Potential Roles of Three ABCB Genes in Quinclorac Resistance Identified in Echinochloa crus-galli var. zelayensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
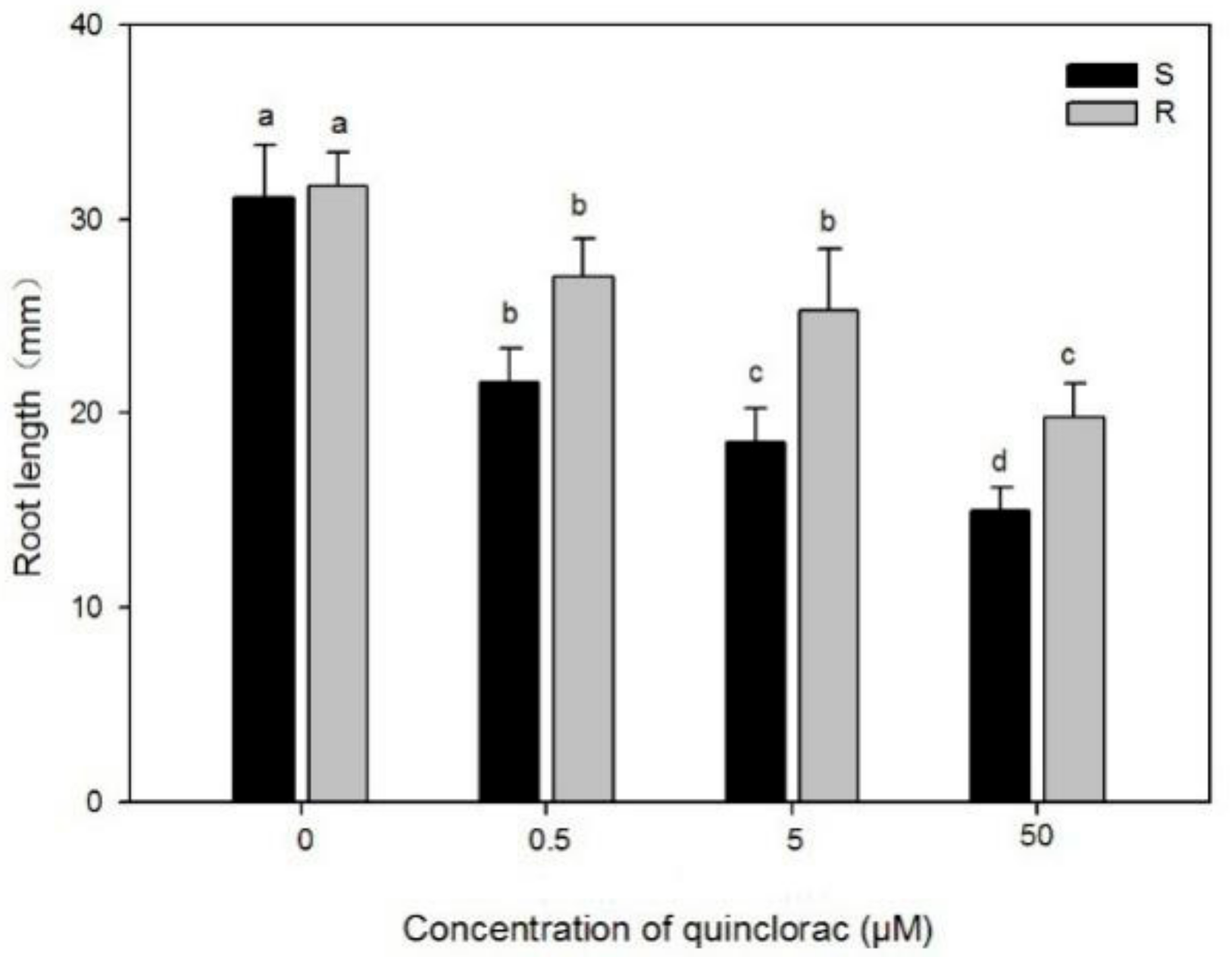
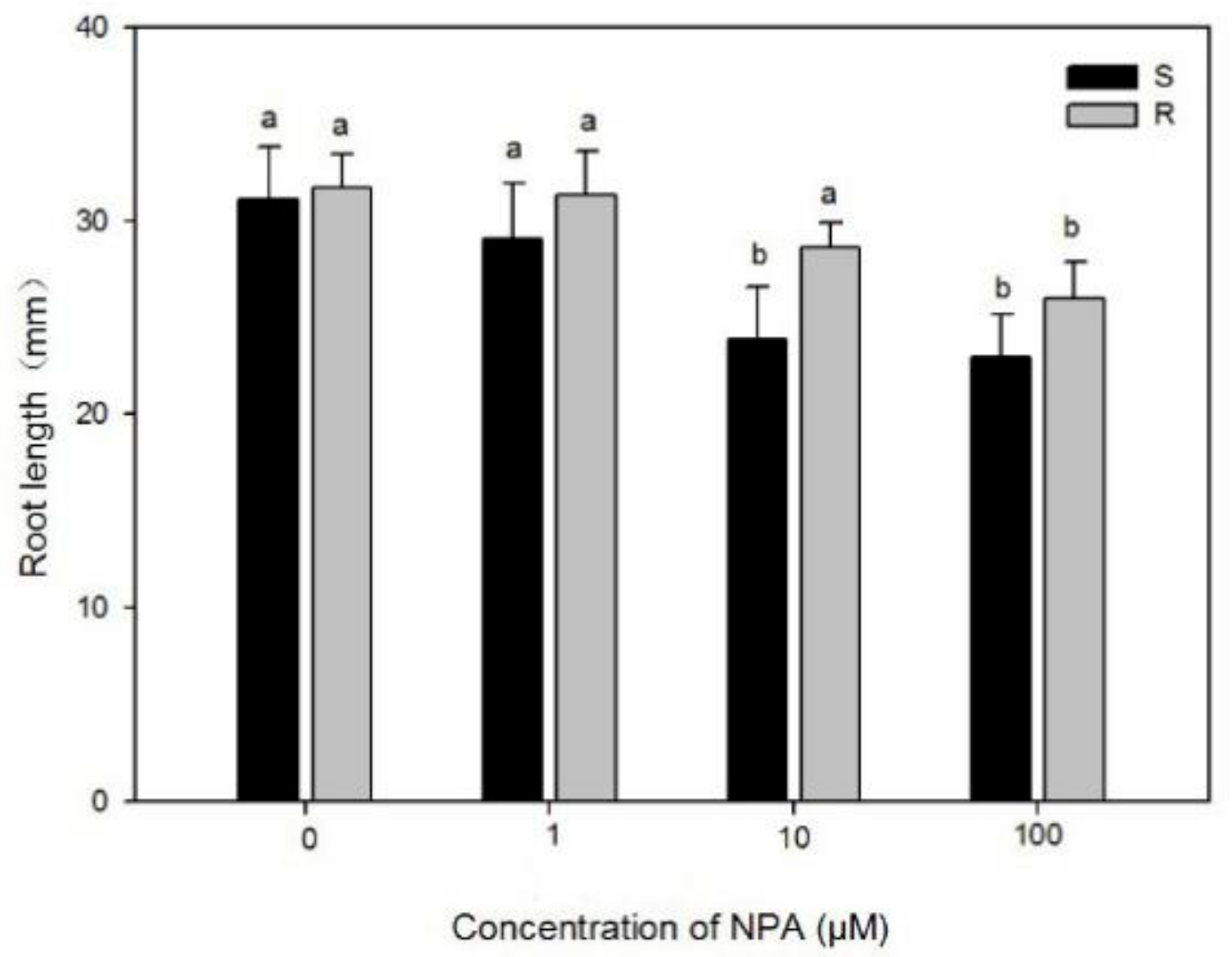
2.1.1. Effects of Quinclorac and NPA on Root Growth of Two Populations
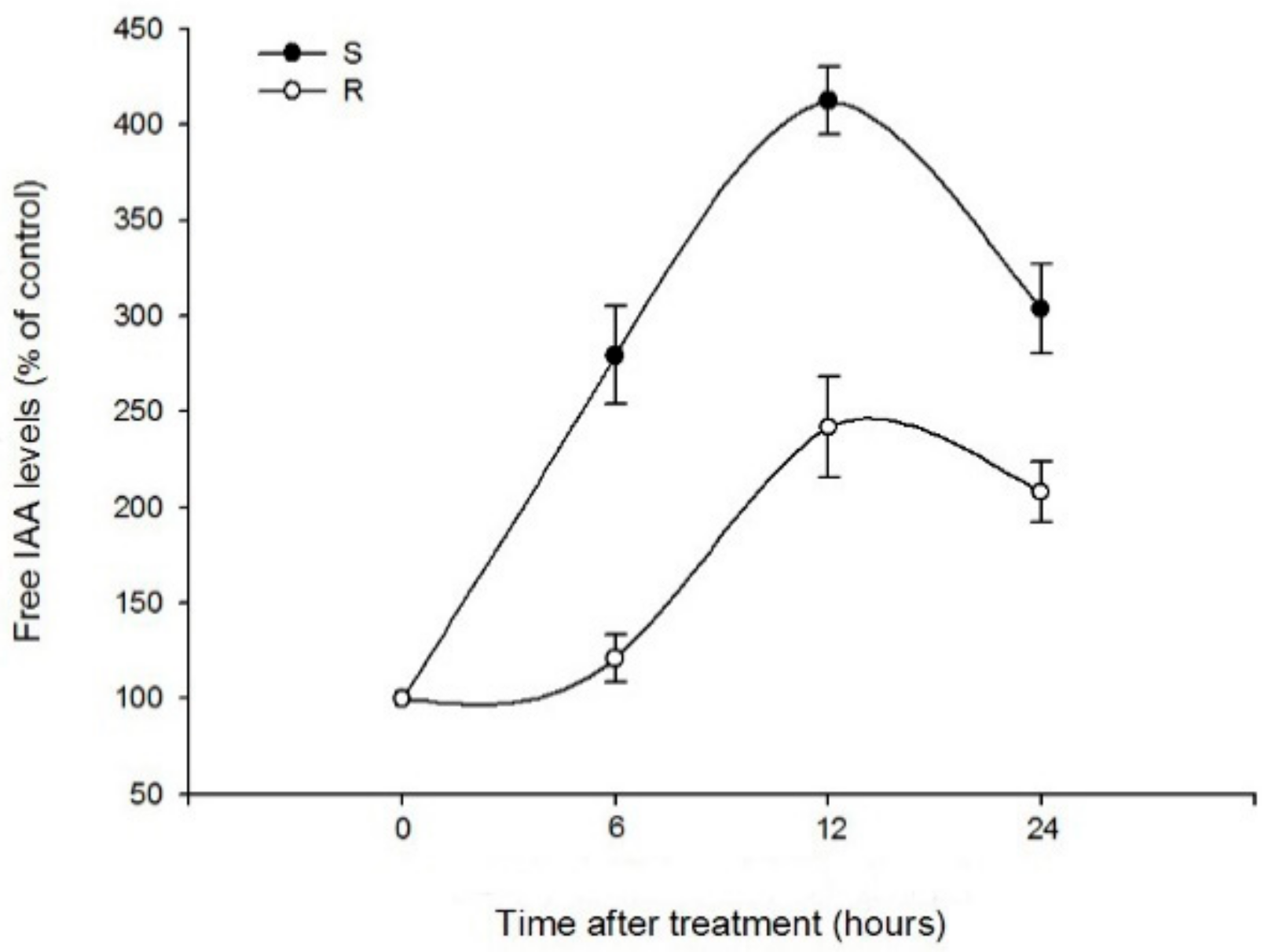
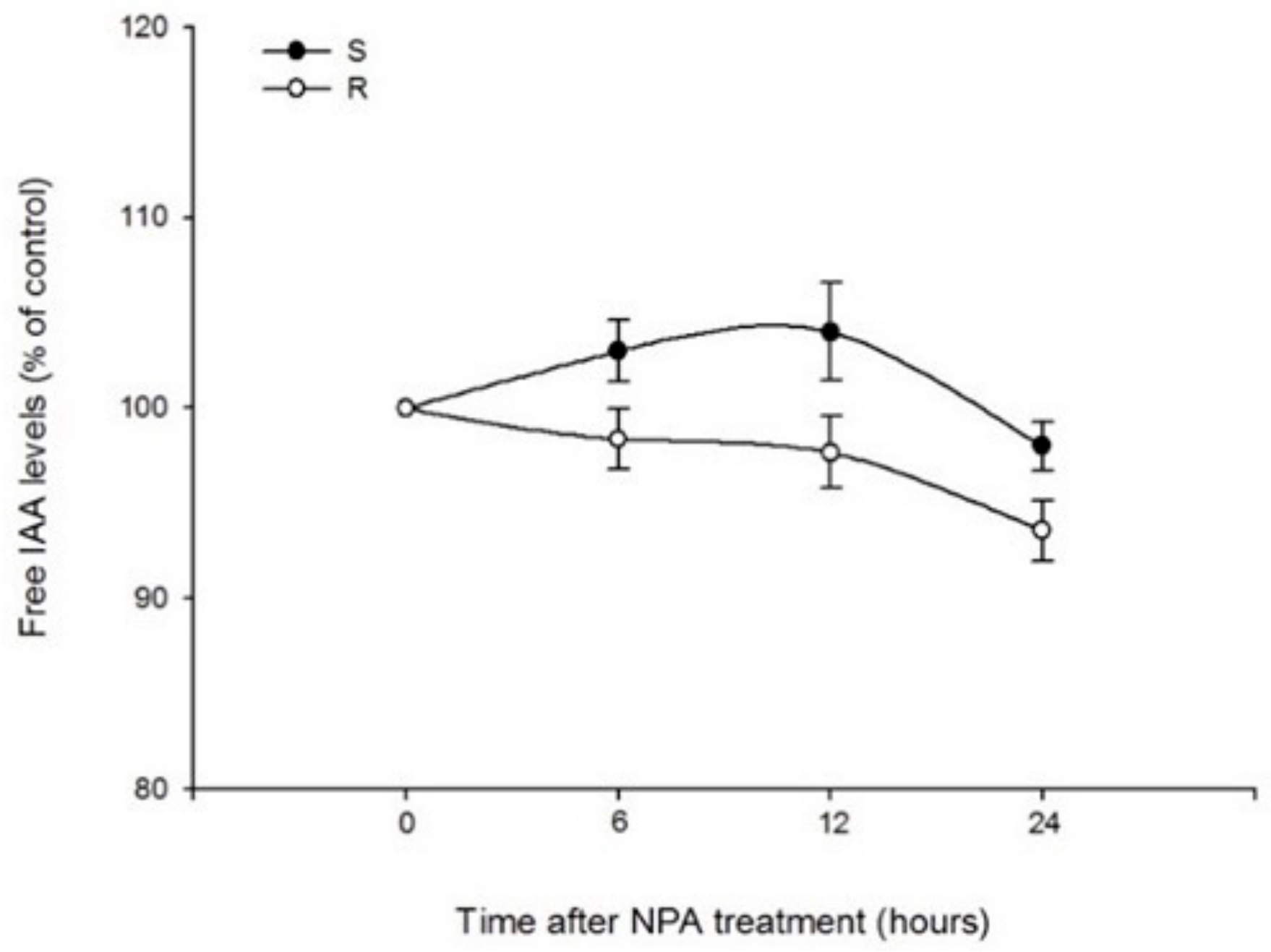
2.1.2. Effects of Quinclorac and NPA on IAA Contents in Roots of Two Populations
2.2. Amplification of ABCB Family Genes
2.3. ABCB Family Genes Expression
2.4. Susceptibility to Quinclorac in Arabidopsis T-DNA Insertional Mutants
2.5. Statistical Analysis
3. Results and Discussion
3.1. Effects of Quinclorac and NPA on Root Growth of Two Populations
3.2. Effects of Quinclorac and NPA on IAA Variation in Roots of Two Populations
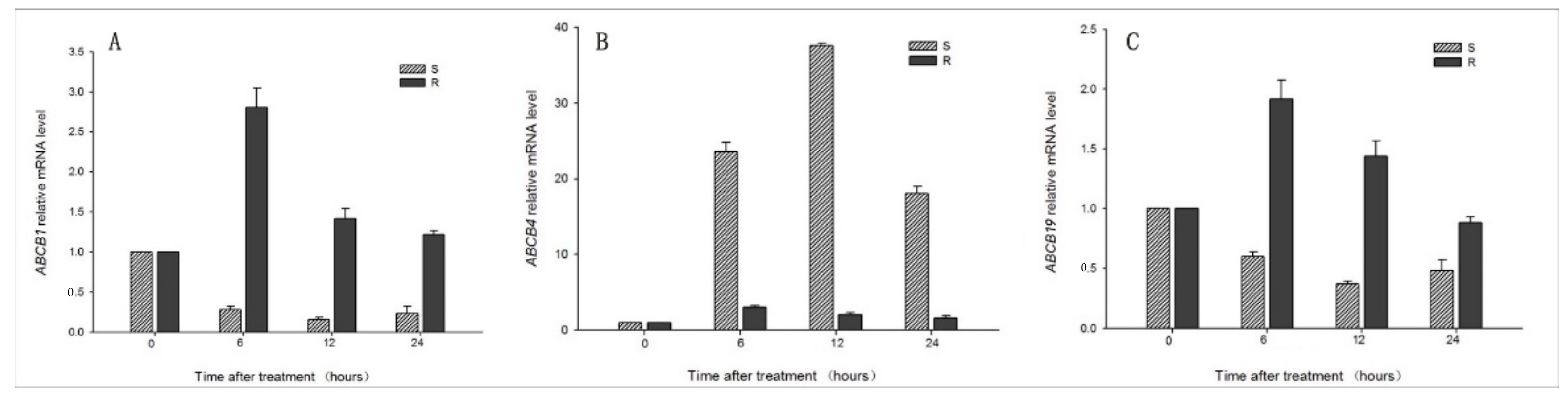
3.3. Sequencing and Expression of ABCBs Genes
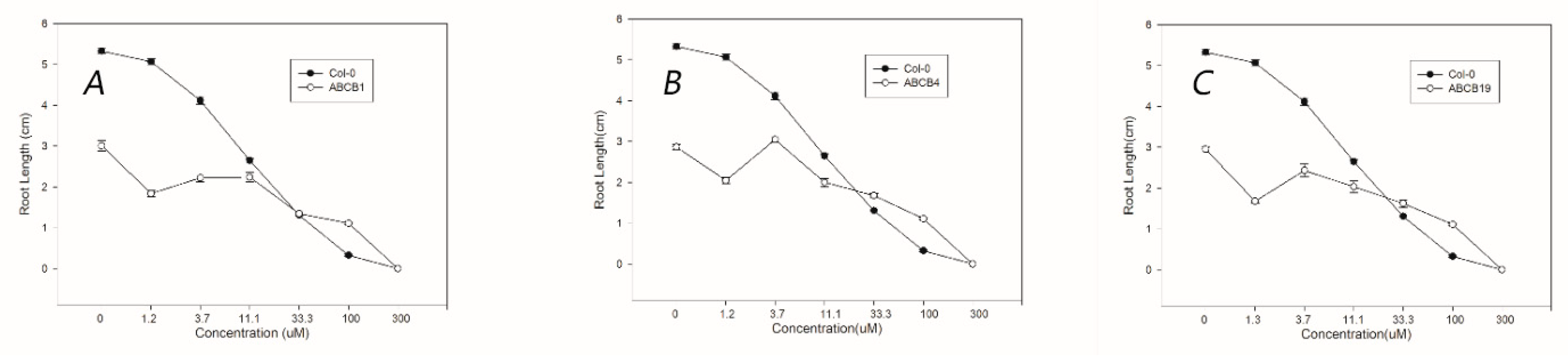
3.4. Susceptibility to Quinclorac in ABCB Mutants of A. thaliana
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xu, J.; Lv, B.; Wang, Q.; Li, J.; Dong, L. A resistance mechanism dependent upon the inhibition of ethylene biosynthesis. Pest Manag. Sci. 2013, 69, 1407–1414. [Google Scholar] [CrossRef]
- Grossmann, K. Quinclorac belongs to a new class of highly selective auxin herbicides. Weed Sci. 1998, 46, 707–716. [Google Scholar] [CrossRef]
- Grossmann, K. Auxin herbicides: Current status of mechanism and mode of action. Pest Manag. Sci. Former. Pestic. Sci. 2010, 66, 113–120. [Google Scholar] [CrossRef]
- Gao, Y.; Li, J.; Pan, X.; Liu, D.; Napier, R.; Dong, L. Quinclorac resistance induced by the suppression of the expression of 1-aminocyclopropane-1-carboxylic acid (ACC) synthase and ACC oxidase genes in Echinochloa crus-galli var. zelayensis. Pestic. Biochem. Physiol. 2018, 146, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, K.; Schmülling, T. The effects of the herbicide quinclorac on shoot growth in tomato is alleviated by inhibitors of ethylene biosynthesis and by the presence of an antisense construct to the 1-aminocyclopropane-1-carboxylic acid (ACC) synthase gene in transgenic plants. Plant Growth Regul. 1995, 16, 183–188. [Google Scholar] [CrossRef]
- Hall, J.C.; Bassi, P.K.; Spencer, M.S.; Vanden Born, W.H. An evaluation of the role of ethylene in herbicidal injury induced by picloram or clopyralid in rapeseed and sunflower plants. Plant Physiol. 1985, 79, 18–23. [Google Scholar] [CrossRef]
- Stepanova, A.N.; Yun, J.; Likhacheva, A.V.; Alonso, J.M. Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 2007, 19, 2169–2185. [Google Scholar] [CrossRef]
- Swarup, R.; Bennett, M. Auxin transport: The fountain of life in plants? Dev. Cell 2003, 5, 824–826. [Google Scholar]
- Sánchez-Fernández, R.; Davies, T.G.E.; Coleman, J.O.; Rea, P.A. The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J. Biol. Chem. 2001, 276, 30231–30244. [Google Scholar] [CrossRef]
- Martinoia, E.; Klein, M.; Geisler, M.; Bovet, L.; Forestier, C.; Kolukisaoglu, U.; Müller-Röber, B.; Schulz, B. Multifunctionality of plant ABC transporters—More than just detoxifiers. Planta 2002, 214, 345–355. [Google Scholar] [CrossRef]
- Petrasek, J.; Friml, J. Auxin transport routes in plant development. Development 2009, 136, 2675–2688. [Google Scholar] [CrossRef] [PubMed]
- Bailly, A.; Yang, H.; Martinoia, E.; Geisler, M.; Murphy, A.S. Plant lessons: Exploring ABCB functionality through structural modeling. Front. Plant Sci. 2012, 2, 108. [Google Scholar] [CrossRef] [PubMed]
- Noh, B.; Murphy, A.S.; Spalding, E.P. Multidrug resistance—Like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 2001, 13, 2441–2454. [Google Scholar] [PubMed]
- Murphy, A.S.; Hoogner, K.R.; Peer, W.A.; Taiz, L. Identification, purification, and molecular cloning of N-1-naphthylphthalmic acid-binding plasma membrane-associated aminopeptidases from Arabidopsis. Plant Physiol. 2002, 128, 935–950. [Google Scholar] [CrossRef] [PubMed]
- Geisler, M.; Kolukisaoglu, H.U.; Bouchard, R.; Billion, K.; Berger, J.; Saal, B.; Frangne, N.; Koncz-Kálmán, Z.; Koncz, C.; Dudler, R.; et al. TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol. Biol. Cell 2003, 14, 4238–4249. [Google Scholar] [CrossRef]
- Terasaka, K.; Blakeslee, J.J.; Titapiwatanakun, B.; Peer, W.A.; Bandyopadhyay, A.; Makam, S.N.; Lee, O.R.; Richards, E.L.; Murphy, A.S.; Sato, F.; et al. PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell 2005, 17, 2922–2939. [Google Scholar] [CrossRef]
- Kamimoto, Y.; Terasaka, K.; Hamamoto, M.; Takanashi, K.; Fukuda, S.; Shitan, N.; Sugiyama, A.; Suzuki, H.; Shibata, D.; Wang, B.; et al. Arabidopsis ABCB21 is a facultative auxin importer/exporter regulated by cytoplasmic auxin concentration. Plant Cell Physiol. 2012, 53, 2090–2100. [Google Scholar] [CrossRef]
- Geisler, M.; Blakeslee, J.J.; Bouchard, R.; Lee, O.R.; Vincenzetti, V.; Bandyopadhyay, A.; Titapiwatanakun, B.; Peer, W.A.; Bailly, A.; Richards, E.L.; et al. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 2005, 44, 179–194. [Google Scholar] [CrossRef]
- Yang, H.; Murphy, A.S. Functional expression and characterization of Arabidopsis ABCB, AUX 1 and PIN auxin transporters in Schizosaccharomyces pombe. Plant J. 2009, 59, 179–191. [Google Scholar] [CrossRef]
- Xu, Y.X.; Liu, Y.; Chen, S.T.; Li, X.Q.; Xu, L.G.; Qi, Y.H.; Jiang, D.A.; Jin, S.H. The B subfamily of plant ATP binding cassette transporters and their roles in auxin transport. Biol. Plant. 2014, 58, 401–410. [Google Scholar] [CrossRef]
- Klein, M.; Geisler, M.; Suh, S.J.; Kolukisaoglu, H.; Azevedo, L.; Plaza, S.; Curtis, M.D.; Richter, A.; Weder, B.; Schulz, B.; et al. Disruption of AtMRP4, a guard cell plasma membrane ABCC-type ABC transporter, leads to deregulation of stomatal opening and increased drought susceptibility. Plant J. 2004, 39, 219–236. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Lee, K.; Lee, J.; Noh, E.W.; Lee, Y. AtPDR12 contributes to lead resistance in Arabidopsis. Plant Physiol. 2005, 138, 827–836. [Google Scholar] [CrossRef] [PubMed]
- Rea, P.A. Plant ATP-binding cassette transporters. Annu. Rev. Plant Biol. 2007, 58, 347–375. [Google Scholar] [CrossRef] [PubMed]
- Crouzet, J.; Roland, J.; Peeters, E.; Trombik, T.; Ducos, E.; Nader, J.; Boutry, M. NtPDR1, a plasma membrane ABC transporter from Nicotiana tabacum, is involved in diterpene transport. Plant Mol. Biol. 2013, 82, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Le Hir, R.; Sorin, C.; Chakraborti, D.; Moritz, T.; Schaller, H.; Tellier, F.; Robert, S.; Morin, H.; Bako, L.; Bellini, C. ABCG 9, ABCG 11 and ABCG 14 ABC transporters are required for vascular development in A. rabidopsis. Plant J. 2013, 76, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Shitan, N.; Dalmas, F.; Dan, K.; Kato, N.; Ueda, K.; Sato, F.; Forestier, C.; Yazaki, K. Characterization of Coptis japonica CjABCB2, an ATP-binding cassette protein involved in alkaloid transport. Phytochemistry 2013, 91, 109–116. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, X.; Shen, Q.; Chen, Y.; Wang, T.; Zhang, F.; Wu, S.; Jiang, W.; Liu, P.; Zhang, L.; et al. Identification of putative Artemisia annua ABCG transporter unigenes related to artemisinin yield following expression analysis in different plant tissues and in response to methyl jasmonate and abscisic acid treatments. Plant Mol. Biol. Report. 2012, 30, 838–847. [Google Scholar] [CrossRef]
- Wilkens, S. Structure and mechanism of ABC transporters. F1000prime Rep. 2015, 7, 14. [Google Scholar] [CrossRef]
- Goggin, D.E.; Cawthray, G.R.; Powles, S.B. 2,4-D resistance in wild radish: Reduced herbicide translocation via inhibition of cellular transport. J. Exp. Bot. 2016, 67, 3223–3235. [Google Scholar] [CrossRef]
- Busi, R.; Goggin, D.E.; Heap, I.M.; Horak, M.J.; Jugulam, M.; Masters, R.A.; Napier, R.M.; Riar, D.S.; Satchivi, N.M.; Torra, J.; et al. Weed resistance to synthetic auxin herbicides. Pest Manag. Sci. 2018, 74, 2265–2276. [Google Scholar] [CrossRef]
- Sunohara, Y.; Shirai, S.; Yamazaki, H.; Matsumoto, H. Involvement of antioxidant capacity in quinclorac tolerance in Eleusine indica. Environ. Exp. Bot. 2011, 74, 74–81. [Google Scholar] [CrossRef]
- Tang, Q.Y.; Zhang, C.X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2013, 20, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, K.; Kwiatkowski, J. Selective induction of ethylene and cyanide biosynthesis appears to be involved in the selectivity of the herbicide quinclorac between rice and barnyardgrass. J. Plant Physiol. 1993, 142, 457–466. [Google Scholar] [CrossRef]
- Grossmann, K.; Scheltrup, F. Selective induction of 1-aminocyclopropane-1-carboxylic acid (ACC) synthase activity is involved in the selectivity of the auxin herbicide quinclorac between barnyard grass and rice. Pestic. Biochem. Physiol. 1997, 58, 145–153. [Google Scholar] [CrossRef]
- Hansen, H.; Grossmann, K. Auxin-induced ethylene triggers abscisic acid biosynthesis and growth inhibition. Plant Physiol. 2000, 124, 1437–1448. [Google Scholar] [CrossRef]
- Abdallah, I.; Fischer, A.J.; Elmore, C.L.; Saltveit, M.E.; Zaki, M. Mechanism of resistance to quinclorac in smooth crabgrass (Digitaria ischaemum). Pestic. Biochem. Physiol. 2006, 84, 38–48. [Google Scholar] [CrossRef]
- Yang, X.; Han, H.; Cao, J.; Li, Y.; Yu, Q.; Powles, S.B. Exploring quinclorac resistance mechanisms in Echinochloa crus-pavonis from China. Pest Manag. Sci. 2021, 77, 194–201. [Google Scholar] [CrossRef]
- Ljung, K.; Hull, A.K.; Celenza, J.; Yamada, M.; Estelle, M.; Normanly, J.; Sandberg, G. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 2005, 17, 1090–1104. [Google Scholar] [CrossRef]
- Mravec, J.; Kubes, M.; Bielach, A.; Gaykova, V.; Petrasek, J.; Skůpa, P.; Chand, S.; Benková, E.; Zazimalova, E.; Friml, J. Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Develpment 2008, 135, 3345–3354. [Google Scholar] [CrossRef]
- Wu, G.; Lewis, D.R.; Spalding, E.P. Mutations in Arabidopsis multidrug resistance-like ABC transporters separate the roles of acropetal and basipetal auxin transport in lateral root development. Plant Cell 2007, 19, 1826–1837. [Google Scholar] [CrossRef]
- Tsuchisaka, A.; Theologis, A. Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol. 2004, 136, 2982–3000. [Google Scholar] [CrossRef] [PubMed]


| Gene | Primer Sequence (5′-3′) | Temperature | Time |
|---|---|---|---|
| EcABCB1 | TTGGTTGCCTCCTTGTTG GCACTCATCTTTGTCGGTCT | 60 °C | 40 s |
| EcABCB4-1 | ATGGCTGGCGAAAGGCAGTCAG TAAGCGCCACAACATCAAGAGGC | 64.3 °C | 2 min |
| EcABCB4-2 | TCGACTACAAGAGACACGTGCTGATG GAGCGGAGCTCTATTAGTGAAGCG | 62.9 °C | 2 min |
| EcABCB19 | TCACCTCCACGGACAATC TGCGCTCTACTTCGTCTACC | 51.7 °C | 1 min |
| Gene | Primer Sequence (5′-3′) |
|---|---|
| EcABCB1 | F: GGGAAGAGCACCGTAGTGTC |
| R: GCAGGTTCTCCTTGATGCTC | |
| EcABCB4 | F: TATAATGACGGGGGCAATGT |
| R: CTCTGGTCTTGCTGGGTAGCT | |
| EcABCB19 | F: TGCTAGGAGTTCGTCGTGTG |
| R: GCTACCAGCGCTTTAGATGC |
| Gene | Primer Sequence (5′-3′) |
|---|---|
| ABCB1 | LP: TTTTTGGTCGAGTCTGATTGG |
| RP: TCCTTGAACCAACCAACAAAG | |
| RP: CTTTCGTGACCTATGTTTCGC | |
| ABCB4 | LP: GATTAAGCCCAAGTTCGATCC |
| RP: ATGGATTCATCAGTGGTCTGC | |
| ABCB19 | LP: AGATCATAACATCAGGCCGTG |
| RP: TTTTTGCAATTTGGTCCTGTC |
| Herbicide | Biotypes | IC50 μM | Regression Equation | Correlation Coefficient | Mutant IC50/Parent IC50 | Confidence Intervals |
|---|---|---|---|---|---|---|
| Quinclorac | Col-0 | 11.77 | Y = 2.46x + 2.63 | 1 | ||
| AtABCB1 | 10.61 | Y = 0.32x + 4.67 | 0.99 | 0.90 | 0.64~1.25 | |
| AtABCB4 | 49.91 * | Y = 0.84x + 3.57 | 0.99 | 4.24 | 3.14~5.71 | |
| AtABCB19 | 44.03 | Y = 0.86x + 3.58 | 0.99 | 3.74 | 3.31~4.22 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, Y.; Guo, Y.; Liu, X.; Gao, Y.; Sun, Y.; Dong, L.; Li, J. Potential Roles of Three ABCB Genes in Quinclorac Resistance Identified in Echinochloa crus-galli var. zelayensis. Agronomy 2022, 12, 1961. https://doi.org/10.3390/agronomy12081961
Qi Y, Guo Y, Liu X, Gao Y, Sun Y, Dong L, Li J. Potential Roles of Three ABCB Genes in Quinclorac Resistance Identified in Echinochloa crus-galli var. zelayensis. Agronomy. 2022; 12(8):1961. https://doi.org/10.3390/agronomy12081961
Chicago/Turabian StyleQi, Yuanlin, Yongli Guo, Xudong Liu, Yuan Gao, Yu Sun, Liyao Dong, and Jun Li. 2022. "Potential Roles of Three ABCB Genes in Quinclorac Resistance Identified in Echinochloa crus-galli var. zelayensis" Agronomy 12, no. 8: 1961. https://doi.org/10.3390/agronomy12081961
APA StyleQi, Y., Guo, Y., Liu, X., Gao, Y., Sun, Y., Dong, L., & Li, J. (2022). Potential Roles of Three ABCB Genes in Quinclorac Resistance Identified in Echinochloa crus-galli var. zelayensis. Agronomy, 12(8), 1961. https://doi.org/10.3390/agronomy12081961







