Temporal Response of Bacterial Community Associated Fe(III) Reduction to Initial pH Shift of Paddy Soils
Abstract
:1. Introduction
2. Materials and Methods
2.1. Soil Sampling
2.2. Treatments and Anaerobic Flooding
2.3. Chemical Analysis
2.4. DNA Extraction and Bacterial Community Analysis
2.5. Statistical Analysis
3. Results
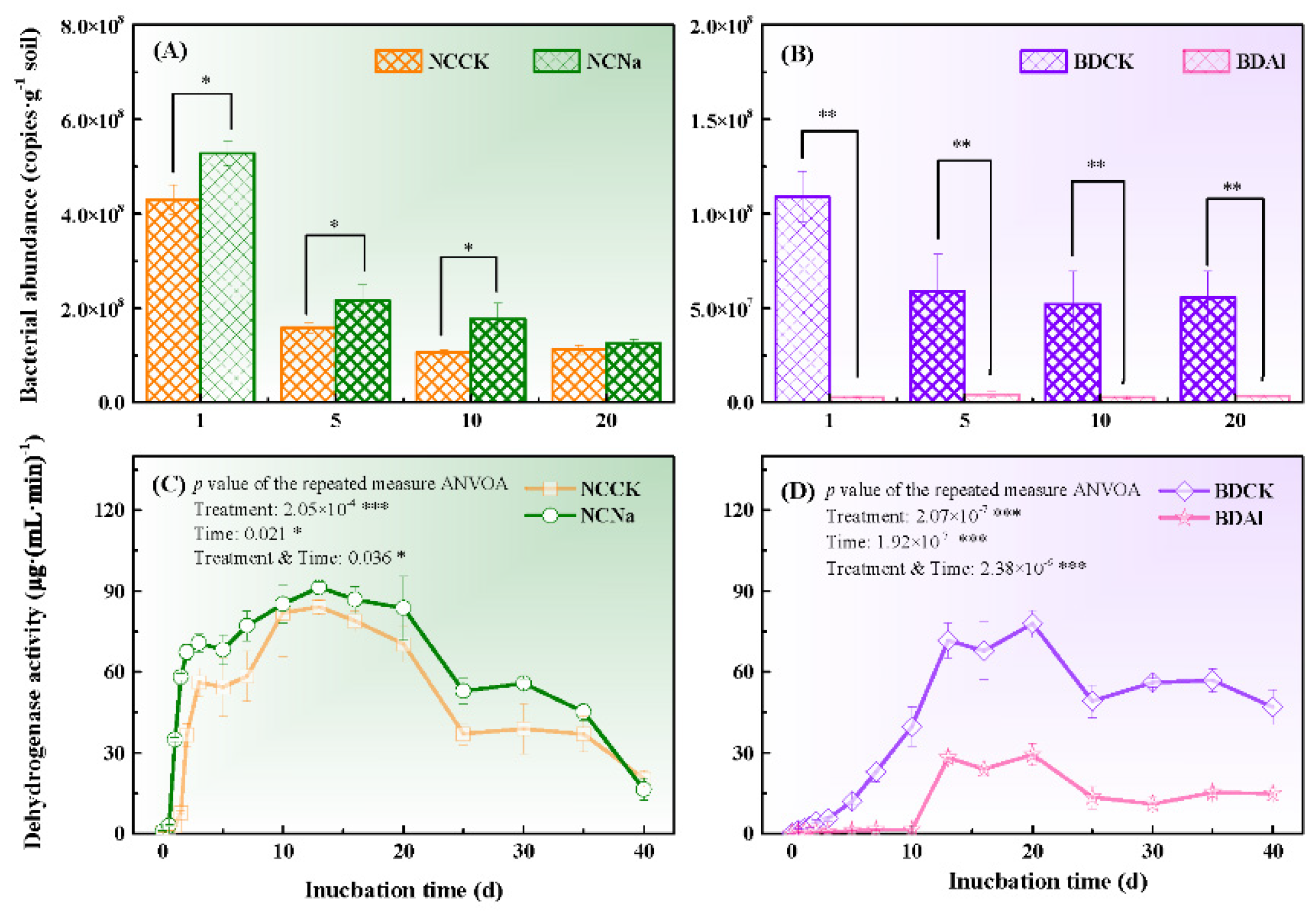
3.1. Bacterial Abundance and Activity
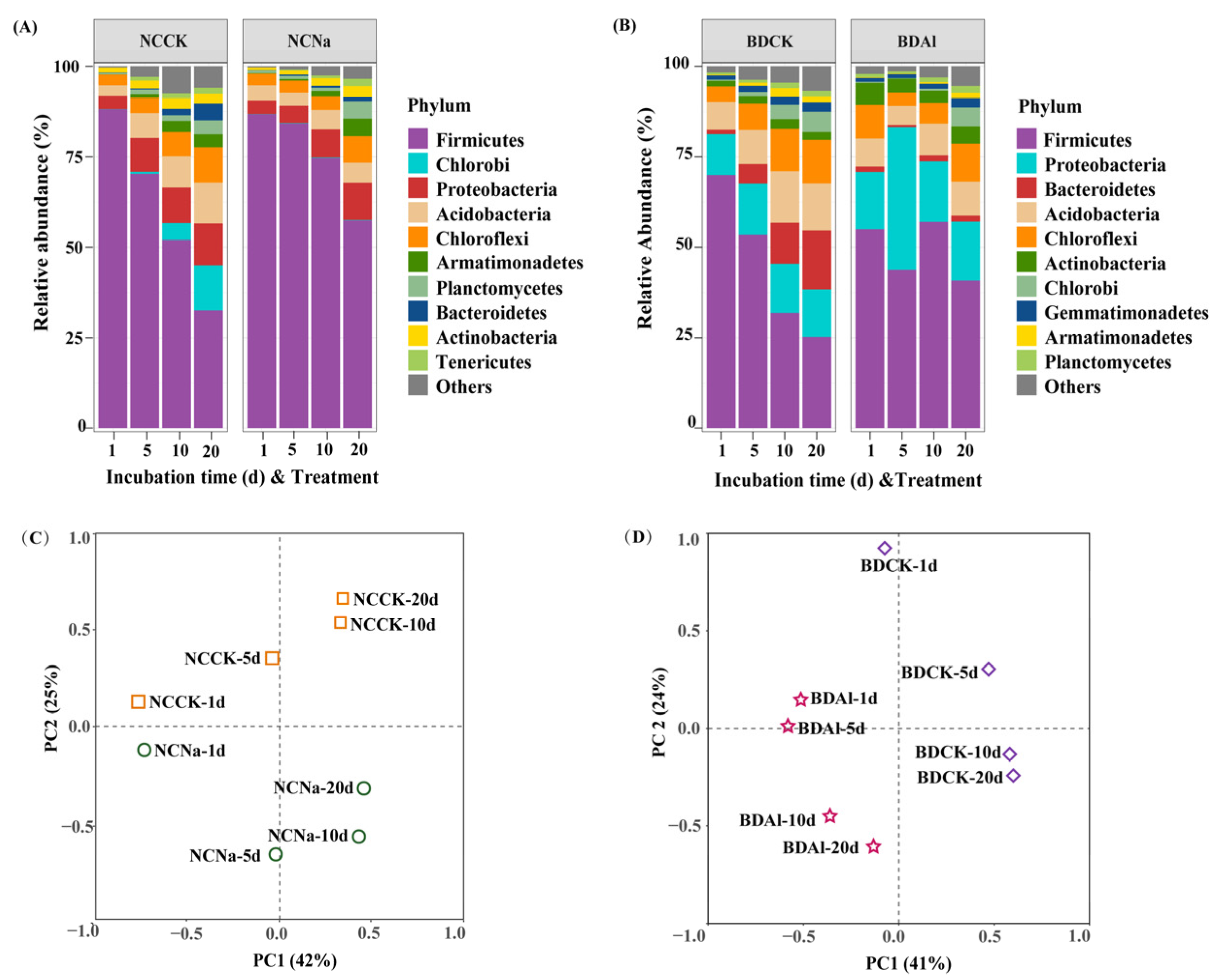
3.2. Bacterial Community Structure and Similarity
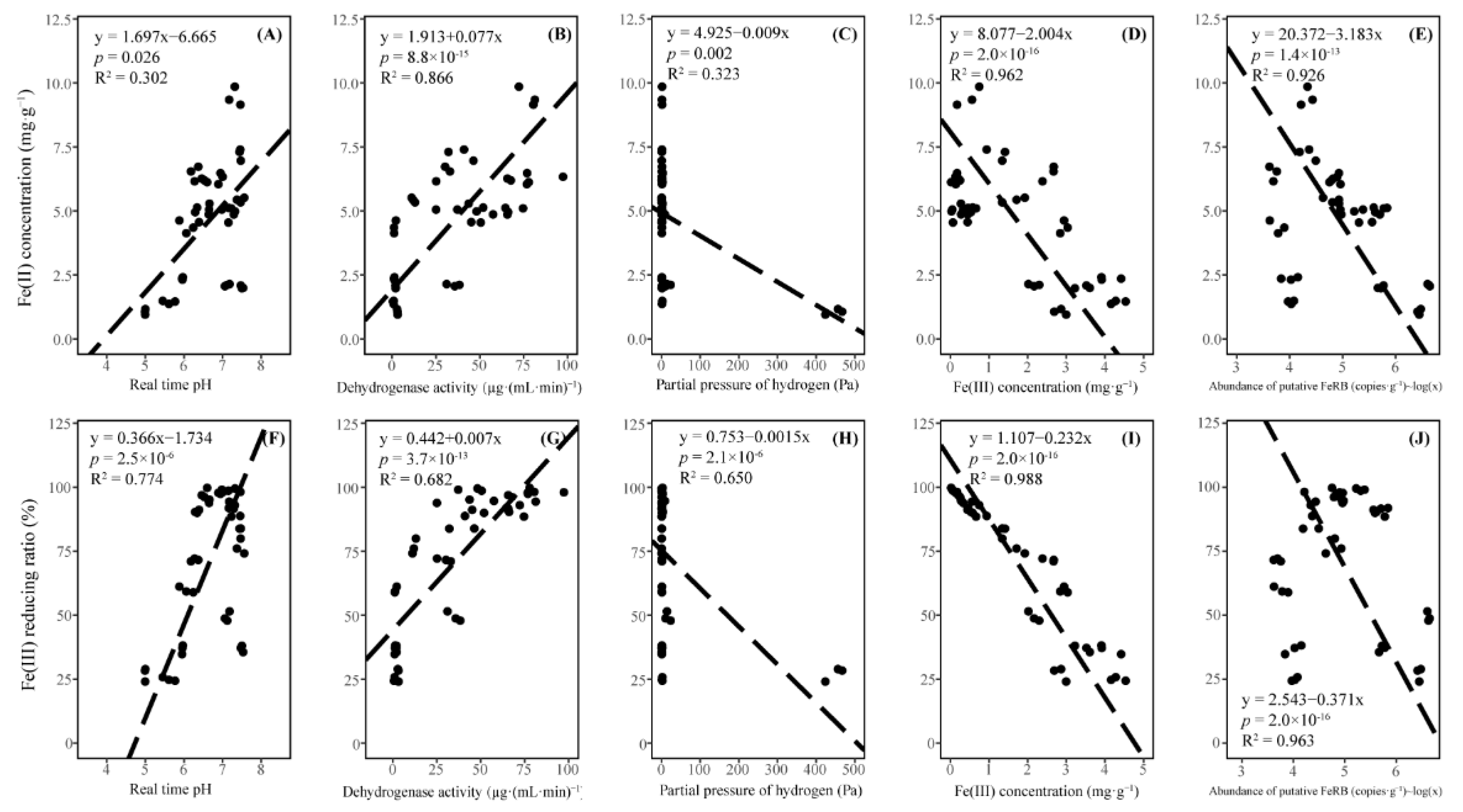
3.3. Fe(III) Reduction Process and Putative Fe(III) Reducing Bacteria
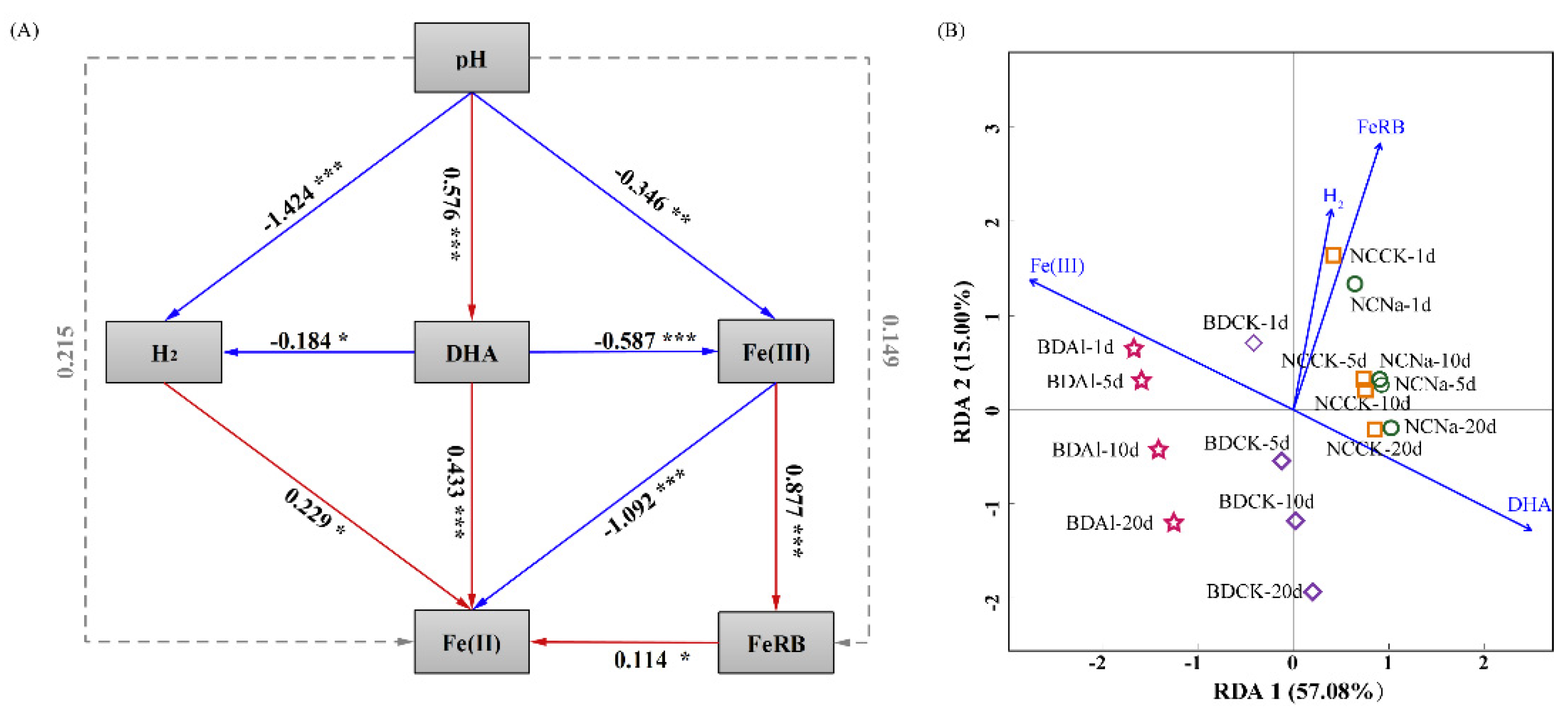
3.4. Contribution of pH Shift-Induced Bacterial Community, Fe(III) Concentration and Biohydrogen to Fe(III) Reduction
4. Discussion
4.1. Bacterial Community Succession in Response to Soil Acidification or Alkalization
4.2. Fe(III) Reduction Associated with the Response of H2 Production and Oxidation to Soil Acidification or Alkalization
4.3. Fe(III) Reduction Associated with the Response of Putative FeRB to Soil Acidification or Alkalization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kögel-Knabner, I.; Amelung, W.; Cao, Z.H.; Fiedler, S.; Frenzel, P.; Jahn, R.; Kalbitz, K.; Kölbl, A.; Schloter, M. Biogeochemistry of paddy soils. Geoderma 2010, 157, 1–14. [Google Scholar] [CrossRef]
- Crowther, T.W.; Hoogen, J.D.; Wan, J.; Mayes, M.A.; Keiser, A.D.; Mo, L.; Averill, C.; Maynard, D.S. The global soil community and its influence on biogeochemistry. Science 2019, 365, eaav0550. [Google Scholar] [CrossRef] [PubMed]
- Luan, L.; Liang, C.; Chen, L.J.; Wang, H.T.; Xu, Q.S.; Jiang, Y.J.; Sun, B. Coupling bacterial community assembly to microbial metabolism across soil profiles. mSystems 2020, 5, e00298-20. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Xu, Y.Q.; Zhang, J.; Hao, X.; Lu, Y.H. Core microbiota in agricultural soils and their potential associations with nutrient cycling. mSystems 2019, 4, e00313-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akter, M.; Deroo, H.; De Grave, E.; Van Alboom, T.; Kader, M.A.; Pierreux, S.; Begum, M.A.; Boeckx, P.; Sleutel, S. Link between paddy soil mineral nitrogen release and iron and manganese reduction examined in a rice pot growth experiment. Geoderma 2018, 326, 9–21. [Google Scholar] [CrossRef]
- Sun, W.M.; Xiao, E.Z.; Pu, Z.L.; Krumins, V.; Dong, Y.R.; Li, B.Q.; Hu, M. Paddy soil microbial communities driven by environment- and microbe-microbe interactions: A case study of elevation-resolved microbial communities in a rice terrace. Sci. Total Environ. 2018, 612, 884–893. [Google Scholar] [CrossRef]
- Yi, B.; Wang, H.H.; Zhang, Q.C.; Jin, H.; Abbas, T.; Li, Y.; Liu, Y.M.; Di, H.J. Alteration of gaseous nitrogen losses via anaerobic ammonium oxidation coupled with ferric reduction from paddy soils in Southern China. Sci. Total Environ. 2019, 652, 1139–1147. [Google Scholar] [CrossRef]
- Zecchin, S.; Corsini, A.; Martin, M.; Romani, M.; Beone, G.M.; Zanchi, R.; Zanzo, E.; Tenni, D.; Fontanella, M.C.; Cavalca, L. Rhizospheric iron and arsenic bacteria affected by water regime: Implications for metalloid uptake by rice. Soil Biol. Biochem. 2017, 106, 129–137. [Google Scholar] [CrossRef]
- Xue, L.L.; Feng, X.; Xu, Y.; Li, X.F.; Zhu, M.; Xu, J.M.; He, Y. The dechlorination of pentachlorophenol under a sulfate and iron reduction co-occurring anaerobic environment. Chemosphere 2017, 182, 166–173. [Google Scholar] [CrossRef]
- Yu, H.Y.; Li, F.B.; Liu, C.S.; Huang, W.; Liu, T.X.; Yu, W.M. Iron redox cycling coupled to transformation and immobilization of heavy metals: Implications for paddy rice safety in the red soil of south china. Adv. Agro. 2016, 137, 279–313. [Google Scholar]
- Lovley, D.R.; Phillips, E.J.P. Novel mode of microbial energy metabolism: Organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 1988, 54, 1472–1480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raiswell, R.; Canfield, D.E. The iron biogeochemical cycle past and present. Geochem. Perspect. 2012, 1, 1–220. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, C.; Behrens, S.; Kappler, A. Ecosystem functioning from a geomicrobiological perspective—A conceptual framework for biogeochemical iron cycling. Environ. Chem. 2010, 7, 399–405. [Google Scholar] [CrossRef] [Green Version]
- Lovley, D.R. Organic matter mineralization with the reduction of ferric iron: A review. Geomicrobiol. J. 1987, 5, 375–399. [Google Scholar] [CrossRef]
- Hamid, Y.; Tang, L.; Hussain, B.; Usman, M.; Gurajala, H.K.; Rashid, M.S.; He, Z.L.; Yang, X.E. Efficiency of lime, biochar, Fe containing biochar and composite amendments for Cd and Pb immobilization in a co-contaminated alluvial soil. Environ. Pollut. 2020, 257, 113609. [Google Scholar] [CrossRef] [PubMed]
- Kemmitt, S.J.; Wright, D.; Goulding, K.W.T.; Jones, D. pH regulation of carbon and nitrogen dynamics in two agricultural soils. Soil Biol. Biochem. 2006, 38, 898–911. [Google Scholar] [CrossRef]
- Zhou, M.; Liu, X.B.; Meng, Q.F.; Zeng, X.N.; Zhang, J.Z.; Li, D.W.; Wang, J.; Du, W.L.; Ma, X.F. Additional application of aluminum sulfate with different fertilizers ameliorates saline-sodic soil of Songnen Plain in Northeast China. J. Soils Sediments 2020, 19, 3521–3533. [Google Scholar] [CrossRef]
- Ponnamperuma, F.N. The chemistry of submerged soils. Adv. Agron. 1972, 24, 29–96. [Google Scholar]
- Pan, Y.Y.; Koopmans, G.F.; Bonten, L.T.C.; Song, J.; Luo, Y.M.; Temminghoff, E.J.M.; Comans, R.N.J. Influence of pH on the redox chemistry of metal (hydr)oxides and organic matter in paddy soils. J. Soils Sediments 2014, 14, 1713–1726. [Google Scholar] [CrossRef]
- Colombo, C.; Palumbo, G.; He, J.Z.; Pinton, R.; Cesco, S. Review on iron availability in soil: Interaction of Fe minerals, plants, and microbes. J. Soils Sediments 2014, 14, 538–548. [Google Scholar] [CrossRef]
- Jiao, S.; Lu, Y.H. Soil pH and temperature regulate assembly processes of abundant and rare bacterial communities in agricultural ecosystems. Environ. Microbiol. 2020, 22, 1052–1065. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.S.; Kirk, M.F. pH as a primary control in environmental microbiology: 1. Thermodynamic perspective. Front. Environ. Sci. 2018, 6, 21. [Google Scholar] [CrossRef]
- Rousk, J.; Baath, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Whittleston, R.A.; Stewart, D.I.; Mortimer, R.J.G.; Burke, I.T. Enhancing microbial iron reduction in hyperalkaline, chromium contaminated sediments by pH amendment. Appl. Geochem. 2013, 28, 135–144. [Google Scholar] [CrossRef] [Green Version]
- Blothe, M.; Akob, D.M.; Kostka, J.E.; Goschel, K.; Drake, H.L.; Kusel, K. pH gradient-induced heterogeneity of Fe(III)-reducing microorganisms in coal mining-associated lake sediments. Appl. Environ. Microb. 2008, 74, 1019–1029. [Google Scholar] [CrossRef] [Green Version]
- Jia, R.; Li, L.N.; Qu, D. pH shift-mediated dehydrogenation and hydrogen production are responsible for microbial iron(III) reduction in submerged paddy soils. J. Soils Sediments 2015, 15, 1178–1190. [Google Scholar] [CrossRef]
- Li, L.N.; Qu, Z.; Wang, B.L.; Jia, R.; Qu, D. The response of metabolically active Clostridium community to initial pH shift is closely correlated with microbial Fe(III) reduction in flooded paddy soils. J. Soils Sediments 2019, 19, 522–532. [Google Scholar] [CrossRef]
- Heděnec, P.; Rui, J.P.; Yao, M.J.; Li, J.B.; Lin, Q.; Frouz, J.; Li, X.Z. Temporal response of soil prokaryotic communities to acidification and alkalization under laboratory conditions. Eur. J. Soil Biol. 2018, 86, 63–71. [Google Scholar] [CrossRef]
- Yang, L.; Lou, J.; Wang, H.Z.; Wu, L.S.; Xu, J.M. Use of an improved high-throughput absolute abundance quantification method to characterize soil bacterial community and dynamics. Sci. Total Environ. 2018, 633, 360–371. [Google Scholar] [CrossRef]
- Zhang, Z.J.; Qu, Y.Y.; Li, S.Z.; Feng, K.; Wang, S.; Cai, W.W.; Liang, Y.T.; Li, H.; Xu, M.Y.; Yin, H.Q. Soil bacterial quantification approaches coupling with relative abundances reflecting the changes of taxa. Sci. Rep. 2017, 7, 4837. [Google Scholar] [CrossRef]
- Laura, R.D. Effects of alkali salts on carbon and nitrogen mineralization of organic matter in soil. Plant Soil 1976, 44, 587–596. [Google Scholar] [CrossRef]
- Nicol, G.W.; Webster, G.; Glover, L.A.; Prosser, J.I. Differential response of archaeal and bacterial communities to nitrogen inputs and pH changes in upland pasture rhizosphere soil. Environ. Microbiol. 2004, 6, 861–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.Z.; Qu, D. Dissimilatory Fe(III) reduction characteristics of paddy soil extract cultures treated with glucose or fatty acids. J. Environ. Sci. 2008, 20, 1103–1108. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Boshier, F.A.T.; Srinivasan, S.; Lopez, A.; Hoffman, N.G.; Proll, S.; Fredricks, D.N.; Schiffer, J.T. Complementing 16S rRNA gene amplicon sequencing with total bacterial load to infer absolute species concentrations in the vaginal microbiome. mSystems 2020, 5, e00777-19. [Google Scholar]
- Lai, J.S.; Zou, Y.; Zhang, J.L.; Peres-Neto, P. Generalizing hierarchical and variation partitioning in multiple regression and canonical analyses using the rdacca.hp R package. Methods Ecol. Evol. 2022, 13, 782–788. [Google Scholar] [CrossRef]
- Liu, W.J.; Graham, E.B.; Zhong, L.H.; Zhang, J.; Li, S.J.; Lin, X.G.; Feng, Y.Z. Long-term stochasticity combines with short-term variability in assembly processes to underlie rice paddy sustainability. Front. Microbiol. 2020, 11, 873. [Google Scholar] [CrossRef]
- Noll, M.; Matthies, D.; Frenzel, P.; Derakshani, M.; Liesack, W. Succession of bacterial community structure and diversity in a paddy soil oxygen gradient. Environ. Microbiol. 2005, 7, 382–395. [Google Scholar] [CrossRef]
- Feng, Y.Z.; Chen, R.R.; Stegen, J.C.; Guo, Z.Y.; Zhang, J.W.; Li, Z.P.; Lin, X.G. Two key features influencing community assembly processes at regional scale: Initial state and degree of change in environmental conditions. Mol. Ecol. 2018, 27, 5238–5251. [Google Scholar] [CrossRef]
- Rui, J.P.; Peng, J.J.; Lu, Y.H. Succession of bacterial populations during plant residue decomposition in rice field soil. Appl. Environ. Microbiol. 2009, 75, 4779–4886. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.F.; Chen, J.H.; Pan, G.X.; Liu, X.Y.; Zhang, X.H.; Li, L.Q.; Sian, R.J.; Cheng, K.; Zheng, J.W. Biochar decreased microbial metabolic quotient and shifted community composition four years after a single incorporation in a slightly acid rice paddy from southwest China. Sci. Total Environ. 2016, 571, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.C.; Zeng, J.; Zhu, Q.H.; Zhang, Z.F.; Lin, X.G. pH is the primary determinant of the bacterial community structure in agricultural soils impacted by polycyclic aromatic hydrocarbon pollution. Sci. Rep. 2017, 7, 40093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, B.M.; Stegen, J.C.; Kim, M.; Dong, K.; Adams, J.M.; Lee, Y.K. Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 2018, 12, 1072–1083. [Google Scholar] [CrossRef] [PubMed]
- Grybos, M.; Davranche, M.; Gruau, G.; Petitjean, P.; Pédrot, M. Increasing pH drives organic matter solubilization from wetland soils under reducing conditions. Geoderma 2009, 154, 13–19. [Google Scholar] [CrossRef]
- Zhang, X.M.; Liu, W.; Zhang, G.M.; Jiang, L.; Han, X.G. Mechanisms of soil acidification reducing bacterial diversity. Soil Biol. Biochem. 2015, 81, 275–281. [Google Scholar] [CrossRef]
- Włodarczyk, T.S.; Stępniewski, W.B.; Brzezińska, M. Dehydrogenase activity, redox potential, and emissions of carbon dioxide and nitrous oxide from Cambisols under flooding conditions. Biol. Fertil. Soils 2002, 36, 200–206. [Google Scholar] [CrossRef]
- Insam, H. Developments in soil microbiology since the mid 1960s. Geoderma 2001, 100, 389–402. [Google Scholar] [CrossRef]
- Lee, H.S.; Vermaas, W.F.; Rittmann, B.E. Biological hydrogen production: Prospects and challenges. Trends Biotechnol. 2010, 28, 262–271. [Google Scholar] [CrossRef]
- Weber, K.A.; Achenbach, L.A.; Coates, J.D. Microorganisms pumping iron: Anaerobic microbial iron oxidation and reduction. Nat. Rev. Microbiol. 2006, 4, 752–764. [Google Scholar] [CrossRef] [Green Version]
- Buyer, J.S.; Teasdale, J.R.; Roberts, D.P.; Zasada, I.A.; Maul, J.E. Factors affecting soil microbial community structure in tomato cropping systems. Soil Biol. Biochem. 2010, 42, 831–841. [Google Scholar] [CrossRef]
- Novara, A.; Catania, V.; Tolone, M.; Gristina, L.; Laudicina, V.A.; Quatrini, P. Cover crop impact on soil organic carbon, nitrogen dynamics and microbial diversity in a Mediterranean semiarid vineyard. Sustainability 2020, 12, 3256. [Google Scholar] [CrossRef] [Green Version]
- Ding, L.J.; Su, J.Q.; Xu, H.J.; Jia, Z.J.; Zhu, Y.G. Long-term nitrogen fertilization of paddy soil shifts iron-reducing microbial community revealed by RNA-13C-acetate probing coupled with pyrosequencing. ISME J. 2015, 9, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.A.; Shaaban, M.; Hu, R.G.; Mo, Y.L.; Wu, Y.P.; Ullah, B. Effects of soluble organic carbon addition on CH4 and CO2 emissions from paddy soils regulated by iron reduction. Soil Res. 2015, 52, 316–323. [Google Scholar] [CrossRef]
- Balashova, V.V.; Zavarzin, G.A. Anaerobic reduction of ferric iron by hydrogen bacteria. Microbiologia 1979, 48, 773–778. [Google Scholar]
- Hori, T.; Aoyagi, T.; Itoh, H.; Narihiro, T.; Oikawa, A.; Suzuki, K.; Ogata, A.; Friedrich, M.W.; Conrad, R.; Kamagata, Y. Isolation of microorganisms involved in reduction of crystalline iron(III) oxides in natural environments. Front. Microbiol. 2015, 6, 386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scala, D.J.; Hacherl, E.L.; Cowan, R.; Young, L.Y.; Kosson, D.S. Characterization of Fe(III)-reducing enrichment cultures and isolatison of Fe(III)-reducing bacteria from the Savannah River site, South Carolina. Res. Microbiol. 2006, 157, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Venkidusamy, K.; Hari, A.R.; Megharaj, M. Petrophilic, Fe(III) reducing exoelectrogen Citrobacter sp. KVM11, isolated from hydrocarbon fed microbial electrochemical remediation systems. Front. Microbiol. 2018, 9, 349–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, W.J.; Wang, B.L.; Qu, D. Diversity of isolates performing Fe(III) reduction from paddy soil fed by different organic carbon sources. Afr. J. Biotechnol. 2012, 11, 4407–4417. [Google Scholar]
- Ramamoorthy, S.; Sass, H.; Langner, H.; Schumann, P.; Kroppenstedt, R.M.; Spring, S.; Overmann, J.; Rosenzweig, R.F. Desulfosporosinus lacus sp nov., a sulfate-reducing bacterium isolated from pristine freshwater lake sediments. Int. J. Syst. Evol. Microbiol. 2006, 56, 2729–2736. [Google Scholar] [CrossRef] [Green Version]
- Shelobolina, E.S.; Nevin, K.P.; Blakeney-Hayward, J.; Johnsen, C.V.; Plaia, T.W.; Krader, P.; Woodard, T.; Holmes, D.E.; VanPraagh, C.G.; Lovley, D.R. Geobacter pickeringii sp nov., Geobacter argillaceus sp nov and Pelosinus fermentans gen. nov., sp nov., isolated from subsurface kaolin lenses. Int. J. Syst. Evol. Microbiol. 2007, 57, 126–135. [Google Scholar] [CrossRef]
- Qiao, S.S. Dynamics of Bacillus and Clostridium Population Abundance and Structure in Flooded Paddy Soil. Ph.D. Thesis, Northwest Agriculture & Forest University, Yangling, China, 2017; pp. 41–43. (In Chinese). [Google Scholar]
- Fernandez-Calvino, D.; Baath, E. Growth response of the bacterial community to pH in soils differing in pH. FEMS Microbiol. Ecol. 2010, 73, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Seuylemezian, A.; Singh, N.K.; Vaishampayan, P.; Venkateswaran, K. Draft genome sequence of Solibacillus kalamii, isolated from an air filter aboard the International Space Station. Genome Announc. 2017, 5, e00696-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, R.; Wang, K.; Li, L.N.; Qu, Z.; Shen, W.S.; Qu, D. Abundance and community succession of nitrogen-fixing bacteria in ferrihydrite enriched cultures of paddy soils is closely related to Fe(III)-reduction. Sci. Total Environ. 2020, 720, 137633. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.J.; Liu, L.P.; Wen, P.; Li, N.; Zong, M.H.; Wu, H. Effects of acetic acid and pH on the growth and lipid accumulation of the oleaginous yeast Trichosporon fermentans. Bioresources 2015, 10, 4152–4166. [Google Scholar] [CrossRef] [Green Version]
- Tibbett, M.; Gil-Martinez, M.; Fraser, T.; Green, I.D.; Duddigan, S.; De Oliveira, V.H.; Raulund-Rasmussen, K.; Sizmur, T.; Diaz, A. Long-term acidification of pH neutral grasslands affects soil biodiversity, fertility and function in a heathland restoration. Catena 2019, 180, 401–415. [Google Scholar] [CrossRef]
- McBride, M. Environmental Chemistry of Soils; Oxford Univ Press: New York, NY, USA, 1994. [Google Scholar]
- Li, Q. Enrichment of phosphate on ferrous iron phases during bio-reduction of ferrihydrite. Int. J. Geosciences. 2012, 3, 314–320. [Google Scholar] [CrossRef] [Green Version]
- Achtnich, C.; Bak, F.; Conrad, R. Competition for electron donors among nitrate reducers, ferric ironreducers, sulfate reducers, and methanogens in anoxic paddy soil. Biol Fert Soils. 1995, 19, 65–72. [Google Scholar] [CrossRef]
- Kwon, M.J.; Boyanov, M.I.; Antonopoulos, D.A.; Brulc, J.M.; Johnston, E.R.; Skinner, K.A.; Kemner, K.M.; O’Loughlin, E.J. Effects of dissimilatory sulfate reduction on FeIII (hydr)oxide reduction and microbial community development. Geochim. Et Cosmochim. Acta 2014, 129, 177–190. [Google Scholar] [CrossRef]
- Lefort, S.; Mucci, A.; Sundby, B. Sediment response to 25 years of persistent hypoxia. Aquat. Geochem. 2012, 18, 461–474. [Google Scholar] [CrossRef]
- Wu, C. Influence of initial pH value on microbial iron reduction in paddy soils (in Chinese). Master Thesis, Northwest A&F university, Yangling, China, 2013; p. 67. [Google Scholar]

| Treatments | Day 1 | Day 5 | Day 10 | Day 20 |
|---|---|---|---|---|
| NCCK | 27.09% ± 2.65% b | 90.47% ± 0.61% a | 94.54% ± 0.65% a | 97.57% ± 1.90% a |
| NCNa | 49.35% ± 1.88% a | 90.66% ± 1.86% a | 97.04% ± 0.45% a | 97.72% ± 0.29% a |
| BDCK | 36.92% ± 1.25% a | 76.72% ± 2.96% a | 85.47% ± 2.84% a | 95.16% ± 2.69% a |
| BDAl | 24.94% ± 0.73% b | 36.68% ± 1.75% b | 59.76% ± 1.20% b | 71.56% ± 0.55% b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, R.; Fan, F.; Li, L.; Qu, D. Temporal Response of Bacterial Community Associated Fe(III) Reduction to Initial pH Shift of Paddy Soils. Agronomy 2022, 12, 1304. https://doi.org/10.3390/agronomy12061304
Jia R, Fan F, Li L, Qu D. Temporal Response of Bacterial Community Associated Fe(III) Reduction to Initial pH Shift of Paddy Soils. Agronomy. 2022; 12(6):1304. https://doi.org/10.3390/agronomy12061304
Chicago/Turabian StyleJia, Rong, Fangmei Fan, Lina Li, and Dong Qu. 2022. "Temporal Response of Bacterial Community Associated Fe(III) Reduction to Initial pH Shift of Paddy Soils" Agronomy 12, no. 6: 1304. https://doi.org/10.3390/agronomy12061304
APA StyleJia, R., Fan, F., Li, L., & Qu, D. (2022). Temporal Response of Bacterial Community Associated Fe(III) Reduction to Initial pH Shift of Paddy Soils. Agronomy, 12(6), 1304. https://doi.org/10.3390/agronomy12061304






