Genetic Characterization of a Plum Landrace Collection from La Palma, Canary Islands
Abstract
:1. Introduction
2. Materials and Methods
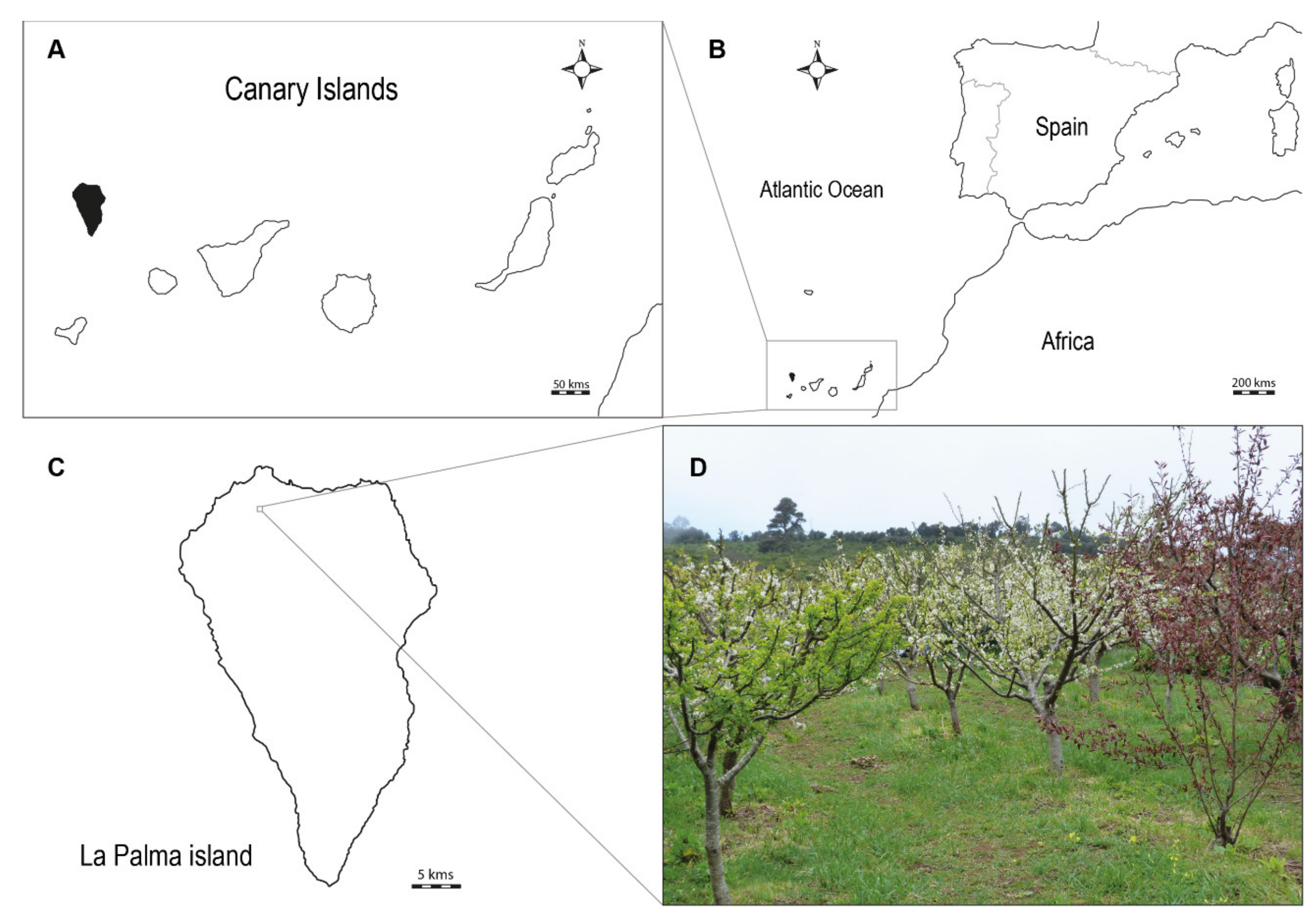
2.1. Plant Material
2.2. Flow Cytometry Analysis
2.3. DNA Extraction and PCR Amplification
2.4. Population Genetic Structure
2.5. Genetic Diversity Analyses
3. Results
3.1. Species Discrimination by Ploidy Level
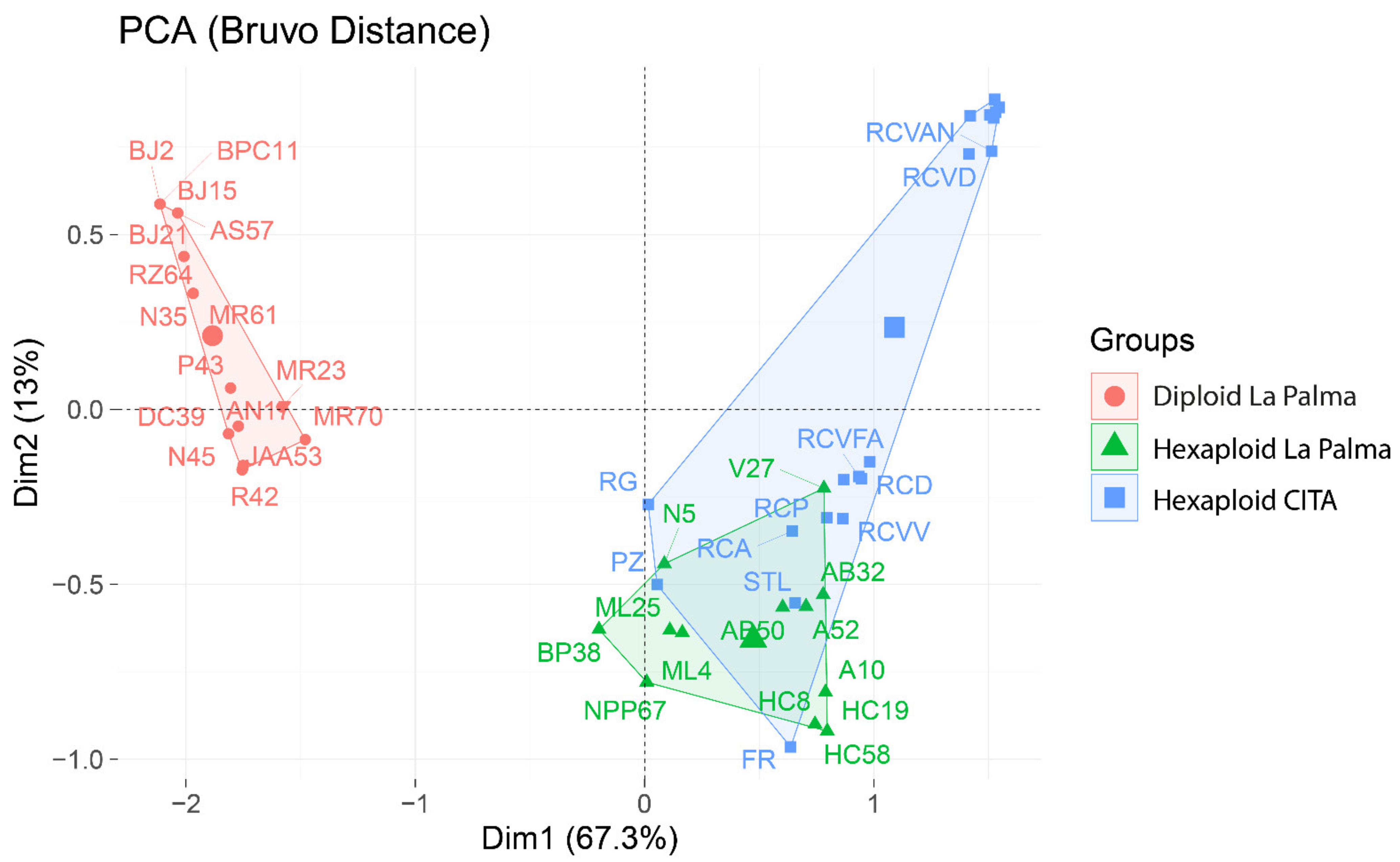
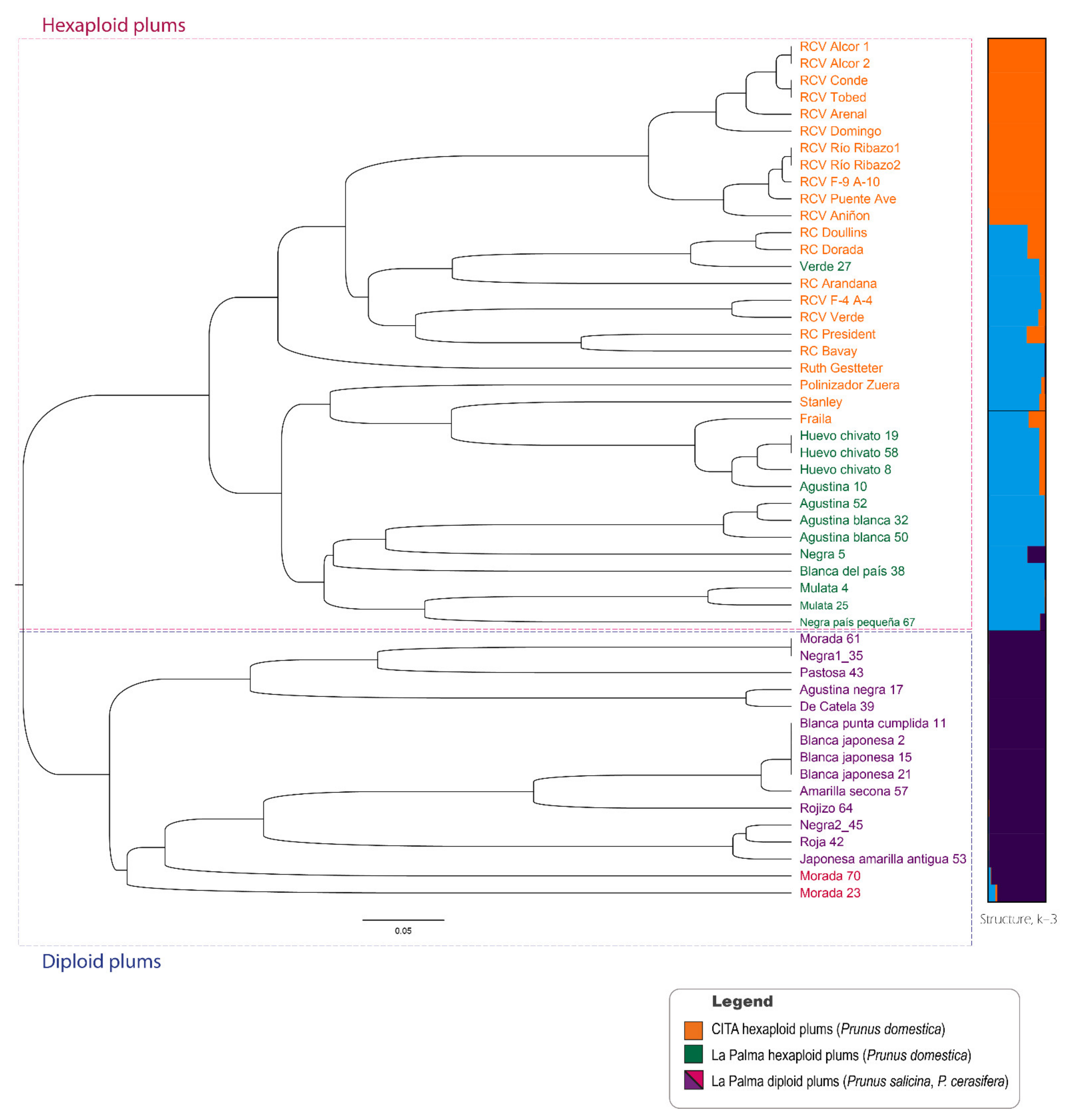
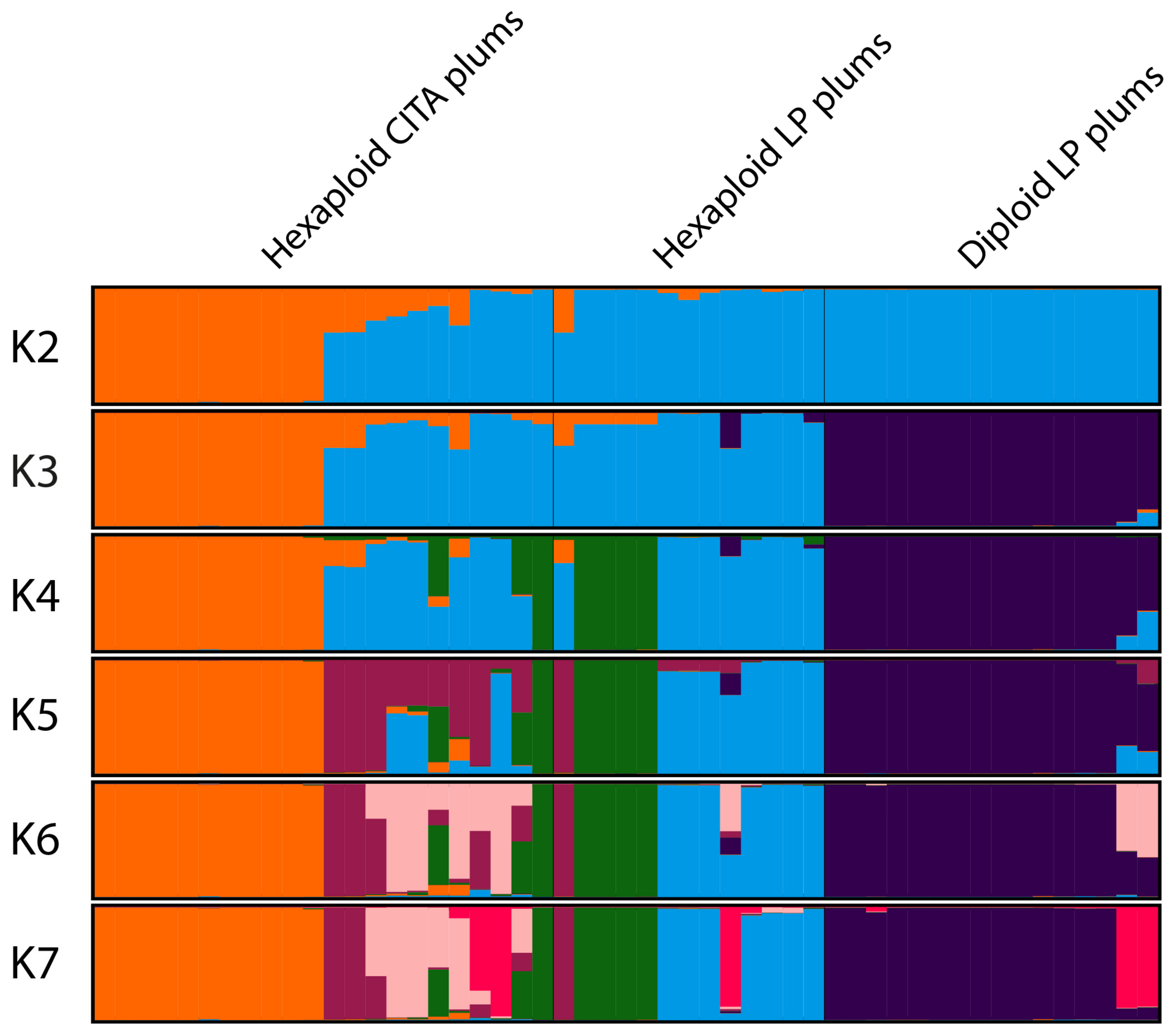
3.2. Genetic Grouping
3.3. SSR Polymorphisms and Molecular Characterization
4. Discussion
4.1. Ploidy Level and Species Assignment
4.2. Genetic Diversity and Population Structure
4.3. Relations between La Palma Plum Accessions
4.4. Genetic Erosion and Conservation of La Palma Plums
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Khoury, C.K.; Bjorkman, A.D.; Dempewolf, H.; Ramirez-Villegas, J.; Guarino, L.; Jarvis, A.; Rieseberg, L.H.; Struik, P.C. Increasing homogeneity in global food supplies and the implications for food security. Proc. Natl. Acad. Sci. USA 2014, 111, 4001–4006. [Google Scholar] [CrossRef] [PubMed]
- Maxted, N.; Hunter, D.; Ortíz, R. Whole Plant, Plantlet and DNA Conservation. In Plant Genetic Conservation; Cambridge University Press: Cambridge, UK, 2020; pp. 368–390. [Google Scholar] [CrossRef]
- Hernández, Y.; Pereira, Â.G.; Barbosa, P. Resilient futures of a small island: A participatory approach in Tenerife (Canary Islands) to address climate change. Environ. Sci. Policy 2018, 80, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Hernández, Á.; Díaz-Díaz, R.; Zumbado, M.; Bernal- Suárez, M.D.M.; Acosta-Dacal, A.; Macías-Montes, A.; Travieso-Aja, M.D.M.; Rial-Berriel, C.; Hernández-Hernández, L.A.; Boada, L.D.; et al. Impact of chemical elements released by the volcanic eruption of La Palma (Canary Islands, Spain) on banana agriculture and European consumers. Chemosphere 2022, 293, 133508. [Google Scholar] [CrossRef] [PubMed]
- Galán-Saúco, V.; Cubero, J.I. Contribution of Spain and Portugal to the exchange and acclimatization of new and old world crops. Acta Hortic. 2011, 916, 71–82. [Google Scholar] [CrossRef]
- Marsal, G.; Méndez, J.J.; Mateo, J.M.; Ferrer, S.; Canals, J.M.; Zamora, F.; Fort, F. Molecular characterization of Vitis vinifera L. local cultivars from volcanic areas (Canary Islands and Madeira) using SSR markers. Oeno One 2019, 53, 667–680. [Google Scholar] [CrossRef]
- Rodríguez-Padrón, C.; Siverio, F.; Pérez-Sierra, A.; Rodríguez, A. Isolation and pathogenicity of Phytophthora species and Phytopythium vexans recovered from avocado orchards in the Canary Islands, including Phytophthora niederhauserii as a new pathogen of avocado. Phytopathol. Mediterr. 2018, 57, 89–106. [Google Scholar] [CrossRef]
- Padilla, G.; Ordás, A. Molecular characterization of almond accessions from the island of La Palma (Canary Islands, Spain) using SSR markers. Plant Genet. Resour. 2014, 12, 323–329. [Google Scholar] [CrossRef]
- Pérez, V.; Larrañaga, N.; Abdallah, D.; Wünsch, A.; Hormaza, J.I. Genetic diversity of local peach (Prunus persica) accessions from La Palma island (Canary Islands, Spain). Agronomy 2020, 10, 457. [Google Scholar] [CrossRef]
- Pereira-Lorenzo, S.; Ramos-Cabrer, A.M.; González-Díaz, A.J.; Díaz-Hernández, M.B. Genetic assessment of local apple cultivars from La Palma, Spain, using simple sequence repeats (SSRs). Sci. Hortic. 2008, 117, 160–166. [Google Scholar] [CrossRef]
- Pereira-Lorenzo, S.; Costa, R.M.L.; Ramos-Cabrer, A.M.; Ciordia-Ara, M.; Ribeiro, C.A.M.; Borges, O.; Barreneche, T. Chestnut cultivar diversification process in the Iberian Peninsula, Canary Islands, and Azores. Genome 2011, 54, 301–315. [Google Scholar] [CrossRef]
- Velázquez-Barrera, M.E.; Ramos-Cabrer, A.M.; Pereira-Lorenzo, S.; Ríos-Mesa, D.J. Genetic pool of the cultivated pear tree (Pyrus spp.) in the Canary Islands (Spain), studied using SSR molecular markers. Agronomy 2022, 12, 1711. [Google Scholar] [CrossRef]
- De Galarreta, J.I.R.; Barandalla, L.; Ríos, D.J.; López, R.; Ritter, E. Genetic relationships among local potato cultivars from Spain using SSR markers. Genet. Resour. Crop Evol. 2011, 58, 383–395. [Google Scholar] [CrossRef]
- Parrilla-González, M.; Rios-Mesa, D.; Méndez-Hernández, C.; Hernández-González, J.Z.; Fernández-Galván, D.; Hernández-Delgado, P.M.; Galán-Saúco, V. Preliminary study of the seedling avocado (Persea americana Mill.) population of Tenerife. Acta Hortic. 2012, 928, 81–86. [Google Scholar] [CrossRef]
- Abdallah, D.; Baraket, G.; Pérez, V.; Ben Mustapha, S.; Salhi-Hannachi, A.; Hormaza, J.I. Analysis of self-incompatibility and genetic diversity in diploid and hexaploid plum genotypes. Front. Plant Sci. 2019, 10, 896. [Google Scholar] [CrossRef] [PubMed]
- Sapir, G.; Stern, R.A.; Shafir, S.; Goldway, M. Full compatibility is superior to semi-compatibility for fruit set in Japanese plum (Prunus salicina Lindl.) cultivars. Sci. Hortic. 2008, 116, 394–398. [Google Scholar] [CrossRef]
- Guerra, M.E.; López-Corrales, M.; Wünsch, A. Improved S-genotyping and new incompatibility groups in Japanese plum. Euphytica 2012, 186, 445–452. [Google Scholar] [CrossRef]
- Guerra, M.E.; Guerrero, B.I.; Casadomet, C.; Rodrigo, J. Self-(in) compatibility, S-RNase allele identification, and selection of pollinizers in new Japanese plum-type cultivars. Sci. Horti. 2020, 261, 109022. [Google Scholar] [CrossRef]
- Socas i Company, R.S.; Kodad, O.; Fernández i Martí, A.; Alonso, J.M. Mutations conferring self-compatibility in Prunus species: From deletions and insertions to epigenetic alterations. Sci. Hortic. 2015, 192, 125–131. [Google Scholar] [CrossRef]
- Fernandez i Marti, A.; Castro, S.; DeJong, T.M.; Dodd, R.S. Evaluation of the S-locus in Prunus domestica, characterization, phylogeny and 3D modelling. PLoS ONE 2021, 16, e0251305. [Google Scholar] [CrossRef]
- Zohary, D.; Hopf, M.; Weiss, E. Domestication of Plants in the Old World: The Origin and Spread of Cultivated Plants in West Asia, Europe and the Nile Valley, 4th ed.; Oxford University Press: New York, NY, USA, 2012; pp. 140–142. [Google Scholar]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef]
- Dufresne, F.; Stift, M.; Vergilino, R.; Mables, B.K. Recent progress and challenges in population genetics of polyploid organisms: An overview of current state-of-the-art molecular and statistical tools. Mol. Ecol. 2014, 23, 40–69. [Google Scholar] [CrossRef] [PubMed]
- Gaši, F.; Sehic, J.; Grahic, J.; Hjeltnes, S.H.; Ordidge, M.; Benedikova, D.; Blouin-Delmas, M.; Drogoudi, P.; Giovannini, D.; Höfer, M.; et al. Genetic assessment of the pomological classification of plum Prunus domestica L. accessions sampled across Europe. Genet. Resour. Crop Evol. 2020, 67, 1137–1161. [Google Scholar] [CrossRef]
- Faust, M.; Timon, B.; Surányi, D.; Nyujtó, F.; Gradziel, T.M. Origin and Dissemination of Prunus Crops: Peach, Cherry, Apricot, Plum and Almond; Janick, J., Ed.; Horticultural Reviews; Gent-Oostakker: Leuven, Belgium, 2011; Volume 17, 241p. [Google Scholar]
- FAOSTAT. Available online: https://www.fao.org/faostat/es/#data/QCL (accessed on 25 May 2022).
- Acuña, C.V.; Rivas, J.G.; Brambilla, S.M.; Cerrillo, T.; Frusso, E.A.; García, M.N.; Villalba, P.V.; Aguirre, N.C.; Sabio y García, J.V.; Martínez, M.C.; et al. Characterization of genetic diversity in accessions of Prunus salicina lindl: Keeping fruit flesh color ideotype while adapting to water stressed environments. Agronomy 2019, 9, 487. [Google Scholar] [CrossRef]
- Guerrero, B.I.; Guerra, M.E.; Herrera, S.; Irisarri, P.; Pina, A.; Rodrigo, J. Genetic diversity and population structure of Japanese plum-Type (hybrids of P. salicina) accessions assessed by SSR markers. Agronomy 2021, 11, 1748. [Google Scholar] [CrossRef]
- Guerrero, B.I.; Guerra, M.E.; Rodrigo, J. Simple Sequence Repeat (SSR)-based genetic diversity in interspecific plumcot-type (Prunus salicina × Prunus armeniaca) hybrids. Plants 2022, 11, 1241. [Google Scholar] [CrossRef]
- Decroocq, V.; Hagen, L.S.; Favé, M.G.; Eyquard, J.P.; Pierronnet, A. Microsatellite markers in the hexaploid Prunus domestica species and parentage lineage of three European plum cultivars using nuclear and chloroplast simple-sequence repeats. Mol. Breed. 2004, 13, 135–142. [Google Scholar] [CrossRef]
- Horvath, A.; Balsemin, E.; Barbot, J.C.; Christmann, H.; Manzano, G.; Reynet, P.; Laigret, F.; Mariette, S. Phenotypic variability and genetic structure in plum (Prunus domestica L.), cherry plum (P. cerasifera Ehrh.) and sloe (P. spinosa L.). Sci. Hortic. 2011, 129, 283–293. [Google Scholar] [CrossRef]
- Gharbi, O.; Wünsch, A.; Rodrigo, J. Characterization of accessions of ‘Reine Claude Verte’ plum using Prunus SRR and phenotypic traits. Sci. Hortic. 2014, 169, 57–65. [Google Scholar] [CrossRef]
- Halapija Kazija, D.; Jelačić, T.; Vujević, P.; Milinović, B.; Čiček, D.; Biško, A.; Pejić, I.; Šimon, S.; Žulj Mihaljević, M.; Pecina, M.; et al. Plum germplasm in Croatia and neighboring countries assessed by microsatellites and DUS descriptors. Tree Genet. Genomes 2014, 10, 761–778. [Google Scholar] [CrossRef]
- Sehic, J.; Nybom, H.; Hjeltnes, S.H.; Gaši, F. Genetic diversity and structure of Nordic plum germplasm preserved ex situ and on-farm. Sci. Hortic. 2015, 190, 195–202. [Google Scholar] [CrossRef]
- Urrestarazu, J.; Errea, P.; Miranda, C.; Santesteban, L.G.; Pina, A. Genetic diversity of Spanish Prunus domestica L. germplasm reveals a complex genetic structure underlying. PLoS ONE 2018, 13, e0195591. [Google Scholar] [CrossRef]
- Manco, R.; Chiaiese, P.; Basile, B.; Corrado, G. Comparative analysis of genomic-and EST-SSRs in European plum (Prunus domestica L.): Implications for the diversity analysis of polyploids. 3 Biotech 2020, 10, 543. [Google Scholar] [CrossRef] [PubMed]
- Tamarzizt, H.B.; Walker, D.; Mustapha, S.B.; Abdallah, D.; Baraket, G.; Hannachi, A.S.; Azzouzi, S.Z. DNA variation and polymorphism in Tunisian plum species (Prunus spp.): Contribution of flow cytometry and molecular markers. Genet. Mol. Res. 2015, 14, 18034–18046. [Google Scholar] [CrossRef] [PubMed]
- Žabka, M.; Ďurišová, Ľ.; Eliáš, P.; Baranec, T. Genome size and ploidy level among wild and cultivated Prunus taxa in Slovakia. Biologia 2018, 73, 121–128. [Google Scholar] [CrossRef]
- Nzweundji, J.G.; Ngwe, M.F.S.N.; Siljak-Yakovlev, S. Insights on cytogenetic of the only strict African representative of genus Prunus (P. africana): First genome size assessment, heterochromatin and rDNA chromosome pattern. Caryologia 2019, 72, 9–19. [Google Scholar] [CrossRef]
- Głowacka, A.; Sitarek, M.; Rozpara, E.; Podwyszyńska, M. Pomological Characteristics and Ploidy Levels of Japanese Plum (Prunus salicina Lindl.) Cultivars Preserved in Poland. Plants 2021, 10, 884. [Google Scholar] [CrossRef]
- Larranaga, N.; Hormaza, J.I. Advances in Genetic Diversity Analysis in Fruit Tree crops. In Progress in Botany 77; Lüttge, U., Cánovas, F., Matyssek, R., Eds.; Springer: Cham, Switzerland, 2016; Volume 77, pp. 245–264. [Google Scholar] [CrossRef]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genomics-assisted breeding for crop improvement. Trends Plant Sci. 2005, 10, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Martín, C.; Viruel, M.A.; Lora, J.; Hormaza, J.I. Polyploidy in fruit tree crops of the genus Annona (Annonaceae). Front. Plant Sci. 2019, 10, 99. [Google Scholar] [CrossRef]
- Hormaza, J.I. Molecular characterization and similarity relationships among apricot (Prunus armeniaca L.) genotypes using simple sequence repeats. Theor. Appl. Genet. 2002, 104, 321–328. [Google Scholar] [CrossRef]
- Wünsch, A. Cross-transferable polymorphic SSR loci in Prunus species. Sci. Hortic. 2009, 120, 348–352. [Google Scholar] [CrossRef]
- Mnejja, M.; Garcia-Mas, J.; Audergon, J.M.; Arús, P. Prunus microsatellite marker transferability across rosaceous crops. Tree Genet. Genomes 2010, 6, 689–700. [Google Scholar] [CrossRef]
- Dirlewanger, E.; Cosson, P.; Tavaud, M.; Aranzana, M.J.; Poizat, C.; Zanetto, A.; Arus, P.; Laigret, F. Development of microsatellite markers in peach [Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.). Theor. Appl. Genet. 2002, 105, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Struss, D.; Boritzki, M.; Karle, R.; Iezzoni, A.F. Microsatellite markers differentiate eight Giessen cherry rootstocks. HortScience 2002, 37, 191–193. [Google Scholar] [CrossRef]
- Cipriani, G.; Lot, G.; Huang, W.G.; Marrazzo, M.T.; Peterlunger, E.; Testolin, R. AC/GT and AG/CT microsatellite repeats in peach Prunus persica (L.) Batsch: Isolation characterisation and cross-species amplification in Prunus. Theor. Appl. Genet. 1999, 9, 65–72. [Google Scholar] [CrossRef]
- Clark, L.V.; Jasieniuk, M. POLYSAT: An R package for polyploid microsatellite analysis. Mol. Ecol. Resour. 2011, 11, 562–566. [Google Scholar] [CrossRef]
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R Package Version 1.0.7. 2020. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 12 September 2022).
- FigTree, Version 1.4.4. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 12 September 2022).
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Hardy, O.J.; Vekemans, X. SPAGeDi: A versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2002, 2, 618–620. [Google Scholar] [CrossRef]
- Martín, C.; Herrero, M.; Hormaza, J.I. Molecular characterization of apricot germplasm from an old stone collection. PLoS ONE 2011, 6, e23979. [Google Scholar] [CrossRef] [Green Version]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. CLUMPAK: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Ronfort, J.; Jenczewski, E.; Bataillon, T.; Rousset, F. Analysis of population structure in autotetraploid species. Genetics 1998, 150, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Slatkin, M. A measure of population subdivision based on microsatellite allele frequencies. Genetics 1995, 139, 457–462. [Google Scholar] [CrossRef]
- Michalakis, Y.; Excoffier, L.A. Genetic estimation of population subdivision using distances between alleles with special reference for microsatellite loci. Genetics 1996, 142, 1061–1064. [Google Scholar] [CrossRef] [PubMed]
- Tortosa, M.; Velasco, P.; Afonso, D.; Padilla, G.; Ríos, D.; Soengas, P. Characterization of a Spanish Brassica oleracea collection by using molecular and biochemical markers. Sci. Hortic. 2017, 219, 344–350. [Google Scholar] [CrossRef]
- Zhebentyayeva, T.; Shankar, V.; Scorza, R.; Callahan, A.; Ravelonandro, M.; Castro, S.; DeJong, T.; Saski, C.A.; Dardick, C. Genetic characterization of worldwide Prunus domestica (plum) germplasm using sequence-based genotyping. Hortic. Res. 2019, 6, 12. [Google Scholar] [CrossRef]
- Lebot, V. Coping with insularity: The need for crop genetic improvement to strengthen adaptation to climatic change and food security in the Pacific. Environ. Dev. Sustain. 2013, 15, 1405–1423. [Google Scholar] [CrossRef]
| SSR Locus | Forward | Reverse | Reference |
|---|---|---|---|
| BPPCT-002 | TCGACAGCTTGATCTTGACC | CAATGCCTACGGAGATAAAAGAC | [47] |
| BPPCT-007 | TCATTGCTCGTCATCAGC | CAGATTTCTGAAGTTAGCGGTA | [47] |
| BPPCT-010 | AAAGCACAGCCCATAATGC | GTACTGTTACTGCTGGGAATGC | [47] |
| BPPCT-012 | ACTTCCATTGTCAGGCATCA | GGAGCAACGATGGAGTGC | [47] |
| BPPCT-028 | TCAAGTTAGCTGAGGATCGC | GAGCTTGCCTATGAGAAGACC | [47] |
| PMS3 | TGGACTTCACTCATTTCAGAGA | ACTGCAGAGAATTTCACAACCA | [48] |
| UDP96-005 | GTAACGCTCGCTACCACAAA | CACCCAGCTCATACACCTCA | [49] |
| UDP96-008 | TTGTACACACCCTCAGCCTG | TGCTGAGGTTCAGGTGAGTG | [49] |
| UDP98-409 | GCTGATGGGTTTTATGGTTTTC | CGGACTCTTATCCTATCAACA | [49] |
| Accession | Abbreviation | Locality | Collection | Ploidy | Species |
|---|---|---|---|---|---|
| Agustina negra 17 1 | AN17 | Puntagorda | LP | Diploid | P. salicina |
| Amarilla secona 57 1 | AS57 | Garafía | LP | Diploid | P. salicina |
| Blanca punta cumplida 11 1 | BPC11 | Garafía | LP | Diploid | P. salicina |
| Blanca japonesa 2 1 | BJ2 | Puntallana | LP | Diploid | P. salicina |
| Blanca japonesa 15 1 | BJ15 | Mazo | LP | Diploid | P. salicina |
| Blanca japonesa 21 1 | BJ21 | Breña Alta | LP | Diploid | P. salicina |
| De Catela 39 1 | DC39 | Garafía | LP | Diploid | P. salicina |
| Japonesa amarilla antigua 53 1 | JAA53 | Garafía | LP | Diploid | P. salicina |
| Morada 23 1 | MR23 | Puntallana | LP | Diploid | P. cerasifera |
| Morada 61 1 | MR61 | Garafía | LP | Diploid | P. salicina |
| Morada 70 1 | MR70 | Garafía | LP | Diploid | P. cerasifera |
| Negra1_35 1 | N35 | Puntallana | LP | Diploid | P. salicina |
| Negra2_45 1 | N45 | Puntallana | LP | Diploid | P. salicina |
| Pastosa 43 1 | P43 | Garafía | LP | Diploid | P. salicina |
| Roja 42 1 | R42 | Garafía | LP | Diploid | P. salicina |
| Rojizo 64 1 | RZ64 | Garafía | LP | Diploid | P. salicina |
| Agustina 10 1 | A10 | Garafía | LP | Hexaploid | P. domestica |
| Agustina 52 1 | A52 | Garafía | LP | Hexaploid | P. domestica |
| Agustina blanca 32 1 | AB32 | Puntagorda | LP | Hexaploid | P. domestica |
| Agustina blanca 50 1 | AB50 | Garafía | LP | Hexaploid | P. domestica |
| Blanca del país 38 1 | BP38 | Garafía | LP | Hexaploid | P. domestica |
| Huevo chivato 8 1 | HC8 | Garafía | LP | Hexaploid | P. domestica |
| Huevo chivato 19 1 | HC19 | Puntagorda | LP | Hexaploid | P. domestica |
| Huevo chivato 58 1 | HC58 | Garafía | LP | Hexaploid | P. domestica |
| Mulata 4 1 | ML4 | Garafía | LP | Hexaploid | P. domestica |
| Mulata 25 1 | ML25 | Garafía | LP | Hexaploid | P. domestica |
| Negra 5 1 | N5 | Garafía | LP | Hexaploid | P. domestica |
| Negra del país pequeña 67 1 | NPP67 | Tijarafe | LP | Hexaploid | P. domestica |
| Verde 27 1 | V27 | Garafía | LP | Hexaploid | P. domestica |
| RC Arandana 2 | RCA | CITA | P. domestica | ||
| RC Bavay 2 | RCB | CITA | P. domestica | ||
| RC Dorada 2 | RCD | CITA | P. domestica | ||
| RC Doullins 2 | RCDL | CITA | P. domestica | ||
| RC President 2 | RCP | CITA | P. domestica | ||
| RCV Alcor 1 1 | RCVA1 | CITA | P. domestica | ||
| RCV Alcor 2 1 | RCVA2 | CITA | P. domestica | ||
| RCV Aniñon 1 | RCVAN | CITA | P. domestica | ||
| RCV Arenal 1 | RCVAR | CITA | P. domestica | ||
| RCV Conde 1 | RCVC | CITA | P. domestica | ||
| RCV Domingo 1 | RCVD | CITA | P. domestica | ||
| RCV F-4 A-4 1 | RCVF4 | CITA | P. domestica | ||
| RCV F-9 A-10 1 | RCVF9 | CITA | P. domestica | ||
| RCV Puente Ave 1 | RCVPA | CITA | P. domestica | ||
| RCV Río Ribazo1 1 | RCVR1 | CITA | P. domestica | ||
| RCV Río Ribazo2 1 | RCVR2 | CITA | P. domestica | ||
| RCV Tobed 1 | RCVT | CITA | P. domestica | ||
| RCV Verde 2 | RCVV | CITA | P. domestica | ||
| Fraila 1 | FR | CITA | P. domestica | ||
| Polinizador Zuera 1 | PZ | CITA | P. domestica | ||
| Ruth Gestteter 2 | RG | CITA | P. domestica | ||
| Stanley 2 | STL | CITA | P. domestica |
| Diversity Index | |||||||
|---|---|---|---|---|---|---|---|
| n | NA | NAe | AR | He | Ho | Fi | |
| Diploid La Palma | 16 | 6.89 | 4.31 | 6.89 | 0.76 | 0.88 | 0.015 |
| Hexaploid La Palma | 13 | 9.33 | 7.07 | 8.55 | 0.85 | 0.74 | 0.018 |
| Hexaploid CITA | 22 | 10.56 | 5.91 | 8.18 | 0.795 | 0.935 | −0.027 |
| Statistic for Pairwise | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Fst | Rho | Rst | |||||||
| Dipl. LP | Hex. LP | Hex. CITA | Dipl. LP | Hex. LP | Hex. CITA | Dipl. LP | Hex. LP | Hex. CITA | |
| Dipl. LP | - | 0.166 | 0.220 | - | 0.346 | 0.469 | - | 0.045 | 0.116 |
| Hex. LP | 0.166 | - | 0.067 | 0.346 | - | 0.228 | 0.045 | - | 0.050 |
| Hex. CITA | 0.220 | 0.067 | - | 0.469 | 0.228 | - | 0.116 | 0.050 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez, V.; Rodrigo, J.; Abdallah, D.; Larranaga, N.; Hormaza, J.I. Genetic Characterization of a Plum Landrace Collection from La Palma, Canary Islands. Agronomy 2022, 12, 2179. https://doi.org/10.3390/agronomy12092179
Pérez V, Rodrigo J, Abdallah D, Larranaga N, Hormaza JI. Genetic Characterization of a Plum Landrace Collection from La Palma, Canary Islands. Agronomy. 2022; 12(9):2179. https://doi.org/10.3390/agronomy12092179
Chicago/Turabian StylePérez, Verónica, Javier Rodrigo, Donia Abdallah, Nerea Larranaga, and José I. Hormaza. 2022. "Genetic Characterization of a Plum Landrace Collection from La Palma, Canary Islands" Agronomy 12, no. 9: 2179. https://doi.org/10.3390/agronomy12092179
APA StylePérez, V., Rodrigo, J., Abdallah, D., Larranaga, N., & Hormaza, J. I. (2022). Genetic Characterization of a Plum Landrace Collection from La Palma, Canary Islands. Agronomy, 12(9), 2179. https://doi.org/10.3390/agronomy12092179







