Mapping of Quantitative Trait Loci Underlying Nodule Traits in Soybean (Glycine max (L.) Merr.) and Identification of Genes Whose Expression Is Affected by the Sinorhizobium fredii HH103 Effector Proteins NopL and NopT
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, and Plant Material
2.2. Generation of the S. fredii HH103ΩNopLΩNopT Mutant
2.3. Nodulation Tests
2.4. QTL Analysis
2.5. Annotation of Genes in QTL Regions
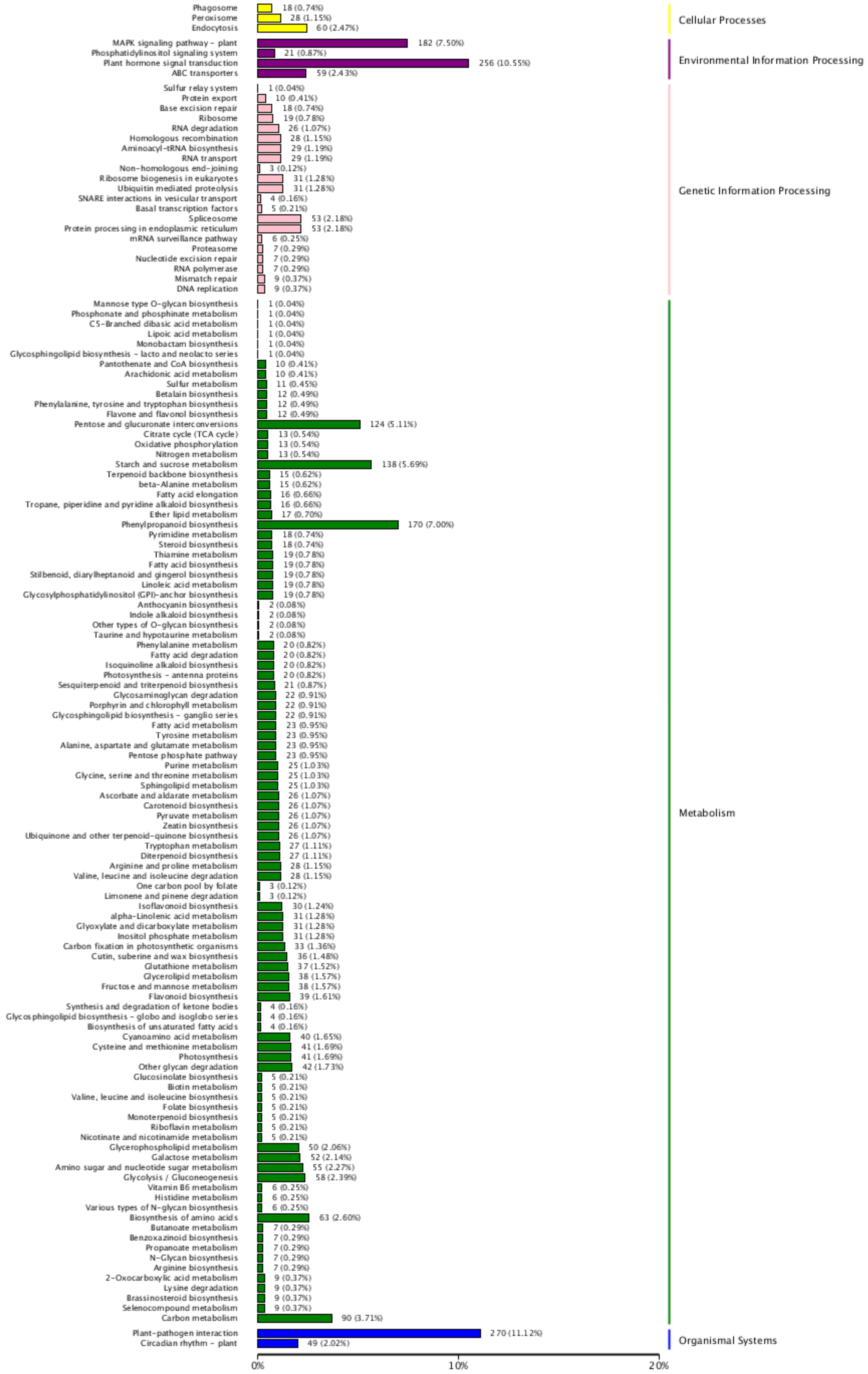
2.6. RNA-Seq KEGG Enrichment Analyses
2.7. RT-qPCR Analysis of Candidate Genes
3. Results
3.1. Effect of NopL and NopT on Various Soybean Germplasms
3.2. Phenotypic Analysis
3.3. QTL Mapping of Nodulation-Related Traits
3.4. Analysis of Candidate Genes
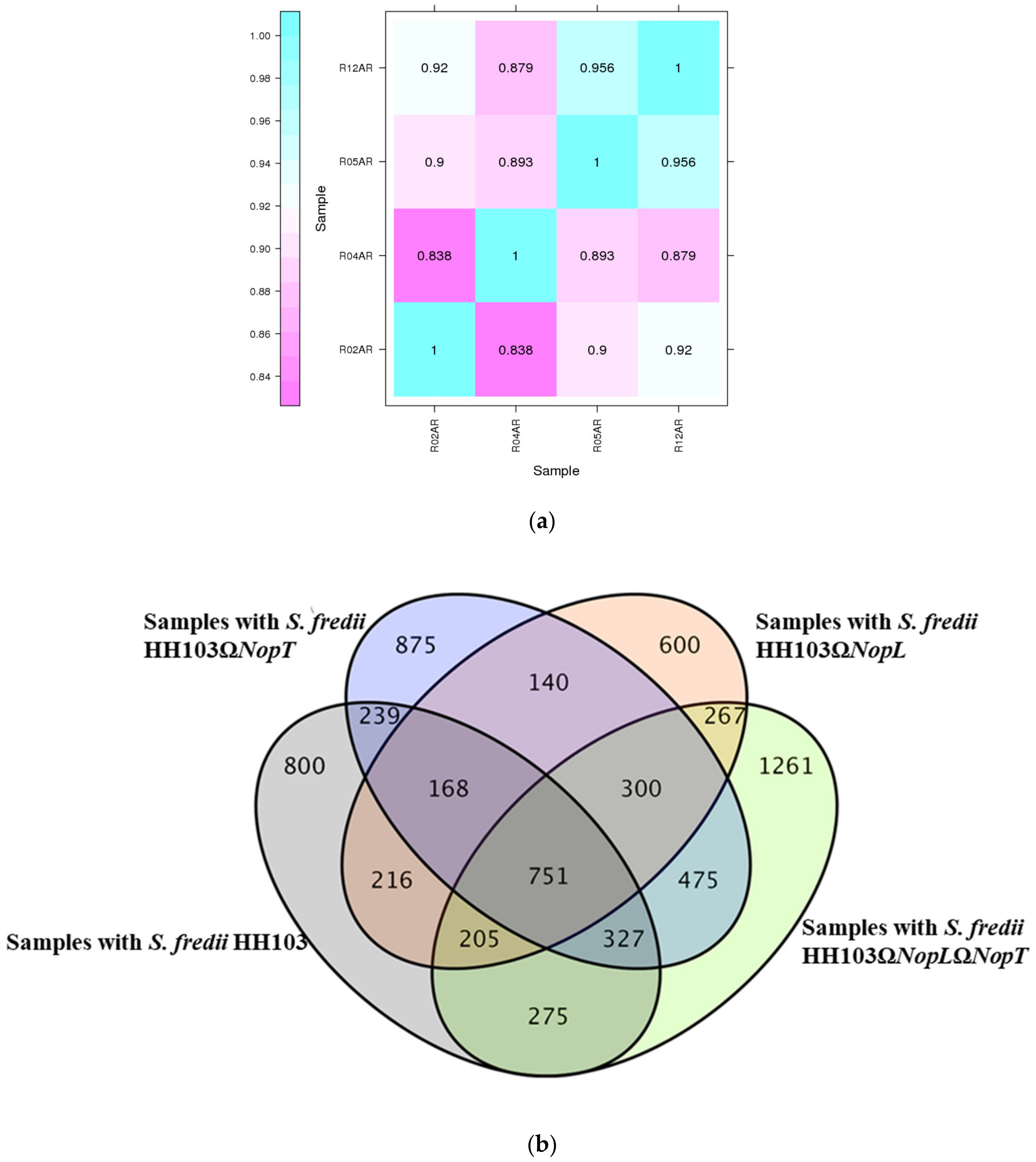
3.5. Differentially Expressed Genes (DEGs) Are Related to the Effectors
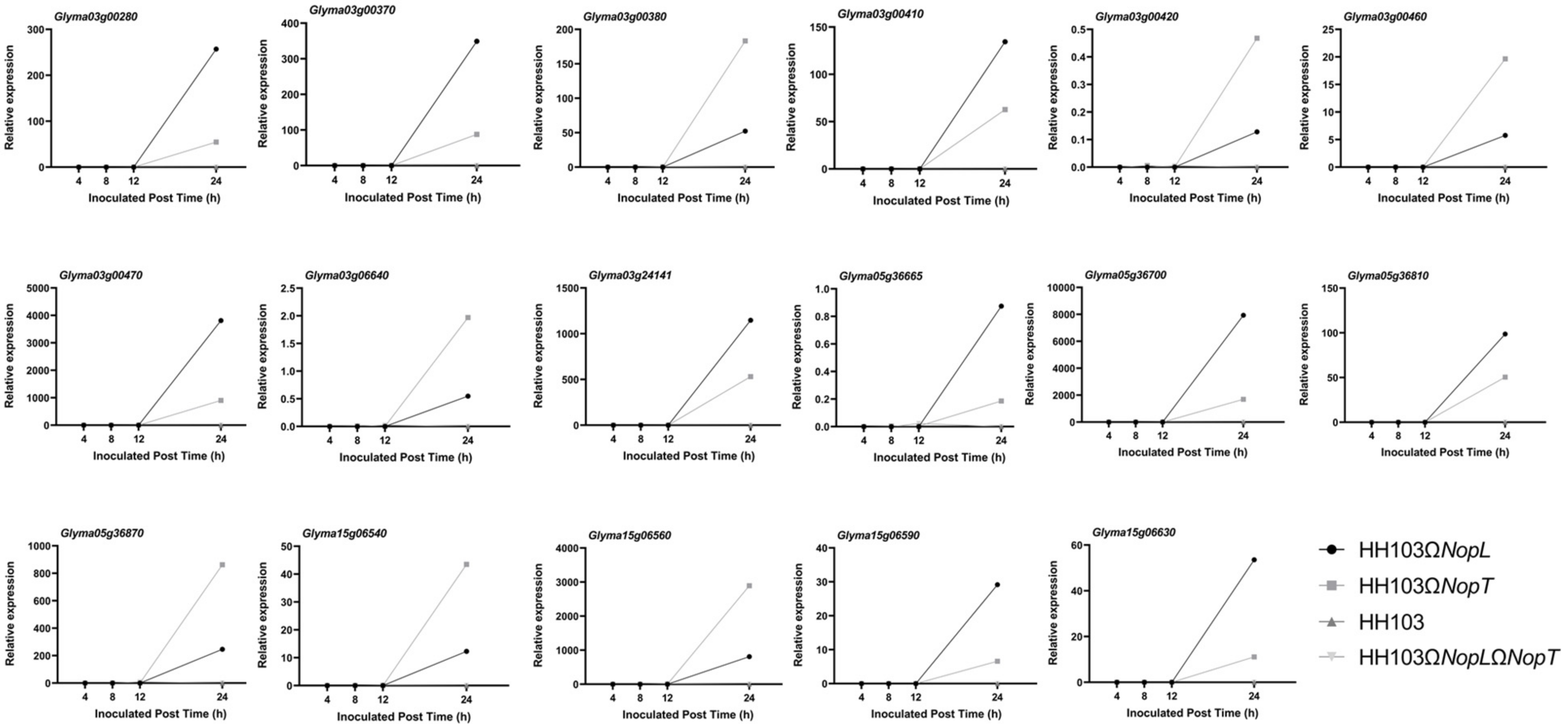
3.6. Validation of Candidate Gene Expression by RT-qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Westhoek, A.; Field, E.; Rehling, F.; Mulley, G.; Webb, I.; Poole, P.S.; Turnbull, L.A. Policing the legume-Rhizobium symbiosis: A critical test of partner choice. Sci. Rep. 2017, 7, 1419. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.; Dickstein, R.; Udvardi, M.K. Celebrating 20 Years of Genetic Discoveries in Legume Nodulation and Symbiotic Nitrogen Fixation. Plant Cell. 2020, 32, 15–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rong, L.; Chen, H.; Yang, Z.; Yuan, S.; Zhou, X. Research status of soybean symbiosis nitrogen fixation. Oil Crop Sci. 2020, 5, 6–10. [Google Scholar] [CrossRef]
- Via, V.D.; Zanetti, M.E.; Blanco, F. How legumes recognize rhizobia. Plant Signal. Behav. 2016, 11, e1120396. [Google Scholar] [CrossRef]
- Schechter, L.M.; Guenther, J.; Olcay, E.A.; Jang, S.; Krishnan, H.B. Translocation of NopP by Sinorhizobium fredii USDA257 into Vigna unguiculata Root Nodules. Appl. Environ. Microbiol. 2010, 76, 3758–3761. [Google Scholar] [CrossRef] [Green Version]
- Okazaki, S.; Kaneko, T.; Sato, S.; Saeki, K. Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proc. Natl. Acad. Sci. USA 2013, 110, 17131–17136. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-Guerrero, I.; Acosta-Jurado, S.; Medina, C.; Ollero, F.J.; Alias-Villegas, C.; Vinardell, J.M.; Pérez-Montaño, F.; López-Baena, F.J. The Sinorhizobium fredii HH103 type III secretion system effector NopC blocks nodulation with Lotus japonicus Gifu. J. Exp. Bot. 2020, 71, 6043–6056. [Google Scholar] [CrossRef]
- Yasuda, M.; Miwa, H.; Masuda, S.; Takebayashi, Y.; Sakakibara, H.; Okazaki, S. Effector-Triggered Immunity Determines Host Genotype-Specific Incompatibility in Legume-Rhizobium Symbiosis. Plant Cell. Physiol. 2016, 57, 1791–1800. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, J.; Ma, C.; Zhou, Z.; Yang, D.; Zheng, J.; Wang, Q.; Li, H.; Zhou, H.; Sun, Z.; et al. QTL Mapping and Data Mining to Identify Genes Associated with the Sinorhizobium fredii HH103 T3SS Effector NopD in Soybean. Front. Plant Sci. 2020, 11, 453. [Google Scholar] [CrossRef]
- Margaret, I.; Crespo-Rivas, J.C.; Acosta-Jurado, S.; Buendía-Clavería, A.M.; Cubo, M.T.; Gil-Serrano, A.; Moreno, J.; Murdoch, P.S.; Rodríguez-Carvajal, M.A.; Rodríguez-Navarro, D.N.; et al. Sinorhizobium fredii HH103 rkp-3 genes are required for K-antigen polysaccharide biosynthesis, affect lipopolysaccharide structure and are essential for infection of legumes forming determinate nodules. MPMI 2012, 25, 825–838. [Google Scholar] [CrossRef] [Green Version]
- Dorival, J.; Philys, S.; Giuntini, E.; Brailly, R.; de Ruyck, J.; Czjzek, M.; Biondi, E.; Bompard, C. Structural and enzymatic characterisation of the Type III effector NopAA (=GunA) from Sinorhizobium fredii USDA257 reveals a Xyloglucan hydrolase activity. Sci. Rep. 2020, 10, 9932. [Google Scholar] [CrossRef] [PubMed]
- Skorpil, P.; Saad, M.M.; Boukli, N.M.; Kobayashi, H.; Ares-Orpel, F.; Broughton, W.J.; Deakin, W.J. NopP, a phosphorylated effector of Rhizobium sp. strain NGR234, is a major determinant of nodulation of the tropical legumes Flemingia congesta and Tephrosia vogelii. Mol. Microbiol. 2005, 57, 1304–1317. [Google Scholar] [CrossRef] [PubMed]
- López-Baena, F.; Monreal, J.; Pérez-Montaño, F.; Guasch-Vidal, B.; Bellogín, R.A.; Vinardell, J.M.; Ollero, F.J. The Absence of Nops Secretion in Sinorhizobium fredii HH103 Increases GmPR1 Expression in Williams Soybean. Mol. Plant-Microbe Interact. 2009, 22, 1445–1454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez-Guerrero, I.; Pérez-Montaño, F.; Medina, C.; Ollero, F.J.; López-Baena, F.J. NopC Is a Rhizobium-Specific Type 3 Secretion System Effector Secreted by Sinorhizobium (Ensifer) fredii HH103. PLoS ONE 2015, 10, e0142866. [Google Scholar]
- Saad, D.M.; Kobayashi, H.; Marie, C.; Brown, I.R.; Mansfield, J.W.; Broughton, W.J.; Deakin, W.J. NopB, a Type III Secreted Protein of Rhizobium sp. Strain NGR234, Is Associated with Pilus-Like Surface Appendages. J. Bacteriol. 2005, 187, 1173–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, J.A.; López-Baena, F.J.; Ollero, F.J.; Vinardell, J.M.; Mdel, R.E.; Bellogín, R.A.; Ruiz-Sainz, J.E.; Thomas, J.R.; Sumpton, D.; Ault, J.; et al. NopM and NopD Are Rhizobial Nodulation Outer Proteins: Identification Using LC-MALDI and LC-ESI with a Monolithic Capillary Column. J. Proteome Res. 2007, 6, 1029–1037. [Google Scholar] [CrossRef] [PubMed]
- Xin, D.W.; Liao, S.; Xie, Z.P.; Hann, D.R.; Steinle, L.; Boller, T.; Staehelin, C. Functional analysis of NopM, a novel E3 ubiquitin ligase (NEL) domain effector of Rhizobium sp. strain NGR234. PLoS Pathog. 2012, 8, e1002707. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-Guerrero, I.; Pérez-Montaño, F.; Medina, C.; Ollero, F.J.; López-Baena, F.J. The Sinorhizobium (Ensifer) fredii HH103 Nodulation Outer Protein NopI Is a Determinant for Efficient Nodulation of Soybean and Cowpea Plants. Appl. Environ. Microbiol. 2017, 83, e02770-16. [Google Scholar] [CrossRef] [Green Version]
- Alexander, V.B.; William, J.D.; Nawal, M.B.; McAlvin, C.B.; Stacey, G.; Malnoe, P.; Broughton, W.J.; Staehelin, C. NopL, an Effector Protein of Rhizobium sp. NGR234, Thwarts Activation of Plant Defense Reactions. Plant Physiol. 2004, 134, 871–879. [Google Scholar]
- Zhang, Y.; Liu, X.; Chen, L.; Fu, Y.; Li, C.; Qi, Z.; Zou, J.; Zhu, R.; Li, S.; Wei, W.; et al. Mining for genes encoding proteins associated with NopL of Sinorhizobium fredii HH103 using quantitative trait loci in soybean (Glycine max Merr.) recombinant inbred lines. Plant Soil 2018, 431, 245–255. [Google Scholar] [CrossRef]
- Dai, W.-J.; Zeng, Y.; Xie, Z.-P.; Staehelin, C. Symbiosis-Promoting and Deleterious Effects of NopT, a Novel Type 3 Effector of Rhizobium sp. Strain NGR234. J. Bacteriol. 2008, 190, 5101–5110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miwa, H.; Okazaki, S. How effectors promote beneficial interactions. Curr. Opin. Plant Biol. 2017, 38, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Liu, D.; Jiao, S.; Liu, S.; Wang, X.; Shen, X.; Wei, G. Identification of Robinia pseudoacacia target proteins responsive to Mesorhizobium amphore CCNWGS0123 effector protein NopT. J. Exp. Bot. 2020, 71, 7347–7363. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, M.; Chen, Q.; Xin, D.; Sun, X. Mapping quantitative trait loci related to nodule number in soybean (Glycine max (L.) Merr.) in response to the Sinorhizobium (Ensifer) fredii HH103 NopT type III effector. J. Plant Interact. 2021, 16, 126–135. [Google Scholar] [CrossRef]
- Zeng, Z.B. QTL Mapping. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: San Diego, MA, USA, 2001; pp. 8–12. [Google Scholar]
- Tominaga, A.; Gondo, T.; Akashi, R.; Zheng, S.; Arima, S.; Suzuki, A. Quantitative trait locus analysis of symbiotic nitrogen fixation activity in the model legume Lotus japonicus. J. Plant Res. 2012, 125, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wang, J.; Li, Q.; Wang, J.; Liu, Y.; Li, J.; Chen, L.; Shi, Y.; Li, S.; Zhang, Y.; et al. QTL analysis of nodule traits and the identification of loci interacting with the type III secretion system in soybean. Mol. Genet. Genom. 2019, 294, 1049–1058. [Google Scholar] [CrossRef]
- Green, R.; Rogers, E.J. Transformation of chemically competent E. coli. Methods Enzym. 2013, 529, 329–336. [Google Scholar]
- Methods: Optimization of culture medium for production of recombinant dengue protein in Escherichia coli. Ind. Biotechnol. 2009, 5, 179–183. [CrossRef]
- Xin, D.; Qi, Z.; Jiang, H.; Hu, Z.; Zhu, R.; Hu, J.; Han, H.; Hu, G.; Liu, C.; Chen, Q. QTL Location and Epistatic Effect Analysis of 100-Seed Weight Using Wild Soybean (Glycine soja Sieb. & Zucc.) Chromosome Segment Substitution Lines. PLoS ONE 2016, 11, e0149380. [Google Scholar]
- Quandt, J.; Hynes, M.F. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 1993, 127, 15–21. [Google Scholar] [CrossRef]
- Figurski, D.H.; Helinski, D.R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 1979, 76, 1648–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pueppke, S.G.; Broughton, W.J. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol. Plant-Microbe Interact. 1999, 12, 293–318. [Google Scholar] [CrossRef] [Green Version]
- Hwang, S.; Ray, J.D.; Cregan, P.B.; King, C.A.; Davies, M.K.; Purcell, L.C. Genetics and mapping of quantitative traits for nodule number, weight, and size in soybean (Glycine max L. [Merr.]). Euphytica 2014, 195, 419–434. [Google Scholar] [CrossRef]
- Yang, J.; Hu, C.; Hu, H.; Yu, R.; Xia, Z.; Ye, X.; Zhu, J. QTLNetwork: Mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 2008, 24, 721–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Zhang, Z.; Wen, Y.; Yu, G.; Zou, J.; Huang, S.; Wang, J.; Zhu, J.; Wang, J.; Chen, L.; et al. RNA Sequencing-Associated Study Identifies GmDRR1 as Positively Regulating the Establishment of Symbiosis in Soybean. Mol. Plant-Microbe Interact. 2020, 33, 798–807. [Google Scholar] [CrossRef]
- Yu, M.; Chen, G.-y.; Zhang, L.-q.; Liu, Y.-x.; Liu, D.-c.; Wang, J.-r.; Pu, Z.-e.; Zhang, L.; Lan, X.-j.; Wei, Y.-m.; et al. QTL Mapping for Important Agronomic Traits in Synthetic Hexaploid Wheat Derived from Aegiliops tauschii ssp. tauschii. J. Integr. Agr. 2014, 13, 1835–1844. [Google Scholar] [CrossRef]
- Jiang, C.; Zeng, Z.B. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 1995, 140, 1111–1127. [Google Scholar] [CrossRef]
- Williams, B.; Kantartzi, S.; Meksem, K.; Grier, R.L., IV; Barakat, A.; Lightfoot, D.A.; Kassem, M.A. Genetic Analysis of Root and Shoot Traits in the ‘Essex’ By ‘Forrest’ Recombinant Inbred Line (RIL) Population of Soybean [Glycine max (L.) Merr.]. J. Plant Biol. 2012, 1, 1–9. [Google Scholar]
- Qi, Z.; Huang, L.; Zhu, R.; Xin, D.; Liu, C.; Han, X.; Jiang, H.; Hong, W.; Hu, G.; Zheng, H.; et al. A high-density genetic map for soybean based on specific length amplified fragment sequencing. PLoS ONE 2014, 9, e104871. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, J.; Liu, C.; Ma, C.; Li, C.; Zhang, Y.; Qi, Z.; Zhu, R.; Shi, Y.; Zou, J.; et al. Identification of Soybean Genes Whose Expression is Affected by the Ensifer fredii HH103 Effector Protein NopP. Int. J. Mol. Sci. 2018, 19, 3438. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, H.; Wang, Q.; Liu, X.; Han, F.; Fang, Z.; Yang, L.; Zhuang, M.; Liu, Y.; Li, Z.; Zhang, Y. Whole-Genome Mapping Reveals Novel QTL Clusters Associated with Main Agronomic Traits of Cabbage (Brassica oleracea var. capitata L.). Front. Plant Sci. 2016, 7, 989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wang, J.; Tian, B.; Li, Q.; Zhu, J.; Liu, X.; Ma, C.; Li, C.; Qi, Z.; Zhu, R.; et al. QTL mapping and gene mining to identify genes on soybean (Glycine max) associated with NopB of Sinorhizobium fredii HH103. Plant Breed. 2019, 138, 677–685. [Google Scholar] [CrossRef]
- Crespo, J.L.; Hall, M.N. Elucidating TOR Signaling and Rapamycin Action: Lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2002, 66, 579–591. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Chen, X.-J.; Lu, H.-B.; Xie, Z.-P.; Staehelin, C. Functional Analysis of the Type 3 Effector Nodulation Outer Protein L (NopL) from Rhizobium sp. NGR234: Symbiotic Effects, Phosphorylation, and Interference with Mitogen-Activated Protein Kinase Signaling. J. Biol. Chem. 2011, 286, 32178–32187. [Google Scholar] [CrossRef] [Green Version]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Anantharaman, V.; Aravind, L. The GOLD domain, a novel protein module involved in Golgi function and secretion. Genome Biol. 2002, 3, research0023. [Google Scholar]
- de Camargos, L.F.; Fraga, O.T.; Oliveira, C.C.; da Silva, J.C.F.; Fontes, E.P.B.; Reis, P.A.B. Development and cell death domain-containing asparagine-rich protein (DCD/NRP): An essential protein in plant development and stress responses. Theor. Exp. Plant Phys. 2019, 31, 59–70. [Google Scholar] [CrossRef]
- Isalan, M. DNA Recognition/Processing | Zinc Fingers: Structure and Design. In Encyclopedia of Biological Chemistry III, 3rd ed.; Jez, J., Ed.; Elsevier: Oxford, UK, 2021; pp. 506–516. [Google Scholar]
- Sumimoto, H.; Kamakura, S.; Ito, T. Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci. Stke Signal Transduct. Knowl. Environ. 2007, 2007, re6. [Google Scholar] [CrossRef]
- Kato, H.; Mutte, S.K.; Suzuki, H.; Crespo, I.; Das, S.; Radoeva, T.; Fontana, M.; Yoshitake, Y.; Hainiwa, E.; van den Berg, W.; et al. Design principles of a minimal auxin response system. Nat. Plants 2020, 6, 473–482. [Google Scholar] [CrossRef]
- Denancé, N.; Szurek, B.; Noël, L.D. Emerging functions of nodulin-like proteins in non-nodulating plant species. Plant Cell Physiol. 2014, 55, 469–474. [Google Scholar] [CrossRef] [PubMed]
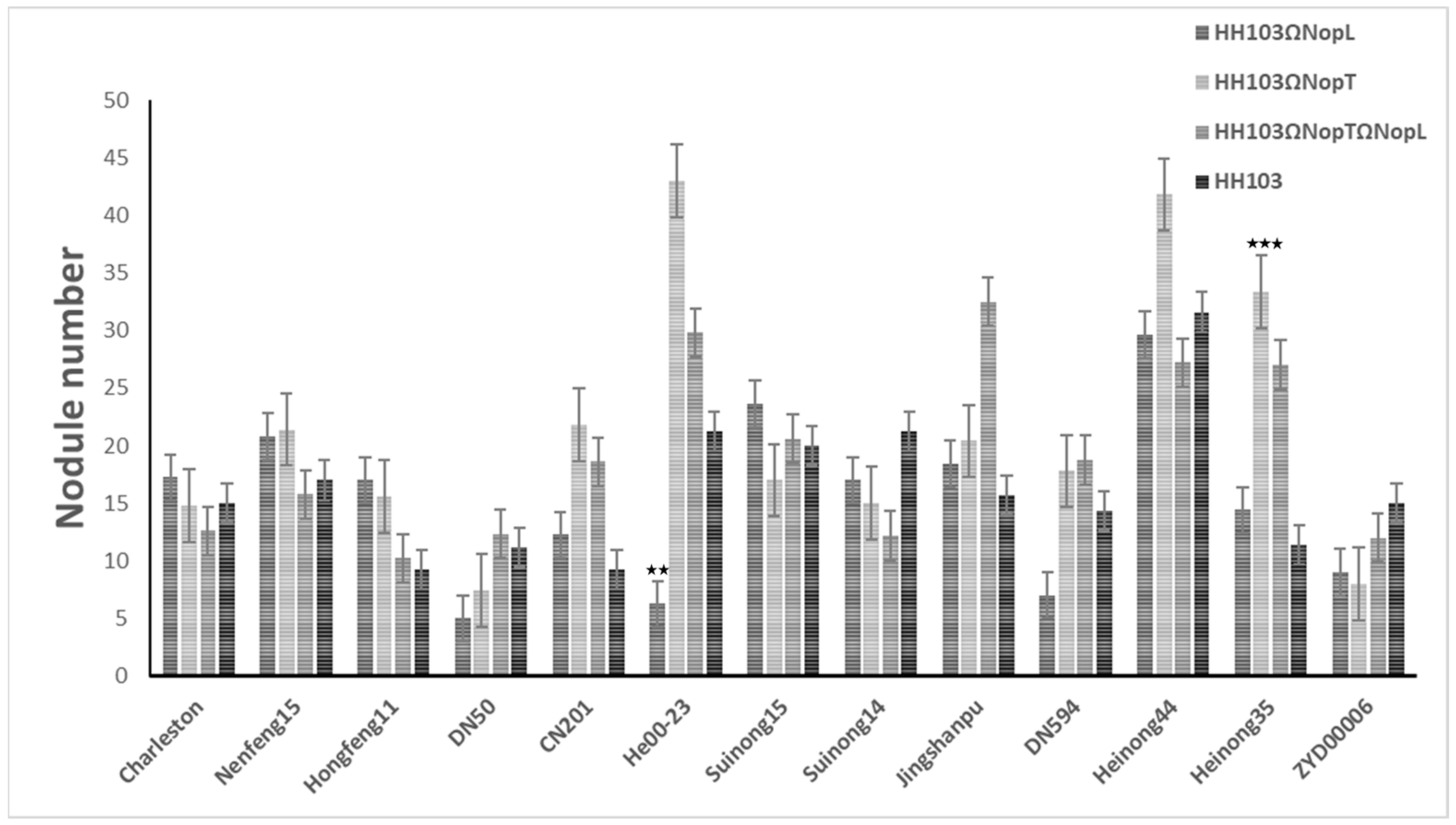
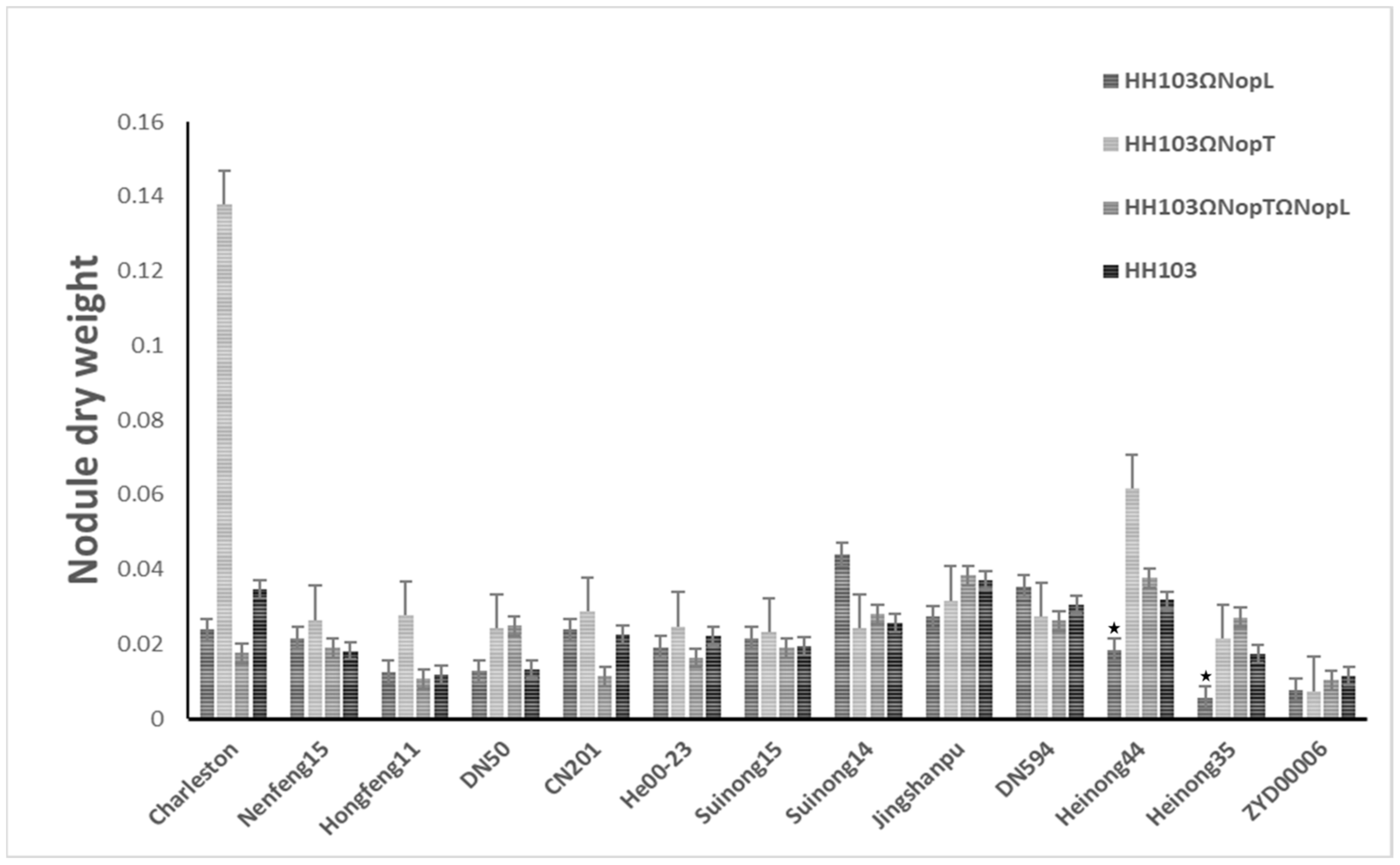
| CSSLs (n = 213) | Parents (Average) | |||||
|---|---|---|---|---|---|---|
| Traits | Average | Standard Deviation | Coefficient of Variation | Suinong14 | ZYD00006 | |
| HH103 | Nodule number | 48.57 | 38.56 | 0.79 | 21.20 ± 19.88 | 14.67 ± 6.51 * |
| Nodule dry weight (g) | 0.04 | 0.03 | 0.69 | 0.03 ± 0.03 | 0.0013 ± 0.0038 | |
| HH103ΩNopL | Nodule number | 13.65 ** | 8.03 | 0.58 | 17.00 ± 6.67 | 10.00 ± 1.00 |
| Nodule dry weight (g) | 0.02 ** | 0.03 | 1.63 | 0.04 ± 0.02 | 0.0077 ± 0.0011 | |
| HH103ΩNopT | Nodule number | 14.01 ** | 10.77 | 0.77 | 15.00 ± 3.61 | 8.33 ± 1.53 |
| Nodule dry weight (g) | 0.02 ** | 0.02 | 1.28 | 0.02 ± 0.02 | 0.0074 ± 0.0038 | |
| HH103ΩNopLΩNopT | Nodule number | 16.77 ** | 11.56 | 0.69 | 12.20 ± 4.71 | 9.00 ± 6.08 |
| Nodule dry weight (g) | 0.02 ** | 0.01 | 0.70 | 0.03 ± 0.02 | 0.0103 ± 0.0075 | |
| Strain | Trait | Gm | Start Position | End Position | PVE (%) | LOD | Additive Effect |
|---|---|---|---|---|---|---|---|
| HH103 | NN | 5 | 39,670,584 | 39,726,296 | 7.80 | 37.9 | 0.053 |
| 5 | 3,976,808 | 39,891,168 | 5.19 | 28.4 | −0.044 | ||
| NDW | 2 | 7,376,054 | 7,504,904 | 8.52 | 12.3 | −0.040 | |
| 2 | 8,200,157 | 8,248,549 | 15.60 | 20.4 | 0.051 | ||
| 3 | 1933 | 234,034 | 4.32 | 6.8 | 0.015 | ||
| HH103ΩNopL | NN | 15 | 4,637,363 | 4,716,807 | 7.40 | 3.4 | 7.851 |
| NDW | 15 | 4,119,708 | 4,302,572 | 1.75 | 2.5 | 0.011 | |
| 19 | 39,031,608 | 39,097,776 | 7.64 | 10.4 | −0.028 | ||
| 19 | 39,194,300 | 39,293,776 | 17.05 | 20.6 | 0.044 | ||
| HH103ΩNopT | NN | 3 | 6,743,461 | 6,777,452 | 17.00 | 8.6 | 5.881 |
| NDW | 1 | 55,304,920 | 55,503,928 | 1.46 | 7.7 | 0.048 | |
| 3 | 2,244,023 | 2,274,724 | 1.18 | 5.8 | 0.010 | ||
| 10 | 39,447,660 | 39,504,396 | 11.34 | 40.2 | 0.067 | ||
| 10 | 39,660,168 | 39,754,196 | 12.85 | 43.2 | 0.058 | ||
| 10 | 40,496,720 | 40,586,292 | 31.55 | 70.7 | −0.128 | ||
| 11 | 10,234,170 | 10,395,019 | 1.04 | 5.7 | 0.009 | ||
| HH103ΩNopLΩNopT | NN | 3 | 1933 | 234,034 | 8.44 | 4.2 | 4.720 |
| 4 | 8,807,067 | 8,829,123 | 5.55 | 2.8 | 5.273 | ||
| 18 | 57,614,440 | 57,682,792 | 4.98 | 2.6 | 4.591 | ||
| NDW | 9 | 41,985,024 | 42,131,960 | 3.55 | 14.7 | 29.446 | |
| 14 | 1,346,935 | 1,422,699 | 1.47 | 6.7 | −23.711 | ||
| 14 | 4,751,415 | 4,955,219 | 22.95 | 56.3 | 104.814 | ||
| 14 | 4,956,367 | 5,109,562 | 17.36 | 47.7 | −104.991 |
| Rhizobium | Chrom. | Gene | Predicted Function |
|---|---|---|---|
| HH103 | 05 | Glyma05g36870 | Zinc finger, RING-type |
| Glyma05g36810 | Phosphatidylinositol 3-/4-kinase, catalytic domain | ||
| Glyma05g36700 | Development/cell death domain | ||
| Glyma05g36665 | Domain of unknown function DUF2439 | ||
| HH103ΩNopL | 15 | Glyma15g06540 | Mg2+ transporter protein, CorA-like/Zinc transport protein ZntB |
| Glyma15g06560 | NET domain | ||
| Glyma15g06590 | PB1 domain | ||
| Glyma15g06630 | CTLH/CRA C-terminal to LisH motif domain | ||
| HH103ΩNopT | 03 | Glyma03g06640 | Concanavalin A-like lectin/glucanase domain |
| HH103ΩNopLΩNopT | 03 | Glyma03g24141 | Nodulin-like |
| Glyma03g00280 | GOLD domain | ||
| Glyma03g00370 | K Homology domain | ||
| Glyma03g00380 | Acid phosphatase (Class B) | ||
| Glyma03g00410 | Thioredoxin | ||
| Glyma03g00420 | TAFII28-like protein | ||
| Glyma03g00470 | RNA recognition motif domain | ||
| Glyma03g00460 | WRKY domain |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, H.; Peng, Y.; Wang, J.; Wang, J.; Yuan, Y.; Fu, T.; Zhu, Z.; Zhang, J.; Pan, X.; Cui, Z.; et al. Mapping of Quantitative Trait Loci Underlying Nodule Traits in Soybean (Glycine max (L.) Merr.) and Identification of Genes Whose Expression Is Affected by the Sinorhizobium fredii HH103 Effector Proteins NopL and NopT. Agronomy 2022, 12, 946. https://doi.org/10.3390/agronomy12040946
Ni H, Peng Y, Wang J, Wang J, Yuan Y, Fu T, Zhu Z, Zhang J, Pan X, Cui Z, et al. Mapping of Quantitative Trait Loci Underlying Nodule Traits in Soybean (Glycine max (L.) Merr.) and Identification of Genes Whose Expression Is Affected by the Sinorhizobium fredii HH103 Effector Proteins NopL and NopT. Agronomy. 2022; 12(4):946. https://doi.org/10.3390/agronomy12040946
Chicago/Turabian StyleNi, Hejia, Yang Peng, Jinhui Wang, Jing Wang, Yantong Yuan, Tingting Fu, Zikun Zhu, Jialin Zhang, Xipeng Pan, Zhuoling Cui, and et al. 2022. "Mapping of Quantitative Trait Loci Underlying Nodule Traits in Soybean (Glycine max (L.) Merr.) and Identification of Genes Whose Expression Is Affected by the Sinorhizobium fredii HH103 Effector Proteins NopL and NopT" Agronomy 12, no. 4: 946. https://doi.org/10.3390/agronomy12040946
APA StyleNi, H., Peng, Y., Wang, J., Wang, J., Yuan, Y., Fu, T., Zhu, Z., Zhang, J., Pan, X., Cui, Z., Liu, C., Chen, Q., & Xin, D. (2022). Mapping of Quantitative Trait Loci Underlying Nodule Traits in Soybean (Glycine max (L.) Merr.) and Identification of Genes Whose Expression Is Affected by the Sinorhizobium fredii HH103 Effector Proteins NopL and NopT. Agronomy, 12(4), 946. https://doi.org/10.3390/agronomy12040946








