Diversity and Potential Function of the Bacterial Rhizobiome Associated to Physalis Ixocarpa Broth. in a Milpa System, in Michoacan, Mexico
Abstract
:1. Introduction
2. Materials and Methods
2.1. Description of the Study Area
2.2. Total DNA Extraction and Metagenomic Sequencing
2.3. Bioinformatic Analyses
3. Results
3.1. Soil Determinations
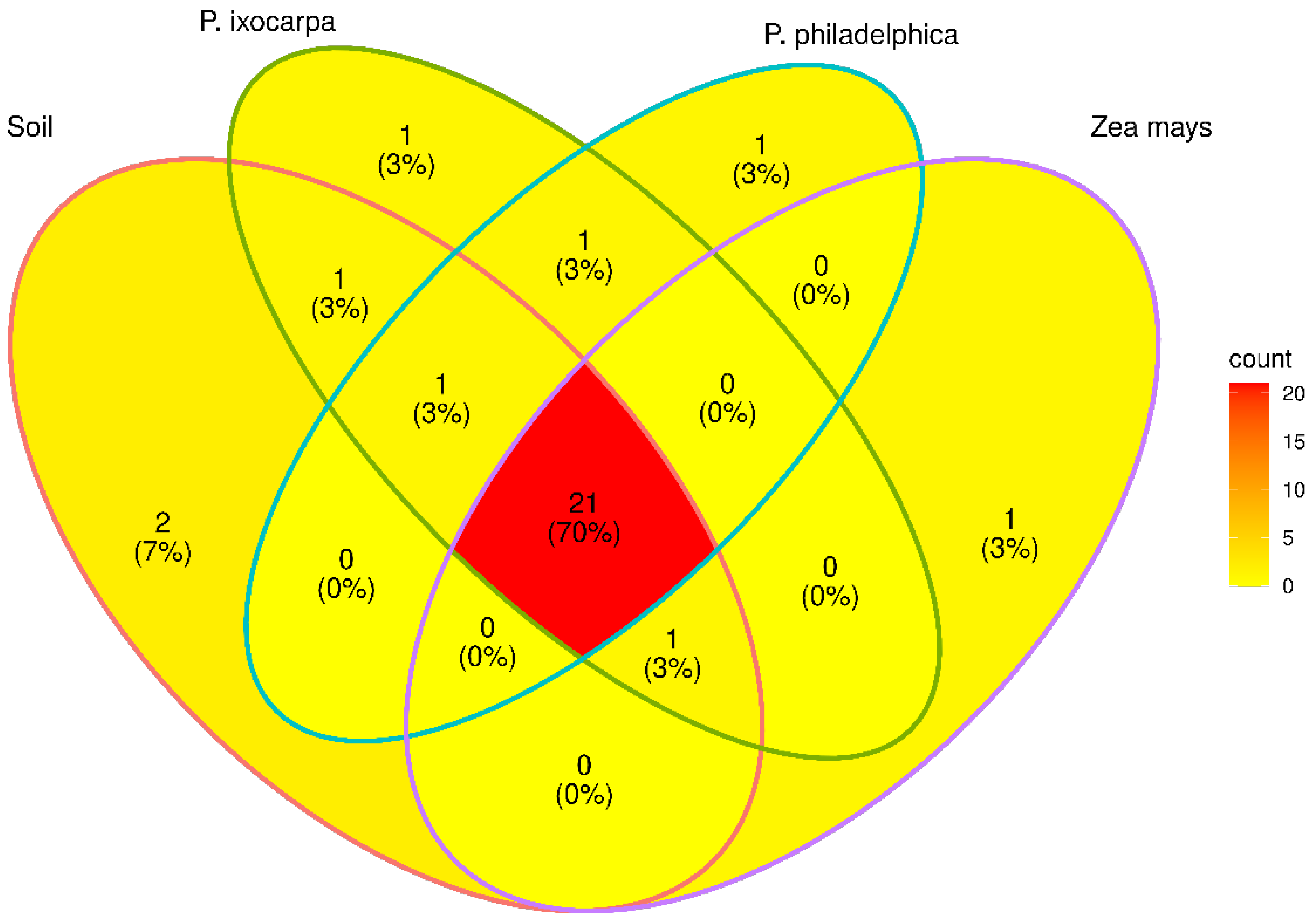
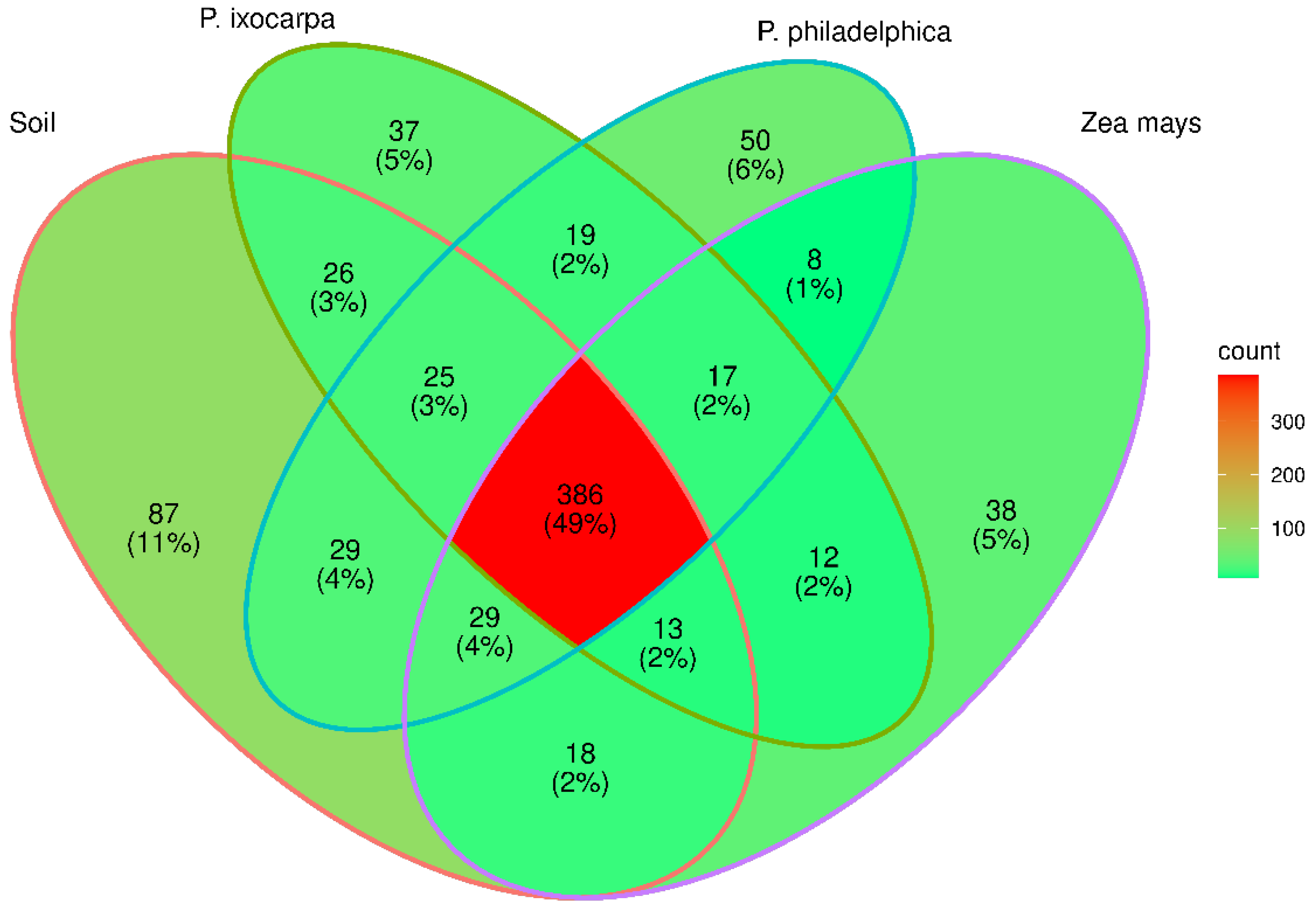
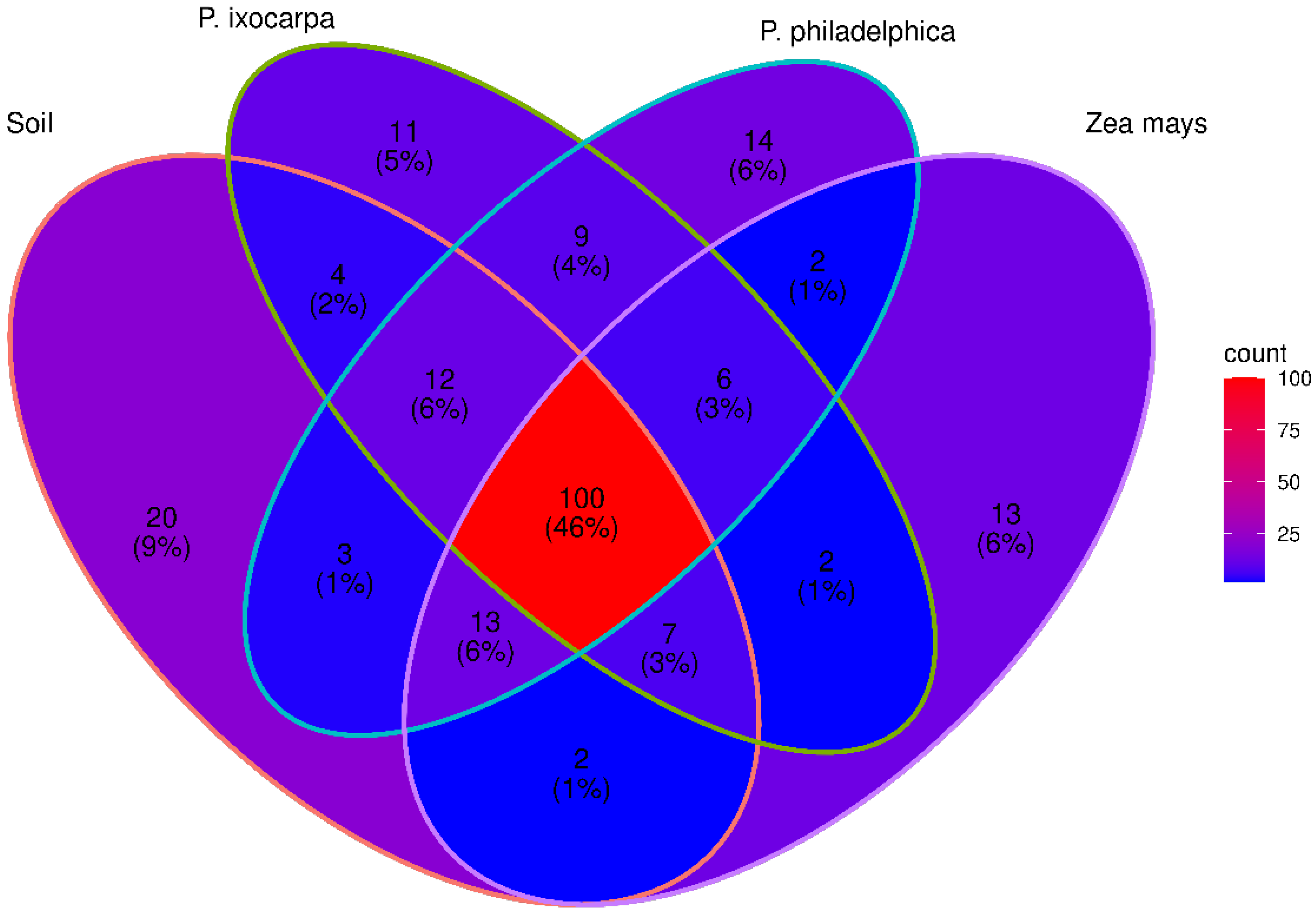
3.2. Structure of Bacterial Communities
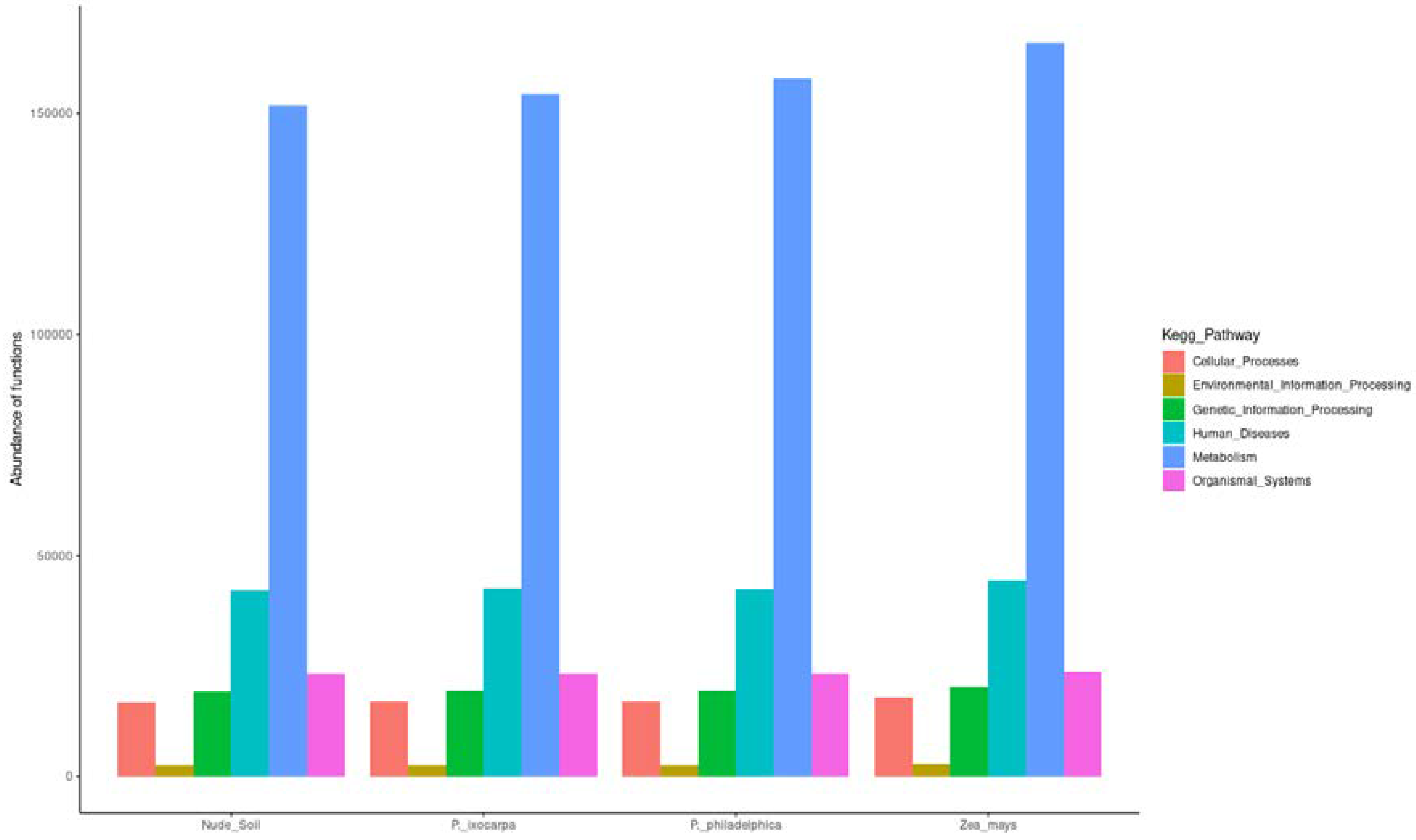
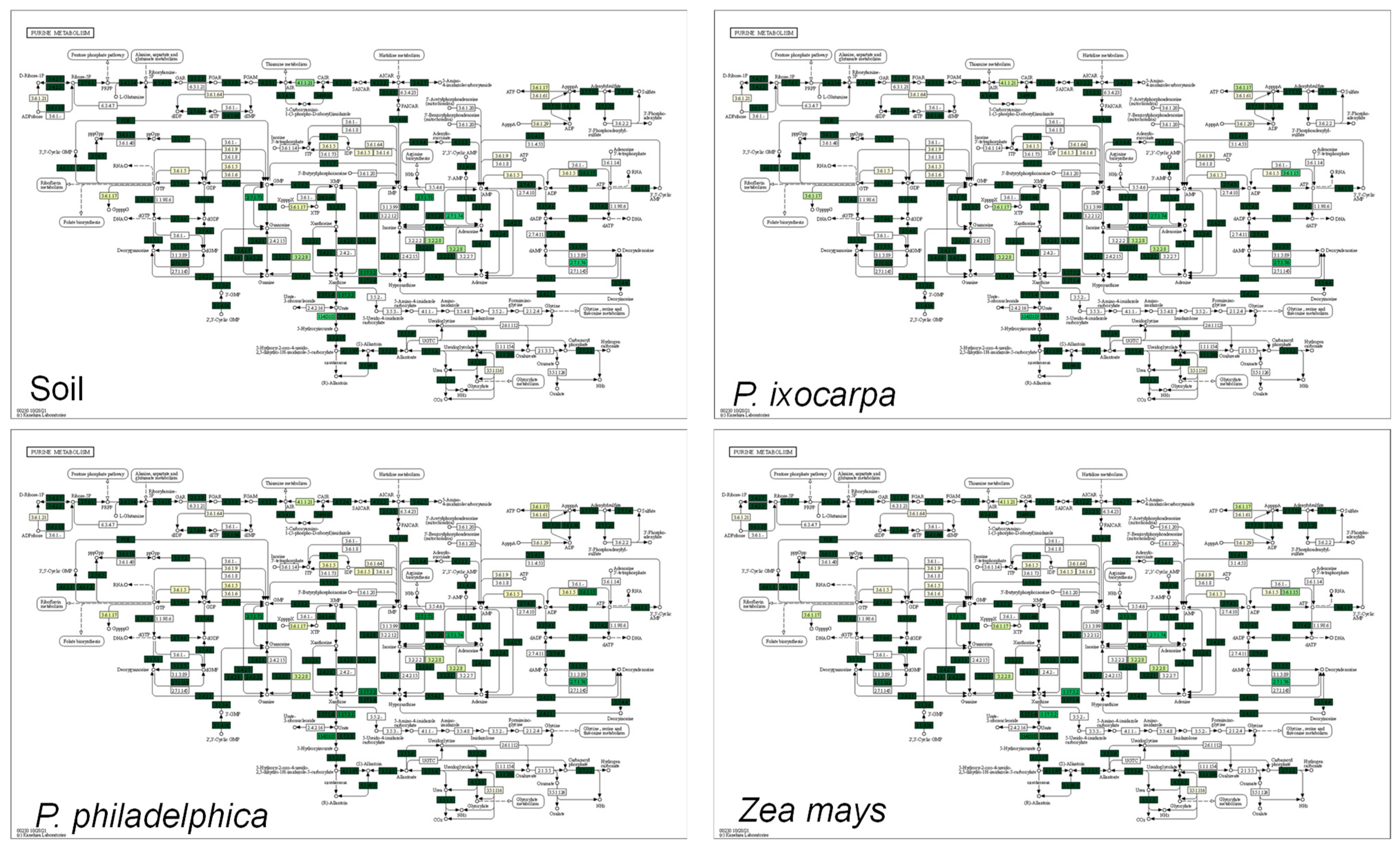
3.3. Inferred Functions of Bacterial on the System
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Villaseñor, J.L.; Ortiz, E.; Juárez, D. Transition zones and biogeographic characterization of endemism in three biogeographic provinces of central Mexico. Bot. Sci. 2021, 99, 938–954. [Google Scholar] [CrossRef]
- Alcalá-Gómez, G.; Pérez-Alquicira, J.; Cabrera-Toledo, D.; Cortés-Cruz, M.; Zamora-Tavares, M.P.; Vargas-Ponce, O. Genetic diversity and structure in husk tomato (Physalis philadelphica Lam.) based on SNPs: A case of diffuse domestication. Genet. Resour. Crop Evol. 2022, 69, 443–459. [Google Scholar] [CrossRef]
- Zizumbo-Villarreal, D.; Colunga-GarcíaMarín, P. Origin of agriculture and plant domestication in West Mesoamerica. Genet. Resour. Crop Evol. 2010, 57, 813–825. [Google Scholar] [CrossRef]
- Casas, A.; Otero-Arnaiz, A.; Pérez-Negrón, E.; Valiente-Banuet, A. In situ Management and Domestication of Plants in Mesoamerica. Ann. Bot. 2007, 100, 1101–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Ridaura, S.; Barba-Escoto, L.; Reyna-Ramirez, C.A.; Sum, C.; Palacios-Rojas, N.; Gerard, B. Maize intercropping in the milpa system. Diversity, extent and importance for nutritional security in the Western Highlands of Guatemala. Sci. Rep. 2021, 11, 3696. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Raina, T.K.; Kumar, A.; Singh, J.; Prasad, R. Plant microbiome: A reservoir of novel genes and metabolites. Plant Gene. 2019, 18, 100177. [Google Scholar] [CrossRef]
- Samaddar, S.; Karp, D.S.; Schmidt, R.; Devarajan, N.; McGarvey, J.A.; Pires, A.F.A.; Scow, K. Role of soil in the regulation of human and plant pathogens: Soils’ contributions to people. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2021, 376, 1834. [Google Scholar] [CrossRef]
- Yachi, S.; Loreau, M. Biodiversity and ecosystem productivity in a fluctuating environment: The insurance hypothesis. Proc. Natl. Acad. Sci. USA 1999, 96, 1463–1468. [Google Scholar] [CrossRef] [Green Version]
- Vargas-Ponce, O.; Pérez-Álvarez, L.F.; Zamora-Tavares, P.; Rodríguez, A. Assessing Genetic Diversity in Mexican Husk Tomato Species. Plant Mol. Biol. Rep. 2011, 29, 733–738. [Google Scholar] [CrossRef]
- Martínez-Vega, A.L.; Oregel-Zamudio, E.; García-Ruíz, I.; Villapando-Arteaga, E.V.; Torres-García, J.R. Genetic and metabolomic differentiation of Physalis ixocarpa Brot. ex Hornem. populations in Michoacan State, Mexico. Genet. Resour. Crop Evol. 2022, 69, 1867–1877. [Google Scholar] [CrossRef]
- Schloss, P.D. Reintroducing mothur: 10 Years Later. Appl. Environ. Microbiol. 2020, 86, e02343-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagpal, S.; Haque, M.M.; Singh, R.; Mande, S.S. IVikodak—A platform and standard workflow for inferring, analyzing, comparing, and visualizing the functional potential of microbial communities. Front. Microbiol. 2019, 10, 3336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 10, e2584. [Google Scholar] [CrossRef] [PubMed]
- Bashir, S.; Zeshan, H.W.; Niazi, N.K.; Bibi, I.; Ahmad, N. Current Approaches for the Assessment of In Situ Remediation of Xenobiotics. In Xenobiotics in the Soil Environment; Hashmi, M., Kumar, V., Varma, A., Eds.; Springer: Cham, Switzerland, 2017; pp. 171–196. [Google Scholar]
- Meena, R.S.; Kumar, S.; Datta, R.; Lal, R.; Vijayakumar, V.; Brtnicky, M.; Sharma, M.P.; Yadav, G.S.; Jhariya, M.K.; Jangir, C.K.; et al. Impact of agrochemicals on soil microbiota and management: A review. Land 2020, 9, 34. [Google Scholar] [CrossRef] [Green Version]
- Kim, N.; Zabaloy, M.C.; Guan, K.; Villamil, M.B. Do cover crops benefit soil microbiome? A meta-analysis of current research. Soil Biol. Biochem. 2020, 142, 107701. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Hastorf, C.A. Rio Balsas most likely region for maize domestication. Proc. Natl. Acad. Sci. USA 2009, 106, 4957–4958. [Google Scholar] [CrossRef] [Green Version]
- Szoboszlay, M.; Lambers, J.; Chappell, J.; Kupper, J.V.; Moe, L.A.; McNear, D.H., Jr. Comparison of root system architecture and rhizosphere microbial communities of Balsas teosinte and domesticated corn cultivars. Soil Biol. Biochem. 2015, 80, 34–44. [Google Scholar] [CrossRef]
- Zamora-Tavares, P.; Vargas-Ponce, O.; Sánchez-Martínez, J.; Dánae, C.T. Diversity and genetic structure of the husk tomato (Physalis philadelphica Lam.) in Western Mexico. Genet. Resour. Crop Evol. 2015, 62, 141–153. [Google Scholar] [CrossRef]
- Zamora-Tavares, M.P.; Martínez, M.; Magallón, S.; Guzmán-Dávalos, L.; Vargas-Ponce, O. Physalis and physaloids: A recent and complex evolutionary history. Mol. Phylogenet. Evol. 2016, 100, 41–50. [Google Scholar] [CrossRef]
- Babich, T.L.; Semenova, E.M.; Sokolova, D.S.; Tourova, T.P.; Bidzhieva, S.K.; Loiko, N.G.; Avdonin, G.I.; Lutsenko, N.I.; Nazina, T.N. Phylogenetic Diversity and Potential Activity of Bacteria and Fungi in the Deep Subsurface Horizons of An Uranium Deposit. Microbiology 2021, 90, 607–620. [Google Scholar] [CrossRef]
- Lupatini, M.; Korthals, G.W.; de Hollander, M.; Janssens, T.K.S.; Kuramae, E.E. Soil microbiome is more heterogeneous in organic than in conventional farming system. Front. Microbiol. 2017, 7, 2064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahram, M.; Hildebrand, F.; Forslund, S.K.; Anderson, J.L.; Soudzilovskaia, N.A.; Bodegom, P.M.; Bengtsson-Palme, J.; Anslan, S.; Coelho, L.P.; Harend, H.; et al. Structure and function of the global topsoil microbiome. Nature 2018, 560, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Miao, Y.; Liu, K.; Ren, Y.; Xu, Z.; Zhang, N.; Feng, H.; Shen, Q.; Zhang, R.; Xun, W. Rhizosphere microbiome assembly involves seed-borne bacteria in compensatory phosphate solubilization. Soil Biol. Biochem. 2021, 159, 108273. [Google Scholar] [CrossRef]
- Carrier, V.; Svenning, M.M.; Gründger, F.; Niemann, H.; Dessandier, P.A.; Panieri, G.; Kalenitchenko, D. The Impact of Methane on Microbial Communities at Marine Arctic Gas Hydrate Bearing Sediment. Front. Microbiol. 2020, 11, 01932. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, J.M.; Zhou, Y.L.; Almeida, A.; Finn, R.D.; Danchin, A.; He, L.S. Phylogenomics of expanding uncultured environmental Tenericutes provides insights into their pathogenicity and evolutionary relationship with Bacilli. BMC Genom. 2020, 21, 408. [Google Scholar] [CrossRef]
- Hugenholtz, P.; Pitulle, C.; Hershberger, K.L.; Pace, N.R. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 1998, 180, 366–376. [Google Scholar] [CrossRef] [Green Version]
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.C.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Grüning, N.M.; Krüger, A.; Alam, M.T.; et al. The return of metabolism: Biochemistry and physiology of the pentose phosphate pathway A. Stincone and others. Biol. Rev. Biol. Proc. Camb. Philos. Soc. 2015, 90, 927–963. [Google Scholar] [CrossRef] [Green Version]
- Becarelli, S.; Chicca, I.; La China, S.; Siracusa, G.; Bardi, A.; Gullo, M.; Petroni, G.; Levin, D.B.; Di Gregorio, S. A New Ciboria sp. for Soil Mycoremediation and the Bacterial Contribution to the Depletion of Total Petroleum Hydrocarbons. Front. Microbiol. 2021, 12, 647373. [Google Scholar] [CrossRef]
- Tourova, T.; Sokolova, D.; Nazina, T.; Grouzdev, D.; Kurshev, E.; Laptev, A. Biodiversity of microorganisms colonizing the surface of polystyrene samples exposed to different aqueous environments. Sustainability 2020, 12, 3624. [Google Scholar] [CrossRef]
- Antonopoulos, D.A.; Huse, S.M.; Morrison, H.G.; Schmidt, T.M.; Sogin, M.L.; Young, V.B. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 2009, 77, 2367–2375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dethlefsen, L.; Huse, S.M.; Sogin, M.L.; Relman, D.A. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. PLoS Biol. 2008, 6, 2383–2400. [Google Scholar] [CrossRef] [PubMed]
- Shade, A.; Handelsman, J. Beyond the Venn diagram: The hunt for a core microbiome. Environ. Microbiol. 2012, 14, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Skourtev, P.; Gehrenfeld, J.; Häggblom, M. Experimental analysis of the effect of exotic and native plant species on the structure and function of soil microbial communities. Soil Biol. Biochem. 2003, 35, 895–905. [Google Scholar] [CrossRef]
- Yu, X.; Yu, D.; Lu, Z.; Ma, K. A new mechanism of invader success: Exotic plant inhibits natural vegetation restoration by changing soil microbe community. Sci. Bull. 2005, 50, 1105–1112. [Google Scholar] [CrossRef]
- Koziol, L.; Bauer, J.T.; Duell, E.B.; Hickman, K.; House, G.L.; Schultz, P.A.; Tipton, A.G.; Wilson, G.W.T.; Bever, J.D. Manipulating plant microbiomes in the field: Native mycorrhizae advance plant succession and improve native plant restoration. J. Appl. Ecol. 2021, 1–10. [Google Scholar] [CrossRef]


| Properties | Value |
|---|---|
| Texture | 40% Clay, 30% Silt, 30% Sand |
| Organic Matter (MO) | 1.6% |
| pH | 6.4 |
| Electric Conductivity | 1.9 dS m−1 |
| Aparent Density | 1.09 g cm−3 |
| Total Nitrogen | 0.096% |
| Phosphorus (Bray y Kurt z) | 17 mg kg−1 |
| Potassium (K) | 0.7 Cmol kg−1 |
| Soil | P. ixocarpa | P. philadelphica | Zea mays L. | |
|---|---|---|---|---|
| Soil | - | 0.001 | 0.001 | 0.001 |
| P. ixocarpa | 0.71 | - | 0.001 | 0.001 |
| P. philadelphica | 0.73 | 0.67 | - | 0.001 |
| Zea mays | 0.8 | 0.76 | 0.76 | - |
| Simpson | Shannon-Wiener | |
|---|---|---|
| Soil | 0.00414 ± 0.000167 b | 6.83 ± 0.0448 a |
| P. ixocarpa | 0.00927 ± 0.00103 a | 6.43 ± 0.0345 b |
| P. philadelphica | 0.00705 ± 0.000403 a | 6.46 ± 0.0625 ab |
| Zea mays | 0.00716 ± 0.000244 a | 6.4 ± 0.123 b |
| Soil | P. ixocarpa | P. philadelphica | Zea mays | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genus | % | Cumulated Percentage | Genus | % | Cumulated Percentage | Genus | % | Cumulated Percentage | Genus | % | Cumulated Percentage |
| Sphingomonas | 12.0 | 12.0 | Sphingobium | 14.5 | 14.5 | Sphingobium | 13.9 | 13.9 | Sphingomonas | 14.9 | 14.9 |
| Nocardioides | 5.6 | 17.5 | Archangium | 4.8 | 19.2 | Pseudorhodoferax | 6.3 | 20.2 | Ramlibacter | 5.5 | 20.4 |
| Streptomyces | 4.8 | 22.4 | Skermanella | 3.8 | 23.0 | Streptomyces | 5.7 | 25.9 | Pseudonocardia | 4.5 | 24.9 |
| Ramlibacter | 3.6 | 26.0 | Pelomonas | 3.6 | 26.6 | Solirubrobacter | 3.9 | 29.8 | Nocardioides | 4.0 | 28.8 |
| Pseudonocardia | 3.6 | 29.5 | Streptomyces | 3.6 | 30.2 | Archangium | 3.8 | 33.6 | Streptomyces | 3.9 | 32.8 |
| Reyranella | 3.0 | 32.5 | Solirubrobacter | 3.5 | 33.8 | Nocardioides | 3.8 | 37.4 | Solirubrobacter | 3.8 | 36.6 |
| Paenibacillus | 3.0 | 35.5 | Nocardioides | 3.4 | 37.1 | Skermanella | 3.4 | 40.8 | Reyranella | 3.4 | 40.0 |
| Solirubrobacter | 2.9 | 38.4 | Nitrospira | 3.2 | 40.3 | Flavisolibacter | 3.3 | 44.1 | Singulisphaera | 3.1 | 43.1 |
| Labilithrix | 2.8 | 41.2 | Tumebacillus | 3.1 | 43.5 | Pseudonocardia | 3.1 | 47.2 | Flavisolibacter | 2.7 | 45.7 |
| Singulisphaera | 2.3 | 43.5 | Flavisolibacter | 3.0 | 46.4 | Nitrospira | 2.6 | 49.8 | Phenylobacterium | 2.6 | 48.3 |
| Massilia | 2.3 | 45.7 | Pseudonocardia | 2.9 | 49.3 | Hamadaea | 2.4 | 52.22 | Labilithrix | 2.6 | 50.9 |
| Chryseolinea | 2.2 | 48.0 | Hamadaea | 2.1 | 51.4 | Tumebacillus | 1.9 | 54.1 | Massilia | 2.4 | 53.3 |
| Lysobacter | 2.2 | 50.2 | Methylobacterium | 1.8 | 53.2 | Methylobacterium | 1.65 | 55.8 | Pseudomonas | 2.2 | 55.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ariza-Mejía, D.; Oyoque-Salcedo, G.; Angóa-Pérez, V.; Mena-Violante, H.G.; Álvarez-Bernal, D.; Torres-García, J.R. Diversity and Potential Function of the Bacterial Rhizobiome Associated to Physalis Ixocarpa Broth. in a Milpa System, in Michoacan, Mexico. Agronomy 2022, 12, 1780. https://doi.org/10.3390/agronomy12081780
Ariza-Mejía D, Oyoque-Salcedo G, Angóa-Pérez V, Mena-Violante HG, Álvarez-Bernal D, Torres-García JR. Diversity and Potential Function of the Bacterial Rhizobiome Associated to Physalis Ixocarpa Broth. in a Milpa System, in Michoacan, Mexico. Agronomy. 2022; 12(8):1780. https://doi.org/10.3390/agronomy12081780
Chicago/Turabian StyleAriza-Mejía, Daniella, Guadalupe Oyoque-Salcedo, Valentina Angóa-Pérez, Hortencia G. Mena-Violante, Dioselina Álvarez-Bernal, and Jesús R. Torres-García. 2022. "Diversity and Potential Function of the Bacterial Rhizobiome Associated to Physalis Ixocarpa Broth. in a Milpa System, in Michoacan, Mexico" Agronomy 12, no. 8: 1780. https://doi.org/10.3390/agronomy12081780
APA StyleAriza-Mejía, D., Oyoque-Salcedo, G., Angóa-Pérez, V., Mena-Violante, H. G., Álvarez-Bernal, D., & Torres-García, J. R. (2022). Diversity and Potential Function of the Bacterial Rhizobiome Associated to Physalis Ixocarpa Broth. in a Milpa System, in Michoacan, Mexico. Agronomy, 12(8), 1780. https://doi.org/10.3390/agronomy12081780







