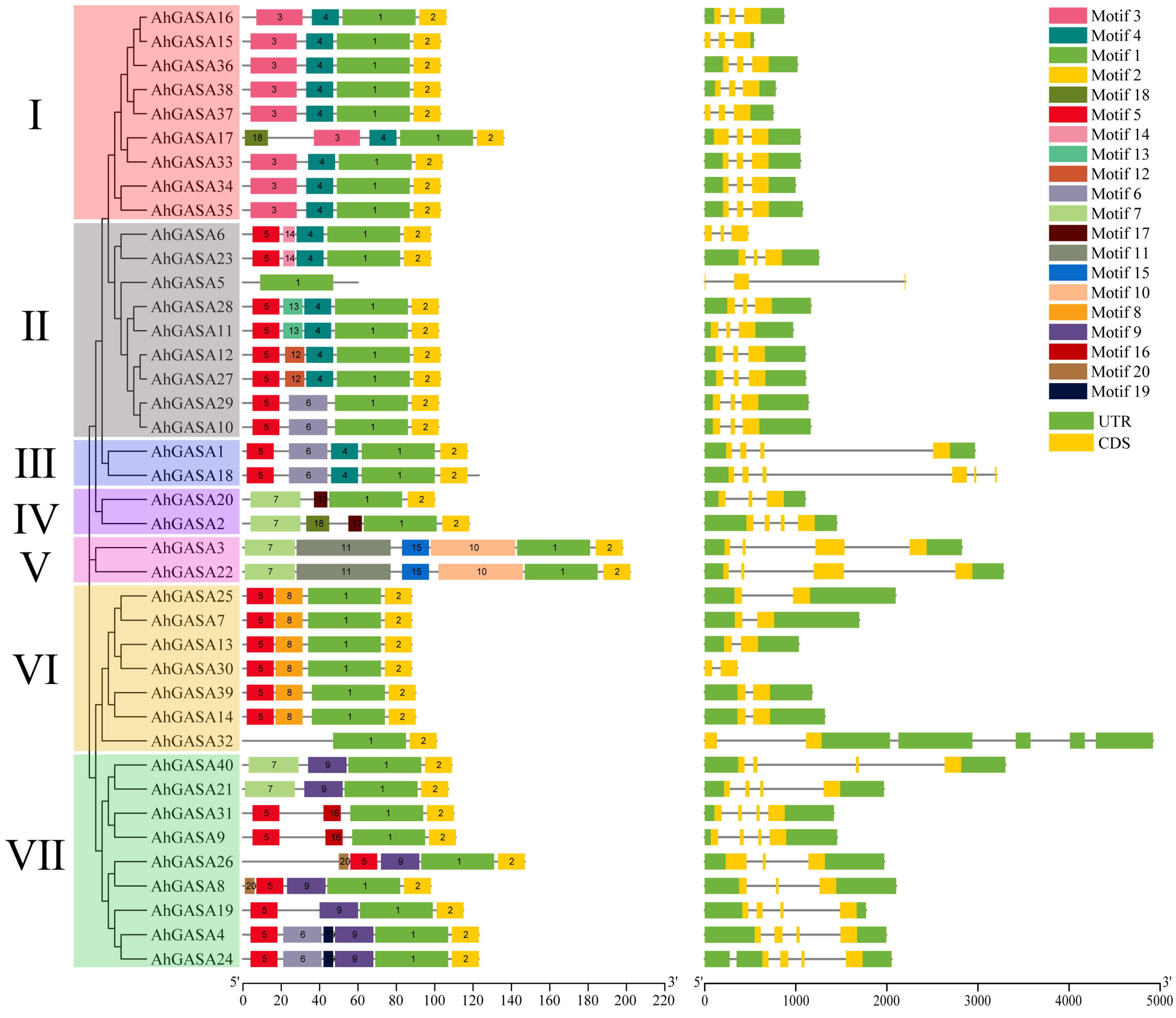
3.3. Correlation Analysis of AhGASA Gene Structure and Motif Composition
3.4. Chromosome Location and Gene Duplication of GASAs
3.5. Duplication Events of GASAs and Collinearity Analysis
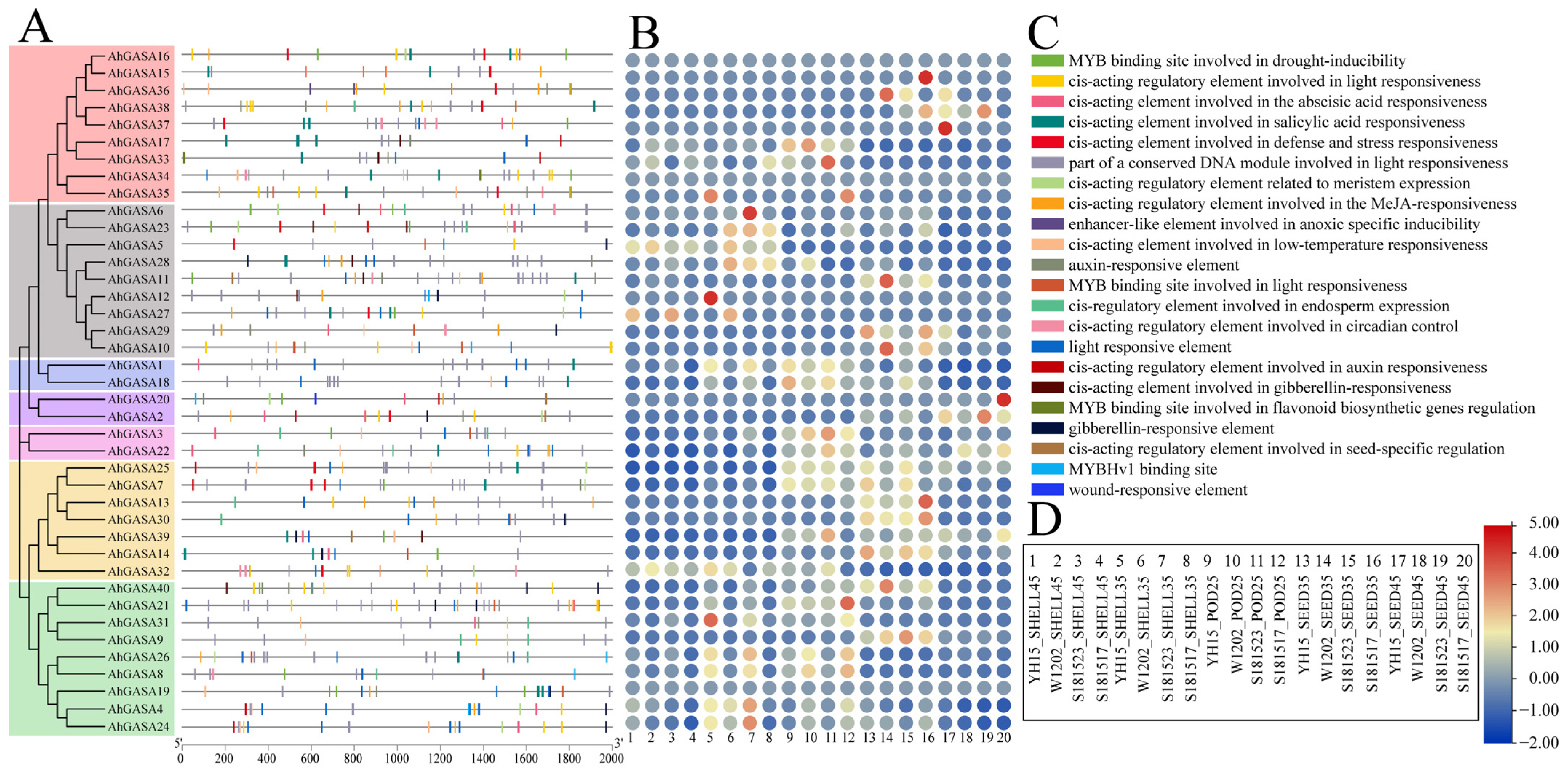
3.6. Analysis of Cis-Acting Elements in AhGASA Promoters
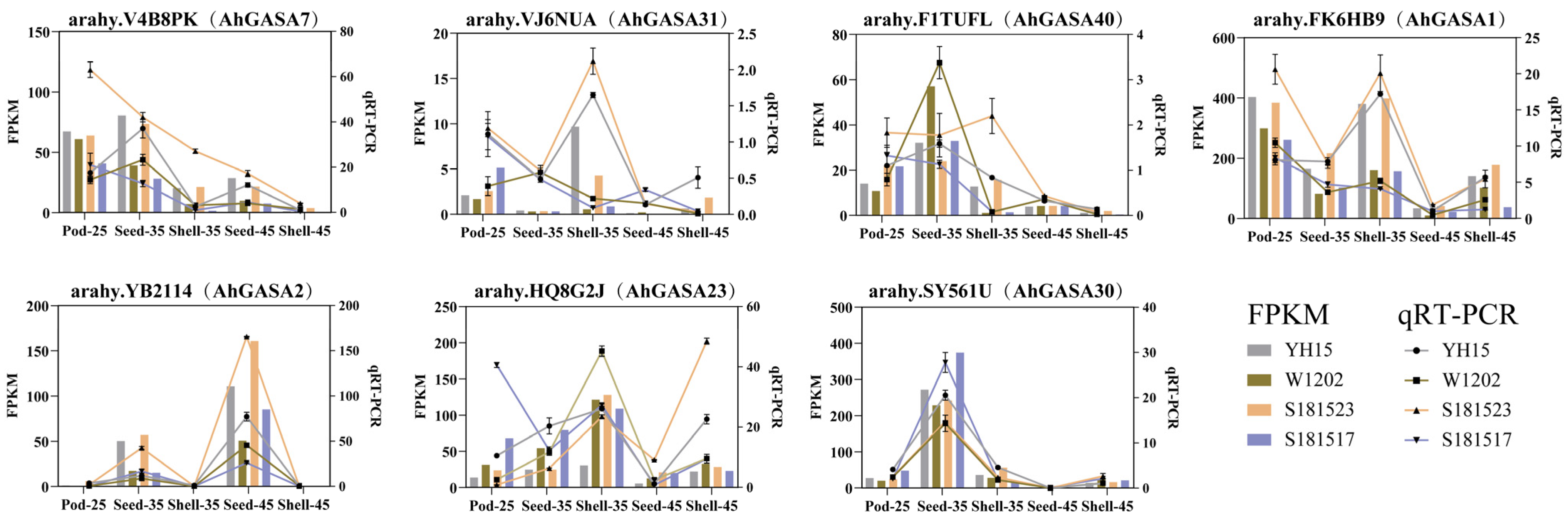
3.7. Expression Analysis of AhGASAs under Different Developmental Stages of Peanut Pod Using RNA-seq
3.8. Expression Analysis of AhGASAs under Different Developmental Stages of Peanut Pod Using RNA-seq
4. Discussion
4.1. Genome-Wide Identification and Characteristics of GASA Gene Families in Cultivated Species
4.2. Evolutionary and Syntenic Relationships among GASA Genes
4.3. AhGASA Gene Expression Profiles and Potential Functions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Roxrud, I.; Lid, S.E.; Fletcher, J.C.; Schmidt, E.D.L.; Opsahl-Sorteberg, H.G. GASA4, One of the 14-Member Arabidopsis GASA Family of Small Polypeptides, Regulates Flowering and Seed Development. Plant Cell Physiol. 2007, 48, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Trapalis, M.; Li, S.F.; Parish, R.W. The Arabidopsis GASA10 gene encodes a cell wall protein strongly expressed in developing anthers and seeds. Plant Sci. 2017, 260, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Zhang, D.; Zhang, L.; Gao, C.; Xin, M.; Tahir, M.M.; Li, Y.; Ma, J.; Han, M. Comprehensive analysis of GASA family members in the Malus domestica genome: Identification, characterization, and their expressions in response to apple flower induction. BMC Genom. 2017, 18, 827. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Gast, R.T.; Gopalraj, M.; Olszewski, N.E. Characterization of a shoot-specific, GA3- and ABA-regulated gene from tomato. Plant J. 1992, 2, 153–159. [Google Scholar] [PubMed]
- Aubert, D.; Chevillard, M.; Dorne, A.M.; Arlaud, G.; Herzog, M. Expression patterns of GASA genes in Arabidopsis thaliana: The GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol. Biol. 1998, 36, 871–883. [Google Scholar] [CrossRef]
- Hou, D.; Bai, Q.; Li, J.; Xie, L.; Li, X.; Cheng, Z.; Gao, J. The Gibberellic Acid-Stimulated Transcript Gene Family in Moso Bamboo: A Genome-Wide Survey and Expression Profiling During Development and Abiotic Stresses. J. Plant Growth Regul. 2018, 37, 1135–1147. [Google Scholar] [CrossRef]
- Muhammad, I.; Li, W.; Jing, X.; Zhou, M.; Shalmani, A.; Ali, M.; Wei, X.; Sharif, R.; Liu, W.; Chen, K. A systematic in silico prediction of gibberellic acid stimulated GASA family members: A novel small peptide contributes to floral architecture and transcriptomic changes induced by external stimuli in rice. J. Plant Physiol. 2019, 234–235, 117–132. [Google Scholar] [CrossRef]
- Lü, L.; Chen, X.; Zhang, Y.; Liu, Q.; Wang, L.; Ma, L.; Li, H. Bioinformatics Identification of GASA Gene Family Expression Profiles in Wheat. Crops 2018, 34, 58. [Google Scholar]
- Ahmad, M.Z.; Sana, A.; Jamil, A.; Nasir, J.A.; Ahmed, S.; Hameed, M.U.; Abdullah. A genome-wide approach to the comprehensive analysis of GASA gene family in Glycine max. Plant Mol. Biol. 2019, 100, 607–620. [Google Scholar] [CrossRef]
- Nahirñak, V.; Rivarola, M.; Gonzalez De Urreta, M.; Paniego, N.; Hopp, H.E.; Almasia, N.I.; Vazquez-Rovere, C. Genome-wide Analysis of the Snakin/GASA Gene Family in Solanum tuberosum cv. Kennebec. Am. J. Potato Res. 2016, 93, 172–188. [Google Scholar] [CrossRef]
- Ahmad, B.; Yao, J.; Zhang, S.; Li, X.; Zhang, X.; Yadav, V.; Wang, X. Genome-Wide Characterization and Expression Profiling of GASA Genes during Different Stages of Seed Development in Grapevine (Vitis vinifera L.) Predict Their Involvement in Seed Development. Int. J. Mol. Sci. 2020, 21, 1088. [Google Scholar] [CrossRef] [PubMed]
- Filiz, E.; Kurt, F. Antimicrobial peptides Snakin/GASA gene family in sorghum (Sorghum bicolor): Genome-wide identification and bioinformatics analyses. Gene Rep. 2020, 20, 100766. [Google Scholar] [CrossRef]
- Qiao, K.; Ma, C.; Lv, J.; Zhang, C.; Ma, Q.; Fan, S. Identification, characterization, and expression profiles of the GASA genes in cotton. J. Cotton Res. 2021, 4, 7. [Google Scholar] [CrossRef]
- Sun, S.; Wang, H.; Yu, H.; Zhong, C.; Zhang, X.; Peng, J.; Wang, X. GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J. Exp. Bot. 2013, 64, 1637–1647. [Google Scholar] [CrossRef]
- Dong, L.; Wang, F.; Liu, T.; Dong, Z.; Li, A.; Jing, R.; Mao, L.; Li, Y.; Liu, X.; Zhang, K.; et al. Natural variation of TaGASR7-A1 affects grain length in common wheat under multiple cultivation conditions. Mol. Breed. 2014, 34, 937–947. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, X. Expression pattern of GASA, downstream genes of DELLA, in Arabidopsis. Sci. Bull. 2008, 53, 3839–3846. [Google Scholar] [CrossRef]
- Li, K.; Bai, X.; Li, Y.; Cai, H.; Ji, W.; Tang, L.; Wen, Y.; Zhu, Y. GsGASA1 mediated root growth inhibition in response to chronic cold stress is marked by the accumulation of DELLAs. J. Plant Physiol. 2011, 168, 2153–2160. [Google Scholar] [CrossRef]
- Furukawa, T.; Sakaguchi, N.; Shimada, H. Two OsGASR genes, rice GAST homologue genes that are abundant in proliferating tissues, show different expression patterns in developing panicles. Genes Genet. Syst. 2006, 81, 171–180. [Google Scholar] [CrossRef]
- Li, N.; Xu, R.; Li, Y. Molecular Networks of Seed Size Control in Plants. Annu. Rev. Plant Biol. 2019, 70, 435–463. [Google Scholar] [CrossRef]
- Shu, K.; Liu, X.D.; Xie, Q.; He, Z.H. Two Faces of One Seed: Hormonal Regulation of Dormancy and Germination. Mol. Plant. 2016, 9, 34–45. [Google Scholar] [CrossRef]
- Nohales, M.A.; Kay, S.A. GIGANTEA gates gibberellin signaling through stabilization of the DELLA proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 21893–21899. [Google Scholar] [CrossRef] [PubMed]
- Pimenta Lange, M.J.; Lange, T. Gibberellin Biosynthesis and the Regulation of Plant Development. Plant Biol. 2006, 8, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Ben-Nissan, G.; Lee, J.; Borohov, A.; Weiss, D. GIP, a Petunia hybrida GA-induced cysteine-rich protein: A possible role in shoot elongation and transition to flowering. Plant J. 2004, 37, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yang, C.; Peng, J.; Sun, S.; Wang, X. GASA5, a regulator of flowering time and stem growth in Arabidopsis thaliana. Plant Mol.Biol. 2009, 69, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Sun, Z.; Fang, Y.; Qi, F.; Liu, H.; Miao, L.; Du, P.; Shi, L.; Gao, W.; Han, S.; et al. Genetic Diversity, Population Structure, and Botanical Variety of 320 Global Peanut Accessions Revealed through Tunable Genotyping-by-Sequencing. Sci Rep 2018, 8, 14500. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nature Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nature Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Liu, Y.; Xiao, L.; Chi, J.; Li, R.; Han, Y.; Cui, F.; Peng, Z.; Wan, S.; Li, G. Genome-wide identification and expression of SAUR gene family in peanut (Arachis hypogaea L.) and functional identification of AhSAUR3 in drought tolerance. BMC Plant Biol. 2022, 22, 178. [Google Scholar] [CrossRef]
- Huang, R.; Xiao, D.; Wang, X.; Zhan, J.; Wang, A.; He, L. Genome-wide identification, evolutionary and expression analyses of LEA gene family in peanut (Arachis hypogaea L.). Bmc Plant Biol. 2022, 22, 155. [Google Scholar] [CrossRef]
- Zhao, N.; He, M.; Li, L.; Cui, S.; Hou, M.; Wang, L.; Mu, G.; Liu, L.; Yang, X. Identification and expression analysis of WRKY gene family under drought stress in peanut (Arachis hypogaea L.). PLoS ONE 2020, 15, e231396. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; Huai, D.; Chen, Y.; Jiang, Y.; Ding, Y.; Kang, Y.; Wang, Z.; Yan, L.; Jiang, H.; et al. Genome-wide identification of peanut PIF family genes and their potential roles in early pod development. Gene 2021, 781, 145539. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Wan, W.; Yu, F.; Meng, L. New Insights into the Molecular Mechanism Underlying Seed Size Control under Drought Stress. J. Agric. Food Chem. 2019, 67, 9697–9704. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sun, Z.; Qi, F.; Tian, M.; Wang, J.; Zhao, R.; Wang, X.; Wu, X.; Shi, X.; Liu, H.; et al. Comparative transcriptomics analysis of developing peanut (Arachis hypogaea L.) pods reveals candidate genes affecting peanut seed size. Front. Plant Sci. 2022, 13, 958808. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, X. One new kind of phytohormonal signaling integrator: Up-and-coming GASA family genes. Plant Signal. Behav. 2017, 12, e1226453. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nature Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lu, Q.; Liu, H.; Zhang, J.; Hong, Y.; Lan, H.; Li, H.; Wang, J.; Liu, H.; Li, S.; et al. Sequencing of Cultivated Peanut, Arachis hypogaea, Yields Insights into Genome Evolution and Oil Improvement. Mol. Plant. 2019, 12, 920–934. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Qu, Y.; Rong, H.; Han, X.; Tian, Y.; Xu, L.A. Identification and Expression Analysis of the Populus trichocarpa GASA-Gene Family. Int. J. Mol. Sci. 2022, 23, 1507. [Google Scholar] [CrossRef]
- Silverstein, K.A.T.; Moskal, W.A.; Wu, H.C.; Underwood, B.A.; Graham, M.A.; Town, C.D.; Vandenbosch, K.A. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007, 51, 262–280. [Google Scholar] [CrossRef]
- Wigoda, N.; Ben-Nissan, G.; Granot, D.; Schwartz, A.; Weiss, D. The gibberellin-induced, cysteine-rich protein GIP2 from Petunia hybrida exhibitsin planta antioxidant activity. Plant J. 2006, 48, 796–805. [Google Scholar] [CrossRef]
- Rubinovich, L.; Weiss, D. The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J. 2010, 64, 1018–1027. [Google Scholar] [CrossRef]
- Moyano-Canete, E.; Bellido, M.L.; Garcia-Caparros, N.; Medina-Puche, L.; Amil-Ruiz, F.; Gonzalez-Reyes, J.A.; Caballero, J.L.; Munoz-Blanco, J.; Blanco-Portales, R. FaGAST2, a strawberry ripening-related gene, acts together with FaGAST1 to determine cell size of the fruit receptacle. Plant Cell Physiol. 2013, 54, 218–236. [Google Scholar] [CrossRef]
- Qu, J.; Kang, S.G.; Hah, C.; Jang, J.C. Molecular and cellular characterization of GA-Stimulated Transcripts GASA4 and GASA6 in Arabidopsis thaliana. Plant Sci. 2016, 246, 550. [Google Scholar] [CrossRef] [PubMed]
- Jafari, M.; Shiran, B.; Rabiei, G.; Ravash, R.; Sayed Tabatabaei, B.E.; Martínez-García, P.J.; Martínez-Gómez, P. Identification of genes associated with kernel size in almond [Prunus dulcis (Mill.) D.A. Webb] using RNA-Seq. Plant Growth Regul. 2022, 97, 357–373. [Google Scholar] [CrossRef]
- Nakano, T.; Fujisawa, M.; Shima, Y.; Ito, Y. Expression profiling of tomato pre-abscission pedicels provides insights into abscission zone properties including competence to respond to abscission signals. Bmc Plant Biol. 2013, 13, 40. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Sun, Z.; Qi, F.; Zhao, M.; Dong, W.; Huang, B.; Zheng, Z.; Zhang, X. Comprehensive Analysis of GASA Family Members in the Peanut Genome: Identification, Characterization, and Their Expressions in Response to Pod Development. Agronomy 2022, 12, 3067. https://doi.org/10.3390/agronomy12123067
Wu Y, Sun Z, Qi F, Zhao M, Dong W, Huang B, Zheng Z, Zhang X. Comprehensive Analysis of GASA Family Members in the Peanut Genome: Identification, Characterization, and Their Expressions in Response to Pod Development. Agronomy. 2022; 12(12):3067. https://doi.org/10.3390/agronomy12123067
Chicago/Turabian StyleWu, Yue, Ziqi Sun, Feiyan Qi, Mingbo Zhao, Wenzhao Dong, Bingyan Huang, Zheng Zheng, and Xinyou Zhang. 2022. "Comprehensive Analysis of GASA Family Members in the Peanut Genome: Identification, Characterization, and Their Expressions in Response to Pod Development" Agronomy 12, no. 12: 3067. https://doi.org/10.3390/agronomy12123067
APA StyleWu, Y., Sun, Z., Qi, F., Zhao, M., Dong, W., Huang, B., Zheng, Z., & Zhang, X. (2022). Comprehensive Analysis of GASA Family Members in the Peanut Genome: Identification, Characterization, and Their Expressions in Response to Pod Development. Agronomy, 12(12), 3067. https://doi.org/10.3390/agronomy12123067





