Abstract
Drought severely limits the growth and development of oat (Avena sativa) seedlings. As an osmotic regulator simulating a drought environment, Polyethylene glycol (PEG) has been widely linked in response to plant drought tolerance. However, the underlying mechanism of oats’ response to PEG stress is still largely unknown. Here, we investigated the physiological and transcriptome variables of the drought-resistant oat variety DA92-2F6, and the drought-susceptible variety Longyan 3 under 15% PEG-6000 drought stress to better understand the underlying drought tolerance molecular mechanisms. The physiological results showed that except for the cell membrane permeability, the antioxidant enzyme, osmotic adjustment substance, and photosynthetic efficiency were significantly higher in the DA92-2F6 after 7 d stress. Further, 12 cDNA libraries and 123,223 unigenes were obtained by RNA-seq. A total of 33,857 differentially expressed genes (DEGs) were detected, of which two co-upregulated and three co-downregulated in four comparisons. We highlighted an analysis of the DEGs in phytohormone signal transduction pathway. The auxin, cytokinin, and brassinosteroid signaling pathways, were suppressed in Longyan 3, while abscisic acid and jasmonic acid signaling pathways were mainly activated in DA92-2F6 under drought stress. The upregulated of PP2C, ABF, SNRK2, GID1, JAZ, and MYC2 genes may enhance the drought tolerance of DA92-2F6. Taken together, these results provided a new transcript resource for the drought tolerance improvement and a reference for oat drought resistance molecular breeding.
1. Introduction
In recent years, environmental problems such as high temperature and drought have become severe with global warming, and have become the principal abiotic stresses limiting plant growth, development, and nutritional quality. Of these problems, the loss of yield caused by drought exceeds the sum of other abiotic stresses [1,2,3]. Drought stress causes plant stomatal closure and chlorophyll degradation, resulting in decreased transpiration rate and photosynthetic efficiency [4]. In addition, the imbalance of plant osmoregulation caused by drought stress leads to the production of malondialdehyde (MDA), which disrupts the structure and function of the membrane, and leads to plant death in severe cases [5]. Plants have produced a series of strategies to resist or adapt to drought stress in the long process of evolution and adaptation to the environment. Sugar, amino acids, and inorganic ions in plants accumulate continuously with the decrease of osmotic potential under drought stress. Meanwhile, the antioxidant enzyme system was activated under drought stress to maintain cell function and inhibit reactive oxygen species (ROS) accumulation [6,7]. Besides, phytohormone also plays a vital role in plant abiotic stress resistance. Under drought stress, the change of abscisic (ABA) concentration significantly affects plant growth and development, stomatal opening and closing, photosynthetic efficiency, and related gene expression [8]. In addition, the impact of drought stress on plants was also related to cell wall hardening, biomass allocation, water use efficiency, stress time, and intensity [9,10,11]. However, the physiological and molecular mechanisms of different plant species to cope with water deficit were different, and the existing regulatory mechanisms of most plants were not sufficient to resist severe and long-term drought stress.
Oat (Avena sativa) is an annual high-quality herbaceous crop in the family Poaceae, widely grown in northwest, southwest, and north China [12,13]. Oat can be used as both human-edible and high-quality forage due to its advantages in cold resistance, wide adaptability, and high nutritional quality. The planting area of this species has also been further expanded in recent years [14]. However, in the current era, the shortage of water resources has become a critical restricting factor for the sustainable development of the social economy in north China. Drought has also caused a severe impact on the oat industry. Understanding oat drought resistance mechanisms is required for cultivation of drought-resistant varieties and high-quality forage production.
Polyethylene glycol (PEG), as a macromolecular compound, cannot be absorbed by plants when its molecular weight is greater than 4000. Due to its high solubility in water, it can decrease water potential in solution, thus simulating a drought stress environment [15]. The physiological changes of oat were studied using PEG to simulate a drought environment. Ultra-dried (4% moisture content) oat seeds were treated with −1.2 MPa PEG solutions for 12 h. The results showed that the seed vigor, mitochondrial antioxidant enzyme activity, and mitochondrial ultrastructure were significantly improved compared with the control plant. At the same time, the mitochondria hydrogen peroxide and malondialdehyde contents were significantly decreased (p < 0.05) [16]. Gao et al. [7] found that exogenously applied 100 μM melatonin could decrease the content of hydrogen peroxide and superoxide anion, and increase the activities of superoxide dismutase, peroxidase, catalase, and ascorbate peroxidase in oat seedlings under 20% PEG-6000 stress.
Transcriptome sequencing could comprehensively and rapidly obtain almost all transcript information of a species during a specific period, with the advantages of wide coverage, high resolution, and identification reliability. It has been more and more applied in the drought resistance research in the family Poaceae in recent years. Lata et al. [17] performed transcriptome sequencing for the seedlings of drought-tolerant foxtail millet (Setaria italica L.) variety under PEG stress for 0.5 h and 6 h, respectively. The results showed that the transcript function involved in both stress periods was related to metabolism, stress, signaling, transcription regulation, translation, and proteolysis. Wu et al. [18] conducted transcriptome sequencing for buckwheat (Fagopyrum esculentum) seedlings under well-watered and PEG-mediated drought stress, respectively. They found that 1329 differentially expressed genes (DEGs) were mainly enriched in plant hormone signal transduction, phenylpropanoid biosynthesis, photosynthesis, and carbon metabolism pathways. After PEG-8000 drought stress, the roots of barley (Hordeum vulgare) were subjected to transcriptome sequencing, and the results showed no or only weak effects of osmotic stress on aquaporin expression. Combined with the results of anatomical and physiological experiments, the sealed apoplast significantly reduces the uncontrolled backflow of water from the root to the medium while maintaining constant water flow through the highly regulated cell-to-cell path to keep normal growth and development under drought stress [19].
At present, the research of underlying drought resistance regulation molecular mechanisms of different oat varieties under PEG stress by transcriptome sequencing technology has not been reported. In this study, we analyzed the physiological and molecular responses of different drought-resistant oat varieties to PEG stress, and focused on the DEGs related to the phytohormone signal transduction pathway. The findings provided a reference for future molecular breeding for oat drought resistance.
2. Materials and Methods
2.1. Plant Material and Experimental Treatments
The experiment was conducted in the greenhouse of the College of Pratacultural Science, Gansu Agricultural University (Lanzhou, China) in 2019. Drought-resistant variety DA92-2F6 and drought-susceptible variety Longyan 3 were selected as experimental materials provided by the College of Pratacultural, Gansu Agricultural University. After disinfection and accelerating germination, oat seeds with consistent germination were sowed in plastic pots (10 cm in diameter and 16 cm in depth) containing fine sand and 15 seeds per pot which were watered and placed in plastic boxes (35 × 25 × 15 cm3) (six pots per box), cultured in a greenhouse with a day and night temperature 25 ± 1 °C/18 ± 1 °C, relative humidity 65–75%, and light intensity 500–700 μmol m−2 s−1. Seedling thinning was carried out when the third leaves emerged, and 10 consistent growth plants were kept per pot. Oat seedlings were irrigated with 1 L Hoagland nutrient solution containing or without 15% PEG-6000 every two days per box for experiment treatment. The first day without irrigating was recorded as 0 d. After treatment 7 and 14 d, the photosynthetic parameters were measured using LI-6800 photosynthesis measurement system (LI-COR, Lincoln, NE, USA), and the water use efficiency was also calculated [20]. Meanwhile, leaf samples from each plant were collected for physiological measurements and transcriptome sequencing. The leaf samples were stored at −80 °C, and three biological replicates for each treatment were performed.
2.2. Physiological Measurements
2.2.1. Antioxidant Characteristics
Catalase (CAT) activity was determined by ultraviolet absorption method [21]; superoxide dismutase (SOD) activity was determined by nitro blue tetrazolium colorimetry method [21]; peroxidase (POD) activity was determined by guaiacol colorimetry method [22].
2.2.2. Osmotic Adjustment Substance
Proline (Pro) content was determined by ninhydrin colorimetry method [23]; soluble sugar content was determined by anthrone colorimetry method [24]; soluble protein content was determined by Coomassie brilliant blue method [25].
2.2.3. Cell Membrane Permeability
Relative electrical conductivity (REC) was determined by the conductivity method [26]; MDA content was determined by the thiobarbituric acid method [27].
2.2.4. Photosynthetic Characteristics
Chlorophyll content was determined by acetone extraction method [28]; water use efficiency = photosynthetic rate/transpiration rate.
2.3. Total RNA Extraction, Library Construction and RNA-Seq
Total RNA of the seedling leaves from two varieties after PEG stress 0 d (CKD2: DA92-2F6, CKL3: Longyan 3) and 7 d (DD2: DA92-2F6, DL3: Longyan 3) were collected using the Tiangen RNA Plant Plus Reagent (Tiangen, Beijing, China) according to the manufacturer’s guidelines. The RNA purity and integrity were analyzed using a Nanophotometer spectrophotometer (Implen, Westlake Village, CA, USA) and an Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). Magnetic beads enriched the mRNA with poly-A tail with OligodT in 0.5 mg of total RNA, and the obtained mRNA was then broken into small fragments by interrupt buffer. Random N6 primers were used for reverse transcription to synthesize the first-strand cDNA. DNA polymerase I and RNaseH were used to synthesize the second-strand cDNA. The synthesized double-stranded cDNA ends were trimmed and phosphorylated at the 5′ end, forming a prominent ′A′ sticky end and connecting a sequencing adaptor with a prominent ′T′ at the 3′ end. Sequencing adaptors were linked to the purified cDNA, and 12 double-strand libraries were obtained by PCR amplification. Paired-end PE150 bp sequencing was performed using the BGISEQ-500 RS sequencing platform at The Beijing Genomics Institute. The raw sequence data have been deposited in the Genome Sequence Archive (CRA005764) at the Beijing Institute of Genomics (BIG) Data Center, Chinese Academy of Sciences. Available online: http://bigd.big.ac.cn/gsa (accessed on 4 January 2022).
2.4. De Novo Transcriptome Assembly
To improve the reliability of the sequencing data, raw data were filtered with SOAPnuke software [29] according to the following rules: (1) remove reads containing the sequencing adapter; (2) remove reads with >5% ambiguous N nucleotides; (3) remove reads with low quality (Q20 > 20%). Clean reads after filter were saved in FASTQ format and aligned to the reference gene sequences with Bowtie 2 software [30]. De novo assembly of the clean reads was performed using Trinity [31] under default parameters, and then Tgicl [32] was used to cluster transcripts to obtain unigenes.
2.5. Unigene Functional Annotation and Classification
Unigenes were used as query sequences against the KEGG (Kyoto Encyclopedia of Genes and Genomes); GO (Gene Ontology); NR (Non-Redundant Protein Sequence Database); NT; Swiss-Prot; Pfam; KOG, and TF databases. Annotations of the best hits were recorded.
2.6. Differentially Expressed Unigene Identification
The expression level of each unigene was calculated and normalized as Fragments Per Kilobase Million (FPKM) values using RSEM software [33]. Identification of DEGs was based on the negative binomial distribution of the DEseq2 package [34,35]. The cut-off for DEGs was fold change ≥1 and adjusted p value ≤ 0.001. DEGs of four comparisons were analyzed, including CKD2/DD2, CKL3/DL3, CKD2/CKL3, and DD2/DL3.
2.7. Real-Time Quantitative PCR Analysis
A total of 20 candidate DEGs involved in phytohormone biosynthesis were selected for qRT-PCR validation. Specific primer pairs for the selected genes were designed and are listed in Table S1. The cDNA was transcribed from 5 μg total RNA using the SuperScript II system (Invitrogen, Shanghai, China) according to the manufacturer’s instructions. The qRT-PCR was carried out with SYBR Premix Ex-Taq (TaKaRa, Dalian, China) on an ABI QuantStudio 7 Flex RT-PCR instrument (Applied Biosystems, Waltham, MA, USA), with reaction volumes of 10 μL that contained 1 μL cDNA, 0.5 μL 2 mM gene-specific primers, 0.5 μL ROX Reference Dye (50×), 5 μL 2 × SYBR Premix Ex-Taq, and 2.5 μL ddH2O. The relative expression level of the selected genes was presented as the fold-change calculated using the −2ΔΔCT method, as previously described [36].
2.8. Statistical Analysis
The data of the physiological parameters were collected by Microsoft Excel. Statistical analyses were performed using a one-way ANOVA followed by Duncan’s tests in SPSS version 25.0 (SPSS, Chicago, IL, USA) [37], and were presented as the means ± standard deviation (SD) from three independent biological experiments (per experiment included 10 plants). Statistical results were shown by Origin software [38].
3. Results
3.1. Physiological Changes under Drought Stress
During the drought stress period, an increase followed by a decline in the CAT, SOD, and POD contents was measured in the DA92-2F6 leaves. However, three antioxidant enzymes in the Longyan 3 leaves showed a continuous upward trend (Figure 1a–c). After being in drought stress for 7 d, the antioxidant enzyme content of DA92-2F6 leaves reached a peak. It was significantly higher than that of Longyan 3 (p < 0.01), indicating stronger stress resistance (Figure 1a–c).

Figure 1.
Physiological changes of Avena sativa seedlings with different drought resistance: (a) catalase content; (b) superoxide dismutase content; (c) peroxidase content; (d) soluble sugar content; (e) soluble protein content; (f) proline content; (g) relative electric conductivity; (h) malondialdehyde content; (i) chlorophyll content, and (j) water use efficiency. Values are expressed as mean ± SD. Differences were assessed by ANOVA and denoted as follows: * p < 0.05; ** p < 0.01 as compared between two groups.
The soluble sugar content of the two varieties showed the same trend, and peaked at 7 days before slowly decreasing under drought stress (Figure 1d). However, the soluble sugar content of DA92-2F6 leaves was significantly higher than that of Longyan 3 from 7 to 14 d (p < 0.01) (Figure 1d). Compared to Longyan 3, DA92-2F6 leaves displayed significantly higher soluble protein content at 7 d (p < 0.01) (Figure 1e). There was no significant difference in proline content between the two varieties at 7 d after stress (Figure 1f). From 7–14 d, the proline content of Longyan 3 leaves continued to increase, while DA92-2F6 had no significant change, resulting in an extremely significant difference at 14 d (p < 0.01) (Figure 1f).
MDA content and relative conductivity were both related to cell membrane permeability. Those two indicators showed an increase followed by a decline in Longyan 3 leaves under drought stress, and were significantly higher than DA92-2F6 leaves at 7 d (p < 0.01) (Figure 1g,h). The MDA content continued to increase under drought stress from 0-14 d, while the relative conductivity decreased slowly when it rapidly increased to 7 d in DA92-2F6 leaves (Figure 1g,h).
Chlorophyll content and WUE showed the same trend under drought stress in two varieties from 0–14 d (Figure 1i,j). However, extremely significant differences at three time points between the two varieties were observed, and DA92-2F6 leaves showed better photosynthetic characteristics, which further proved the difference in drought resistance between the two varieties (p < 0.01) (Figure 1i,j).
3.2. Assembly of RNA-Seq and De Novo Transcriptomes
Comparative transcriptomic sequencing of the oat leaves was performed to reveal the molecular mechanism and identification of the key genes of different drought resistance oat seedlings in response to PEG stress. After being in drought stress for 0 and 7 d, DA92-2F6 and Longyan 3 leaves were sampled, and 12 cDNA libraries were constructed using the BGISEQ-500 sequencing platform; 548.6 M raw data were generated from all samples (Table S2). After filtering reads, a total of 510.36 M clean reads and 76.54 Gb clean bases were obtained (Table S2). After assembling and clustering clean reads, a total number of 123,223 unigenes with a total length of 162,582,089 bp and an average GC content of 48.51% were generated (Figure S1, Table S3). Principal component analysis was performed on the three biological replicates of each sample based on gene expression level, and the results also proved the accuracy of the transcriptome sequencing (Figure S2).
3.3. Unigene Functional Annotation, Classification, and TF Prediction
All unigenes were BLASTed against the seven public databases to functionally annotate the oat transcriptome. A total of 85,581 (69.45%) unigenes provided significant blast hits in the NR database (Table 1), of which the closest species matched to unigene was Aegilops tauschii subsp. Tauschii, followed by Brachypodium distachyon, Hordeum vulgare subsp. Vulgare, Triticum aestivum, and Triticum Urartu (Figure S3). In addition, 75,713 (61.44%), 62,215 (50.49%), 66,782 (54.20%), 62,777 (50.95%), 62,825 (50.98%), and 65,800 (53.40%) unigenes were annotated in the NT, Swiss-Prot, KEGG, KOG, Pfam, and GO databases, respectively (Table 1). The number of unigenes that provided significant blast hits in at least one database and either database was 91,457 and 37,353, respectively (Table 1).

Table 1.
Summary of annotations for the assembled Avena sativa unigenes in public databases.
Gene ontology is an international standard database to describe the function of genes at the molecular, cellular, and organizational levels which were also used to classify the function of the predicted oat unigenes. The most common assignments in the biological process category were cellular processes (19,297, 15.66%). The most common assignments in the cellular components were cell (25,172, 20.43%), followed by membrane part (17,939, 14.56%). The most common assignments in the molecular function were binding (34,345, 27.88%), followed by catalytic activity (30,946, 25.11%) (Figure S4). The KEGG database can analyze the links between the genomes, biological pathways, and chemicals. To identify the biological pathways activated in oat under drought stress, we mapped the unigenes to the canonical reference pathway in the KEGG database. The most enriched secondary biological pathways under the five classification systems were transport and catabolism (2944, 2.39%), signal transduction (3882, 3.15%), translation (7080, 5.75%), global and overview maps (14,592, 11.84%), and environmental adaptation (2955, 2.40%), respectively (Figure S5).
A total of 2436 unigenes belonging to 55 transcription factor families were detected after predicting the unigenes with the ability to encode transcription factors. The MYB family contained the most unigenes, 294, followed by the bHLH family (239) and AP2-EREBP family (194), and the least were ULT family (one) (Figure S6). These results showed that a large number of biological pathways and transcription factors were activated under drought stress.
3.4. Analysis of Differentially Expressed Genes
To investigate the difference in gene expression between oat varieties with different drought resistance, the gene expression of all unigenes was calculated by the RSEM software package. A total of 33,857 DEGs were detected in four comparisons, of which 15,442 were upregulated, and 18,415 were downregulated (Figure 2a). In the CKD2/DD2 comparison, we found 186 upregulated and 205 downregulated DEGs, whereas 365 were upregulated and 804 were downregulated in the CKL3/DL3 comparison, indicating that drought stress had a greater effect on Longyan 3 (Figure 2a). In addition, 12,056 and 20,241 DEGs were detected in CKD2/CKL3 and DD2/DL3 comparison, respectively, which indicated that PEG stress could induce more genes to cope with drought stress (Figure 2a). The overlapped DEGs among the different comparisons were also analyzed. Two co-upregulated and three co-downregulated genes were detected in the four comparisons, respectively. These genes may be the key genes that caused the drought resistance difference between the two varieties (Figure 2b,c). A total of 20 phytohormone synthesis-related DEGs were selected for qRT-PCR verification, and the results showed the high accuracy and reproducibility of our RNA-seq data (Figure S7).
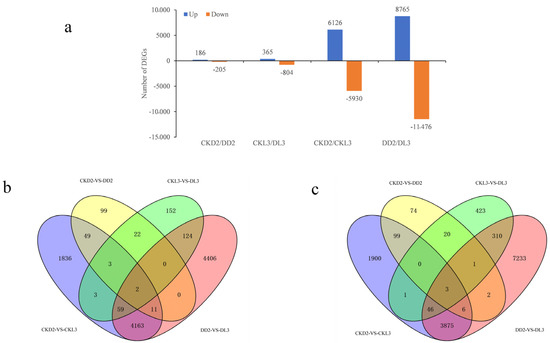
Figure 2.
Analysis of differentially expressed genes (DEGs): (a) DEGs in four comparisons; (b) Overlapping upregulated DEGs in four comparisons; (c) Overlapping downregulated DEGs in four comparisons.
GO enrichment analyses were performed on the DEGs in different comparisons. The number of GO terms assigned into the four comparisons were 39 (CKD2/DD2) (Figure 3a), 47 (CKL3/DL3) (Figure 3b), 52 (CKD2/CKL3) (Figure 3c), and 54 (DD2/DL3) (Figure 3d), respectively. The most enriched GO terms under the three main GO classification systems were the cellular process, cell, and binding in the CKD2/CKL3, CKD2/DD2, and DD2/DL3 comparisons, while in CKL3/DL3 were cellular process, cell, and catalytic activity. The enrichment of DEGs in different GO terms clearly indicated the molecular and cellular events in different oat varieties under drought stress. A KEGG enrichment analysis of DEGs was performed to reveal further the changes in the biological pathways of different oat varieties in response to drought stress. The results showed that the main enriched pathways in CKD2/DD2 comparison were phenylpropanoid, MAPK signaling pathway-plant, and plant hormone signal transduction (Figure 4a). CKL3/DL3 comparisons were plant hormone signal transduction, pyruvate metabolism, and carbon fixation in photosynthetic organisms (Figure 4b). CKD2/CKL3 comparisons were mRNA monitoring pathway, RNA transport, and phenylpropanoid biosynthesis (Figure 4c). DD2/DL3 comparisons were glyoxylate and dicarboxylate metabolism, carbon fixation in photosynthesis organisms, and RNA transport (Figure 4d). The above-enriched pathways may be essential for oat survival under drought stress.
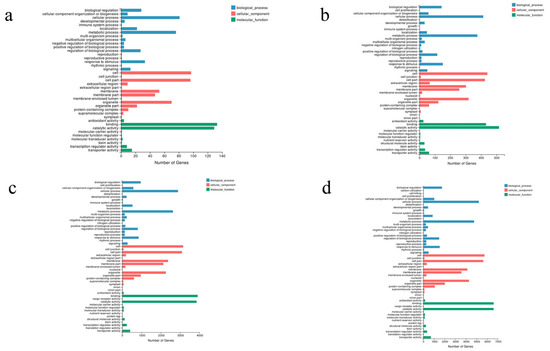
Figure 3.
Gene ontology (GO) classifications of differential expression genes in four comparisons. Results are summarized in three main GO categories: biological process, cellular component, and molecular function: (a) CKD2/DD2; (b) CKL3/DL3; (c) CKD2/CKL3, and (d) DD2/DL3.
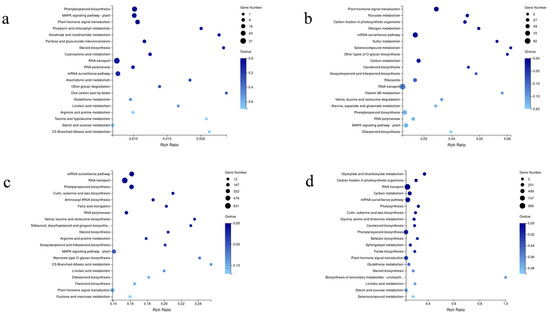
Figure 4.
Summary statistics for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of differential expression genes. The vertical axis shows the pathway, and the horizontal axis shows the enrichment factor. The size of the dot represents the number of DEGs in the pathway. Dot color corresponds to the range of Q-value: (a) CKD2/DD2; (b) CKL3/DL3; (c) CKD2/CKL3, and (d) DD2/DL3.
3.5. Genes Involved in Phytohormone Signal Transduction
Phytohormone signal transduction pathway was enriched in both CKL3/DL3 and CKD2/DD2 comparison, indicating a potentially important role in drought stress response. Therefore, we dissected the profiles of the genes involved in the phytohormone signal transduction pathway to elucidate the possible key regulators and the molecular mechanisms of the drought tolerance difference between the two varieties. In the CKL3/DL3 comparison, 65 DEGs were found, with 45 downregulated and 20 upregulated. While 20 DEGs were identified in the CKD2/DD2 comparison, with 18 upregulated and two downregulated (Figure 5).

Figure 5.
Drought-stress induced changes in phytohormone signaling pathway gene expression profiles in two comparisons: (a) auxin signal; (b) cytokinin signal; (c) gibberellin signal; (d) abscisic acid signal; (e) ethylene signal; (f) salicylic acid; (g) brassinosteroid signal, and (h) jasmonic acid signal.
Four downregulated SAUR (auxin responsive protein) DEGs were identified (three in the CKL3/DL3 comparison and one in the CKD2/DD2 comparison) in the auxin (IAA) signal transduction pathway (Figure 5a).
We highlighted the DEGs in the cytokinin (CTK) signal transduction pathway, 17 DEGs in the A-ARR (A-ARR: Two-component response regulator ARR-A family) pathway and two DEGs in the B-ARR (B-ARR: Two-component response regulator ARR-B family) pathway were downregulated in the CKL3/DL3 comparison. At the same time, there was only one downregulated gene (A-ARR) in the CKD2/DD2 comparison (Figure 5b).
In the gibberellin (GA) signal transduction pathway, one GID1 (gibberellin receptor GID1) gene was upregulated in the CKD2/DD2 comparison. Two DELLA (DELLA protein) genes and four TF (two PIF3 and two PIF4: phytochrome-interacting factor 4) genes were identified with inconsistent expression patterns in the CKL3/DL3 comparison (Figure 5c).
In the CKD2/DD2 comparison, one PP2C (protein phosphatase 2C) gene and one ABF (ABA-responsive element binding factor) gene were upregulated in the abscisic acid (ABA) signal transduction pathway. A total of five SnRK2 (serine/threonine-protein kinase SRK2) genes were identified in the two comparisons, with one upregulated in the CKD2/DD2 comparison and four downregulated in the CKL3/DL3 comparison (Figure 5d).
Upregulated DEGs were consistently detected in the ethylene (ETH) and the salicylic acid (SA) signal transduction pathway. One CTR1 (serine/threonine-protein kinase CTR1) gene, one ERF1/2 (ethylene-responsive transcription factor 1) gene, and one NPR1 (regulatory protein NPR1) gene were upregulated in the CKL3/DL3 comparison (Figure 5e,f). Interestingly, no ETH signal-related DEG was detected in the CKD2/DD2 comparison (Figure 5e).
No DEG was identified in the CKD2/DD2 comparison, while 15 DEGs identified in the CKL3/DL3 comparison were downregulated, including the BRI1 (protein brassinosteroid insensitive 1) gene and TCH4 (xyloglucan: xyloglucosyl transferase TCH4) gene in the brassinosteroid (BR) signal transduction pathway (Figure 5g).
In the jasmonic acid (JA) signal transduction pathway, eight and seven JAZ- (jasmonate ZIM domain-containing protein) related DEGs were identified in the CKL3/DL3 comparison and the CKD2/DD2 comparison, respectively. Four same JAZ-related DEGs were identified in both comparisons, but the CKD2/DD2 comparison expression was higher than those in the CKL3/DL3 comparison (Figure 5h). Seven MYC2-related DEGs (transcription factor MYC2) were identified in both comparisons and were upregulated except for CL1282.Contig3. All six same MYC2-related DEGs were identified in both comparisons, while the CKD2/DD2 comparison expression was also higher than that in the CKL3/DL3 comparison (Figure 5h).
4. Discussion
Oat is an annual herbaceous forage crop mainly cultivated in northwest China. Drought severely limits its growth, development, and productivity [12]. The differences in physiological responses of different drought-resistant oat varieties under drought stress were analyzed in our study. The results showed that the antioxidant enzyme activities of drought-resistant varieties DA92-2F6 were significantly higher than those in Longyan 3 after 7 d stress, indicating that DA92-2F6 had better stress tolerance under moderate stress (Figure 1a–c). In addition, some studies have shown that the oat Asmap1 and Aspk11 gene and MYB transcription factors in the MAPK signaling pathway were upregulated with the increased antioxidant enzyme activity under drought stress. The expression or activation of these genes or pathways may be the main factor for the difference in antioxidant enzyme activity between the two varieties [7]. Soluble sugar and soluble protein showed a significant difference between the two varieties after 7 d stress (Figure 1d,e), which was consistent with Du’s results [39], and may be related to the activation of the carbohydrate metabolism pathway under drought stress. Significant differences were identified in chlorophyll content and WUE between the two varieties at all time points, and DA92-2F6 could maintain higher photosynthetic capacity by reducing chlorophyll degradation after stress (Figure 1i,j). Studies have shown that some genes, including chlorophyll enzyme (CHLASE), demagnesium chlorophyll enzyme (PPH), and demagnesium chlorophyll-a-oxygenase (PAO), were involved in chlorophyll degradation and will be stimulated expression after drought stress [40]. In this study, the expression of CL11666. Contig3_All gene, which encodes chlorophyll enzyme in Longyan 3 was significantly higher than that in DA92-2F6 in CKD2/CKL3 and DD2/DL3 comparisons after drought stress, and may be the key gene causing the difference of photosynthetic capacity and drought resistance between the two varieties.
In recent years, de novo assembly supported by RNA-seq has become an effective tool for functional gene mining, resistance gene identification, and stress mechanism analysis of plants without high-quality reference genomes [41,42,43]. Previous studies on oat drought resistance mainly focused on morphology, physiology, or cell level [44,45,46]. The molecular level was still relatively rare. The lack of genetic information leads to the research on abiotic stress tolerance, and breeding of resistant varieties lags far behind other Gramineae plants. In this study, two oat varieties with different drought resistance were identified through physiological indicators. A total of 33,857 DEGs were found in the four comparisons, and the number of upregulated genes (15,442) was less than that of downregulated (18,415). The same results were also observed in CKD2/DD2 and CKL3/DL3 comparisons (Figure 2a), consistent with the research in Aegilops tauschii, Giant Juncao, and pearl millet [47,48,49]. Combined with the KEGG enrichment results of DEGs, we found only the phytohormone signal transduction pathway was significantly enriched in CKD2/DD2 and CKL3/DL3 comparisons (Figure 4a,b), indicating that the normal life activities of oat plants may be maintained by downregulating more phytohormone-related genes under moderate drought stress. In addition, more DEGs were detected in CKL3/DL3 comparison compared with CKD2/DD2, indicating that drought stress had a greater effect on Longyan 3 (Figure 2a). The four comparisons detected two co-upregulated and three co-downregulated DEGs (Figure 2b,c). Among the co-upregulated genes, CL1579.Contig5_All was annotated as transcription factor bHLH35-like protein. Studies have shown that OsbHLH148 regulates rice drought tolerance by participating in the jasmonic acid signaling pathway [50]. CL13605.Contig4_All was annotated as the BURP protein family. The family name was derived from the initials of BNM2, USP, RD22, and PGIβ proteins, which were representative subfamily members with the same domain. It also plays a key role in plant growth, development, and stress response [51]. Among the co-downregulated genes, Unigene17147_All was annotated as auxin responsive protein of SAUR family protein, and Unigene26661_All was annotated as peroxidase. These co-DEGs may play an important role in oat response to drought stress.
Phytohormones are simple small molecular compounds, but they can cause complex and diverse physiological effects, and induce stress response gene expression by triggering a series of signal events [52]. It is recognized that ABA can improve plants’ drought resistance and salt tolerance [53]. SnRK2 (sucrose non-fermenting1-related protein kinase) is a kind of Ser/Thr protein kinase that plays an important role in stress resistance physiology. The interaction between upstream PYR/PYL (ABA receptor) and PP2C can reduce the inhibitory effect on SnRK2 [54]. Previous reports showed that overexpression wheat TaSnRK2.4 gene could significantly increase Arabidopsis drought resistance compared with the control plant [55]. In our study, an upregulated PP2C gene (CL3379. Contig7_All) (Figure 5d) was detected in the CKD2/DD2 comparison. The upregulated expression of this gene may promote the SnRK2 gene (CL10999. Contig3_All), thereby enhancing the drought resistance of DA92-2F6.
IAA was generally considered a negative regulator of plant drought resistance [56]. SAUR is an early auxin response gene family. In rice, overexpression of OsSAUR39 inhibits the growth of stems and roots. It increases the contents of anthocyanin, abscisic acid, sugar, and starch, indicating that its stress resistance was improved, and this gene plays a negative regulatory in auxin synthesis and transport [57]. In our study, three downregulated SAUR genes (Figure 5a) were found in CKL3/DL3 comparison, indicating that the downregulation of SAUR genes may lead to the decrease of drought resistance of Longyan 3 under drought stress.
CTK and IAA have a synergistic effect to regulate the growth and development of plant cells jointly [58]. In the cytokinin signal transduction pathway, A-ARR and B-ARR are response-regulated proteins in Arabidopsis thaliana, and the latter can induce the former gene expression. Studies have shown that overexpression of ARR5 can induce ABA sensitivity and drought resistance [59]. Our study found a large number of downregulated Arabidopsis response regulator proteins in the CKL3/DL3 comparison, and these genes may lead to the decrease of drought resistance in Longyan 3 (Figure 5b).
It was generally believed that GA has an antagonistic effect on ABA. Drought stress leads to the upregulation of the GA nuclear receptor GID1 in the CKD2/DD2 comparison, which was consistent with the findings of Du et al. [60], and may have a positive regulatory effect on the drought resistance of DA92-2F6. However, two DELLA proteins and four TF genes with inconsistent expression patterns were detected in the CKL3/DL3 comparison, and their specific regulatory mechanisms still need further study (Figure 5c).
Drought stress can induce the expression of the ERF gene in the ethylene pathway, resulting in an increase in drought sensitivity [61]. In this study, an upregulated ERF gene was detected in the CKL3/DL3 comparison, which may lead to the decreased drought resistance of Longyan 3 (Figure 5e). An upregulated CTR1 gene was also detected in the CKL3/DL3 comparison. Studies have shown that the CTR1 gene plays a negative regulatory role in the ethylene signal transduction pathway. The upregulation of this gene may lead to the reduction of ethylene synthesis and thus affect plant drought resistance [62].
NPRI1 gene is a key regulator of salicylic acid-mediated disease resistance response. It was also involved in various pathways such as the auxin signaling pathway, jasmonic acid signaling pathway, ethylene response, and auxin metabolism. Studies have shown that OsNPR1 functions as a positive regulator of rice cell death, and OsCUL3a’s interaction with OsNPR1 could promote the degradation of OsNPR1 through the 26S proteasome [63]. In our study, the upregulation of the NPR1 gene in the CKL3/DL3 comparison may decrease the drought resistance of Longyan 3 by accelerating the cell death process (Figure 5f).
Studies have shown that both JA- and SA-mediated signaling pathways were closely related to plant resistance [62]. JAZ and MYC2 were two important gene families in the JA signal transduction pathway, and studies have shown that they mainly function by interacting with EIN3/EIN1 in the ethylene signaling pathway. Treatment of Arabidopsis thaliana with JA can induce the expression of JA-responsive genes, promote the development of root hairs, and inhibit root elongation. However, the above phenomena were inhibited in EIN3/EIN1 double mutants, indicating that EIN3/EIL1 plays a positive regulatory role in JA responses [64]. The main transcriptional repressor JAZ in the JA signaling pathway could interact with EIN3/EIL1 to inhibit its transcriptional activity. In addition, studies have shown that EIN3 and MYC2 have a mutual inhibitory effect, and upregulation of MYC2 can enhance JA-mediated plant resistance to herbivores [65,66]. Both JAZ and MYC2 were repressors of EIN3 transcriptional activity. In our study, JAZ and MYC2 genes’ upregulation was observed in CKL3/DL3 and CKD2/DD2 comparison, but the upregulation was higher in DA92-2F6 (Figure 5h). It was speculated that DA92-2F6 increased drought resistance through higher upregulation of JAZ and MYC2 genes under drought stress.
BRI1 is a cell membrane surface receptor kinase and a receptor for BR signaling. It could bind to the co-receptor BAK1 to form a heterodimer, and activate the BR pathway by autophosphorylation or mutual phosphorylation [67]. Our study found that a BRI1 gene was downregulated in the CKL3/DL3 comparison under drought stress, which may lead to the decreased drought resistance of Longyan 3 (Figure 5g). Studies have shown that the TCH4 gene family in the BR pathway encodes a xyloglucan endoglycosyltransferase, which was closely related to the formation and degradation of cell walls and also played an important role in plant stress resistance, and its expression behavior was different from other anti-stress genes, as slight stimulation such as leaf touching, shaking, and other behaviors can lead to dramatic changes of their expression [68]. In this study, 14 downregulated TCH4 genes were found in the CKL3/DL3 comparison, which may also be one of the reasons for the decreased drought resistance of Longyan 3 (Figure 5g).
5. Conclusions
In our study, DA92-2F6 was demonstrated to be more tolerant in drought stress compared with Longyan 3. Higher activity of the antioxidase enzyme (SOD, POD, and CAT), fewer destroyed of cell membrane permeability (REC and MDA), more accumulation of osmotic adjustment substance (soluble sugar and soluble protein), and better photosynthetic characteristics (chlorophyll content and water use efficiency) were found in the tolerance variety DA92-2F6 under 7 d drought stress. Further, the first comprehensive transcriptome analysis and identification of DEGs in Avena sativa under drought stress were conducted; 123,223 unigenes and 33,857 differentially expressed genes (DEGs) were obtained by RNA-seq. These DEGs were mainly involved in plant hormone signal transduction, pyruvate metabolism and carbon fixation in photosynthetic organisms, and MAPK signaling pathway-plant. In addition, we identified the expression profile of the phytohormone signaling transduction pathways in oat seedlings with different drought tolerance under drought stress. Notably, several well-characterized phytohormone signaling pathways, for example, the IAA, CTK, and BR signaling pathways, were suppressed in Longyan 3, while ABA and JA signaling pathways were mainly activated in DA92-2F6 under drought stress. We speculated that DA92-2F6 may enhance drought tolerance under drought stress through mediating the expression of PP2C, ABF, SNRK2, GID1, JAZ, and MYC2 genes in the phytohormone signaling pathways. In summary, our findings provided new insights for drought tolerance molecular mechanisms research, and abundant transcript information for molecular breeding of oat.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/agronomy12051005/s1, Figure S1: Length distribution of unigenes, Figure S2: Principal component analysis among the 12 samples of Avena sativa, Figure S3: Similarities of Avena sativa to other species, Figure S4: Annotation of the assembled unigenes in Avena sativa transcriptome in GO database, Figure S5: Annotation of the assembled unigenes in Avena sativa transcriptome in KEGG database, Figure S6: Annotation of the assembled unigenes in Avena sativa transcriptome in TF database, Figure S7: The relative expression levels of 20 DEGs identified in the comparison between RNA-Seq and qRT-PCR, Table S1: Specific primer pairs for qRT-PCR, Table S2: Overview of the RNA-sequencing reads generated from each sample, Table S3: Unigene statistics.
Author Contributions
Conceptualization, G.Z., W.G. and Z.J.; methodology, W.G. and Z.J.; investigation, W.G., Z.J., D.L. and W.S.; data curation, W.G. and Z.J.; writing—original draft preparation, W.G.; writing—review and editing, G.Z., W.G., X.Z. and J.C.; project administration, G.Z.; funding acquisition, G.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Key Laboratory of Superior Forage Germplasm in the Qinghai-Tibetan Plateau, grant number 2020-ZJ-Y03, and the Science and Technology Program of Gansu Province, grant number 19ZD2NA002.
Data Availability Statement
All the sequence data used in the study have been deposited in the Genome Sequence Archive (CRA005764) in Beijing Institute of Genomics (BIG) Data Center, Chinese Academy of Sciences. Available online: http://bigd.big.ac.cn/gsa (accessed on 4 January 2022). The other data generated in the study were included in this published article and its additional files.
Acknowledgments
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sallam, A.; Alqudah, A.M.; Dawood, M.F.A.; Baenziger, P.S.; Börner, A. Drought stress tolerance in wheat and barley: Advances in physiology, breeding and genetics research. Int. J. Mol. Sci. 2019, 20, 3137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, F.; Kuromori, T.; Sato, H.; Shinozaki, K. Regulatory Gene Networks in Drought Stress Responses and Resistance in Plants. Adv. Exp. Med. Biol. 2018, 1081, 189–214. [Google Scholar] [CrossRef] [PubMed]
- Ahkami, A.H.; Wang, W.; Wietsma, T.W.; Winkler, T.; Lange, I.; Jansson, C.; Lange, B.M.; McDowell, N.G. Metabolic shifts associated with drought-induced senescence in Brachypodium. Plant Sci. 2019, 289, 110278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xie, Z.; Wang, L.; Li, M.; Lang, D.; Zhang, X. Silicon alleviates salt and drought stress of Glycyrrhiza uralensis seedling by altering antioxidant metabolism and osmotic adjustment. J. Plant Res. 2017, 130, 611–624. [Google Scholar] [CrossRef] [PubMed]
- Torun, H. Time-course analysis of salicylic acid effects on ROS regulation and antioxidant defense in roots of hulled and hulless barley under combined stress of drought, heat and salinity. Physiol. Plant. 2018, 165, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Zhang, Y.; Feng, Z.; Bai, Q.; He, J.; Wang, Y. Effects of Melatonin on Antioxidant Capacity in Naked Oat Seedlings under Drought Stress. Molecules 2018, 23, 1580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef]
- Hughes, N.M.; Reinhardt, K.; Feild, T.S.; Gerardi, A.R.; Smith, W.K. Association between winter anthocyanin production and drought stress in angiosperm evergreen species. J. Exp. Bot. 2010, 61, 1699–1709. [Google Scholar] [CrossRef] [Green Version]
- Eziz, A.; Yan, Z.; Tian, D.; Han, W.; Tang, Z.; Fang, J. Drought effect on plant biomass allocation: A meta-analysis. Ecol. Evol. 2017, 7, 11002–11010. [Google Scholar] [CrossRef]
- Ma, Y.; Dias, M.C.; Freitas, H. Drought and Salinity Stress Responses and Microbe-Induced Tolerance in Plants. Front. Plant Sci. 2020, 11, 591911. [Google Scholar] [CrossRef] [PubMed]
- Raguindin, P.F.; Itodo, O.A.; Stoyanov, J.; Dejanovic, G.M.; Gamba, M.; Asllanaj, E.; Minder, B.; Bussler, W.; Metzger, B.; Muka, T.; et al. A systematic review of phytochemicals in oat and buckwheat. Food Chem. 2020, 338, 127982. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.Q.; Ju, Z.L.; Chai, J.K.; Jiao, T.; Jia, Z.F.; Casper, D.P.; Zeng, L.; Wu, J.P. Effects of silage additives and varieties on fermentation quality, aerobic stability, and nutritive value of oat silage. J. Anim. Sci. 2018, 96, 3151–3160. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Guo, G.; Yuan, X.J.; Zhang, J.; Wen, A.Y.; Sun, X.H.; Shao, T. Effect of ensiling whole crop oat with lucerne in different ratios on fermentation quality, aerobic stability and in vitro digestibility on the Tibetan plateau. J. Anim. Physiol. Anim. Nutr. 2017, 101, e144–e153. [Google Scholar] [CrossRef]
- Zeid, I.; Shedeed, Z. Alterations in Nitrogen Metabolites after Putrescine Treatment in Alfalfa under Drought Stress. Pak. J. Biol. Sci. 2007, 10, 1513–1518. [Google Scholar] [CrossRef] [Green Version]
- Xia, F.; Wang, M.; Chen, L.; Cheng, H.; Sun, Y.; Li, M.; Dong, K.; Zhao, X.; Mao, P. Responses of mitochondrial ultrastructure and physiological variations to PEG-priming on ultra-dried oat (Avena sativa L.) seeds after ageing. Seed Sci. Technol. 2017, 45, 622–637. [Google Scholar] [CrossRef]
- Lata, C.; Sahu, P.P.; Prasad, M. Comparative transcriptome analysis of differentially expressed genes in foxtail millet (Setaria italica L.) during dehydration stress. Biochem. Biophys. Res. Commun. 2010, 393, 720–727. [Google Scholar] [CrossRef]
- Wu, Q.; Zhao, G.; Bai, X.; Zhao, W.; Xiang, D.; Wan, Y.; Wu, X.; Sun, Y.; Tan, M.; Peng, L. Characterization of the transcriptional profiles in common buckwheat (Fagopyrum esculentum) under PEG-mediated drought stress. Electron. J. Biotechnol. 2019, 39, 42–51. [Google Scholar] [CrossRef]
- Kreszies, T.; Shellakkutti, N.; Osthoff, A.; Yu, P.; Baldauf, J.A.; Zeisler-Diehl, V.V.; Ranathunge, K.; Hochholdinger, F.; Schreiber, L. Osmotic stress enhances suberization of apoplastic barriers in barley seminal roots: Analysis of chemical, transcriptomic and physiological responses. New Phytol. 2018, 221, 180–194. [Google Scholar] [CrossRef] [Green Version]
- Stinziano, J.R.; Morgan, P.B.; Lynch, D.J.; Saathoff, A.J.; McDermitt, D.K.; Hanson, D.T. The rapid A-Ci response: Photosynthesis in the phenomic era. Plant Cell Environ. 2017, 40, 1256–1262. [Google Scholar] [CrossRef] [Green Version]
- Elavarthi, S.; Martin, B. Spectrophotometric assays for antioxidant enzymes in plants. Methods Mol. Biol. 2010, 639, 273–281. [Google Scholar] [PubMed]
- Harauchi, T.; Yoshizaki, T. A fluorimetric guaiacol method for thyroid peroxidase activity. Anal. Biochem. 1982, 126, 278–284. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Wang, Q.; Su, Z.; Zhang, S.; Li, Y. Soluble sugar content of clonal plant Neosinocalamus affinis at module and ramet levels. J. Appl. Ecol. 2004, 15, 1994–1998. [Google Scholar]
- Ahsan, N.; Lee, D.-G.; Lee, S.-H.; Kang, K.Y.; Bahk, J.D.; Choi, M.S.; Lee, I.-J.; Renaut, J.; Lee, B.-H. A comparative proteomic analysis of tomato leaves in response to waterlogging stress. Physiol. Plant. 2007, 131, 555–570. [Google Scholar] [CrossRef]
- Yu, X.; Peng, Y.H.; Zhang, M.H.; Shao, Y.J.; Su, W.A.; Tang, Z.C. Water relations and an expression analysis of plasma membrane intrinsic proteins in sensitive and tolerant rice during chilling and recovery. Cell Res. 2006, 16, 599–608. [Google Scholar] [CrossRef] [Green Version]
- Kumar, G.; Knowles, N.R. Changes in Lipid Peroxidation and Lipolytic and Free-Radical Scavenging Enzyme Activities during Aging and Sprouting of Potato (Solanum tuberosum) Seed-Tubers. Plant Physiol. 1993, 102, 115–124. [Google Scholar] [CrossRef] [Green Version]
- Inskeep, W.P.; Bloom, P.R. Extinction coefficients of chlorophyll a and B in n,n-dimethylformamide and 80% acetone. Plant Physiol. 1985, 77, 483–485. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yuxin, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience 2017, 7, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Langdon, W.B. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min. 2015, 8, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genom. Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genom. Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Yi, D.; Yang, J.; Liu, X.; Pang, Y. Genome-Wide Identification, Expression Analysis and Functional Study of CCT Gene Family in Medicago truncatula. Plants 2020, 9, 513. [Google Scholar] [CrossRef]
- Park, E.; Cho, M.; Ki, C.-S. Correct Use of Repeated Measures Analysis of Variance. Ann. Lab. Med. 2009, 29, 1–9. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Du, Y.; Zhao, Q.; Chen, L.; Yao, X.; Zhang, W.; Zhang, B.; Xie, F. Effect of drought stress on sugar metabolism in leaves and roots of soybean seedlings. Plant Physiol. Biochem. 2019, 146, 1–12. [Google Scholar] [CrossRef]
- Wang, P.; Sun, X.; Li, C.; Wei, Z.; Liang, D.; Ma, F. Long-term exogenous application of melatonin delays drought-induced leaf senescence in apple. J. Pineal Res. 2012, 54, 292–302. [Google Scholar] [CrossRef]
- Zhao, Y.; Wei, X.; Long, Y.; Ji, X. Transcriptional analysis reveals sodium nitroprusside affects alfalfa in response to PEG-induced osmotic stress at germination stage. Protoplasma 2020, 257, 1345–1358. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Hu, Y.; Huo, P.; Zhang, Q.; Chen, X.; Zhang, Z. Transcriptome analysis of hexaploid hulless oat in response to salinity stress. PLoS ONE 2017, 12, e0171451. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Ma, L.; Gong, P.; Liu, X.; Wang, Z.; Zhao, G. Development and application of EST–SSRs markers for analysis of genetic diversity in erect milkvetch (Astragalus adsurgens Pall.). Mol. Biol. Rep. 2018, 46, 1323–1326. [Google Scholar] [CrossRef] [PubMed]
- Punia, S.; Sandhu, K.S.; Dhull, S.B.; Siroha, A.K.; Purewal, S.S.; Kaur, M.; Kidwai, M.K. Oat starch: Physico-chemical, morphological, rheological characteristics and its applications-A review. Int. J. Biol. Macromol. 2020, 154, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Idziak-Helmcke, D.; Warzecha, T.; Sowa, M.; Warchoł, M.; Dziurka, K.; Czyczyło-Mysza, I.; Skrzypek, E. 3-D nucleus architecture in oat × maize addition lines. Int. J. Mol. Sci. 2020, 21, 4280. [Google Scholar] [CrossRef] [PubMed]
- Davies, P.A.; Sidhu, P.K. Oat Doubled Haploids Following Maize Pollination. Methods Mol. Biol. 2017, 1536, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Bai, S.; Li, L.; Han, X.; Li, J.; Zhu, Y.; Fang, Y.; Zhang, D.; Li, S. Comparative Transcriptome Analysis of Two Aegilops tauschii with Contrasting Drought Tolerance by RNA-Seq. Int. J. Mol. Sci. 2020, 21, 3595. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, S.; Shi, W.; David-Schwartz, R.; Li, S.; Yang, F.; Lin, Z. Transcriptome profiling reveals the effects of drought tolerance in Giant Juncao. BMC Plant Biol. 2021, 21, 2. [Google Scholar] [CrossRef]
- Shivhare, R.; Asif, M.H.; Lata, C. Comparative transcriptome analysis reveals the genes and pathways involved in terminal drought tolerance in pearl millet. Plant Mol. Biol. 2020, 103, 639–652. [Google Scholar] [CrossRef]
- Seo, J.-S.; Joo, J.; Kim, M.-J.; Kim, Y.-K.; Nahm, B.H.; Song, S.I.; Cheong, J.-J.; Lee, J.S.; Kim, J.-K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef]
- Wang, L.; Wu, N.; Zhu, Y.; Song, W.; Zhao, X.; Li, Y.; Hu, Y. The divergence and positive selection of the plant-specific BURP -containing protein family. Ecol. Evol. 2015, 5, 5394–5412. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Yu, B.; Wu, Q.; Min, Q.; Zeng, R.; Xie, Z.; Huang, J. OsMADS23 phosphorylated by SAPK9 confers drought and salt tolerance by regulating ABA biosynthesis in rice. PLoS Genet 2021, 17, e1009699. [Google Scholar] [CrossRef] [PubMed]
- Belda-Palazón, B.; Adamo, M.; Valerio, C.; Ferreira, L.J.; Confraria, A.; Reis-Barata, D.; Rodrigues, A.; Meyer, C.; Rodriguez, P.L.; Baena-González, E. A dual function of SnRK2 kinases in the regulation of SnRK1 and plant growth. Nat. Plants 2020, 6, 1345–1353. [Google Scholar] [CrossRef]
- Mao, X.; Zhang, H.; Tian, S.; Chang, X.; Jing, R. TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. J. Exp. Bot. 2009, 61, 683–696. [Google Scholar] [CrossRef] [Green Version]
- Naser, V.; Shani, E. Auxin response under osmotic stress. Plant Mol. Biol. 2016, 91, 661–672. [Google Scholar] [CrossRef]
- Kant, S.; Rothstein, S. Auxin-responsiveSAUR39gene modulates auxin level in rice. Plant Signal. Behav. 2009, 4, 1174–1175. [Google Scholar] [CrossRef] [Green Version]
- Shao, R.; Wang, K.; Shangguan, Z. Cytokinin-induced photosynthetic adaptability of Zea mays L. to drought stress associated with nitric oxide signal: Probed by ESR spectroscopy and fast OJIP fluorescence rise. J. Plant Physiol. 2010, 167, 472–479. [Google Scholar] [CrossRef]
- Kang, N.Y.; Cho, C.; Kim, N.Y.; Kim, J. Cytokinin receptor-dependent and receptor-independent pathways in the dehydration response of Arabidopsis thaliana. J. Plant Physiol. 2012, 169, 1382–1391. [Google Scholar] [CrossRef]
- Du, H.; Chang, Y.; Huang, F.; Xiong, L. GID1 modulates stomatal response and submergence tolerance involving abscisic acid and gibberellic acid signaling in rice. J. Integr. Plant Biol. 2014, 57, 954–968. [Google Scholar] [CrossRef] [Green Version]
- Wan, L.; Zhang, J.; Zhang, H.; Zhang, Z.; Quan, R.; Zhou, S.-R.; Huang, R. Transcriptional Activation of OsDERF1 in OsERF3 and OsAP2-39 Negatively Modulates Ethylene Synthesis and Drought Tolerance in Rice. PLoS ONE 2011, 6, e25216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Ning, Y.; Zhang, Y.; Yu, N.; Zhao, C.; Zhan, X.; Wu, W.; Chen, D.; Wei, X.; Wang, G.-L.; et al. OsCUL3a Negatively Regulates Cell Death and Immunity by Degrading OsNPR1 in Rice. Plant Cell 2017, 29, 345–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; Mu, A.; Jiang, Z.; Kim, J.M.; To, T.K.; Li, W.; et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Huang, H.; Gao, H.; Wang, J.; Wu, D.; Liu, X.; Yang, S.; Zhai, Q.; Li, C.; Qi, T.; et al. Interaction between MYC2 and ETHYLENE INSENSITIVE3 Modulates Antagonism between Jasmonate and Ethylene Signaling in Arabidopsis. Plant Cell 2014, 26, 263–279. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhu, Z.; An, F.; Hao, D.; Li, P.; Song, J.; Yi, C.; Guo, H. Jasmonate-activated MYC2 represses ETHYLENE INSEN-SITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell 2014, 26, 1105–1117. [Google Scholar] [CrossRef] [Green Version]
- Jaillais, Y.; Vert, G. Brassinosteroid signaling and BRI1 dynamics went underground. Curr. Opin. Plant Biol. 2016, 33, 92–100. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Purugganan, M.M.; Polisensky, D.H.; Antosiewicz, D.M.; Fry, S.C.; Braam, J. Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 1995, 7, 1555–1567. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).