Abstract
Plastid DNA holds a substantial amount of plant genetic information, including maternal ancestry information. It helps to uncover interrelations between a wide variety of tuberous species of the genus Solanum to search for promising sources of high-yielding potato varieties resistant to bio- and abiotic stressors. This paper demonstrated the opportunities of de novo assembly of potato plastid DNA and its phylogenetic and genome type identification based only on Oxford Nanopore Technologies (ONT) long reads. According to our results, of 28 potato varieties developed at the Ural Research Institute of Agriculture, 16 varieties had one of the most primitive W-type plastomes. Ten varieties’ plastomes belonged to the T-type of cultivated Solanum tuberosum subsp. tuberosum. The varieties Legenda and 15-27-1 were the closest to the wild species Solanum chacoense plastome. Using long-sequencing reads, we confirmed the presence of two isoforms of the plastid genome differing in the orientation of SSC region. We should note that irrespective of sequencing depth and improvements in software for working with ONT reads, a correct de novo plastome assembly and its annotation using only long-reads is impossible. The most problematic regions are homopolymers longer than 5 bp—they account for all detected indels, leading to a change in the reading frame or the deletion of entire genes.
1. Introduction
Plastid DNA or plastome is an essential part of the genetic information of all photosynthetic plants. In most terrestrial plants, plastome is a circular DNA molecule from 115 to 165 kb long, contains an average of 110–130 genes, and is divided into four parts: a large single-copy region, a small single-copy region, and two inverted repeats [1]. Compared with nuclear and mitochondrial genomes, plastid DNA remains the most conservative in terms of gene structure and location [2]. However, single nucleotide polymorphisms (SNP), changes in microsatellite composition (SSR), insertions and deletions, including species-specific ones, are found in the plastome [3,4,5]. Solanum plastomes range from 154 to 156 kb, including four segments: a large single-copy (LSC) region ~86 kb, a small single-copy (SSC) region ~18 kb, and two inverted repeats between them (IRa and IRb) ~25 kb [3,4]. The plastome of S. tuberosum contains 81 protein coding, 30 tRNAs, and 4 rRNAs genes [6].
As a group of tuber-forming plants of the genus Solanum, the potato has four cultivated and 107 wild species [7]. The use of the plastome to establish phylogenetic relationships within the potato has its own difficulties. Even though large clades obtained from plastome and nuclear genome data coincide, their topology can differ significantly; the reason for this lies in introgression and hybridization [8]. However, this discrepancy between the nuclear genome and the plastome may provide important information about evolutionary relationships.
The plastid genome of potatoes is usually divided into five main types W, T, C, A, and S, depending on the restriction fragments [9]. The W-type is the earliest and is considered the ancestor of the rest, more common in wild types [10]. A single nucleotide substitution in the rps11 gene in LSC, which causes a change in PvuII restriction site [6], defines the W2-type. A large deletion of 241 nucleotides distinguishes the T-type from W-type. The T-type is most characteristic of the cultivated Chilean potato Solamun tuberosum subsp. tuberosum [6,9]. SNP between the cemA and petA genes leading to a change in the BamHI restriction site is characteristic of types C, S, and A. At the same time, two latter types derived from the C-type: the S-type appears by a deletion of 48 nucleotides, while the A-type appears by a nucleotide change in the ccsA gene and as a result of a change in another BamHI restriction site. These three types are found in Solanum tuberosum subsp. andigena [6,9]. A phylogenetic analysis demonstrated that Solanum plastomes clustered according to the types [6].
In this paper, we (1) assembled plastomes of 28 potato varieties bred at Ural Research Institute of Agriculture using ONT long-reads, (2) considered their phylogenetic diversity, and (3) showed a cost-efficient way to confirm the variety origin.
2. Materials and Methods
2.1. DNA Extraction and Sequencing
DNA was isolated from the leaves and stems of sterilized young potato shoots grown on an MSO [11] with the addition of gibberellin A1 at a concentration of 0.2 mg/L. Using an MP-24 homogenizer, we homogenized about 100 mg of leaves with 1 mm zirconium beads in 2-mL tubes. Extraction from homogenized leaves was performed using the innuPREP Plant DNA Extraction Kit (Analytik Jena, Jena, Germany) following Protocol #3. At the stage of nucleic acid binding to the column membrane, RNase Cocktail™ Enzyme Mix (Invitrogen, Carlsbad, CA, USA) was used to degrade RNA. The eluate was further purified by AMPure XP magnetic particles (Beckman Coulter, Bray, CA, USA). Sequencing was performed following with the ONT 1D (SQK-LSK109) protocol and Native Barcoding Expansion (EXP-NBD104, EXP-NBD114) onto the FLO-MIN106 cell (Oxford Nanopore Technologies, Oxford, UK) on the sequencer MinION Mk1C.
2.2. Quality Control of Reads
Basecalling was performed using Guppy v6.0.1 [12]. We trimmed 80 bp from the start and end of the reads and filtered them using Nanofilt v2.7.1 [13]. We kept only reads >5000 bp long and with quality >7 on the Phred quality score for further analysis.
2.3. Plastid Genome Assembly and Quality Assessment
The sequences related to the Solanum tuberosum plastome were extracted by alignment to the MT120865 reference plastome using the NGMLR v0.2.7 [14], Samtools v1.9 [15], and bedtools v2.30.0 [16].
For de novo assembly of the draft genome, Flye v2.9 [17], Unicycler v0.4.8 [18], and Raven v1.7.0 [19] were used in the Trycycler v0.5.3 [20] pipeline, which is capable of combining the results of several sequence-assembly algorithms into one consensus assembly. Regions of the resulting draft contigs were manually trimmed for the contig matching stage inside the Trycycler pipline. De novo assembly polishing of draft plastomes was performed using medaka v1.5.0 [21]. The final assembly was evaluated by the quality of the read mapping to the corresponding genome by the program Qualimap v2.2.1 [22]. The call of structural variants for determining the number of SNPs and indels was carried out using the programs msa2vcf [23] and RTG Tools v3.9.1 [24].
2.4. Alignment and Phylogenetic Identification
Alignment of the resulting plastome assemblies with reference-based sequences was performed using FMAlign [25]. For phylogenetic tree construction and visualization, we used fasttree v2.1.10 [26] and iTOL v5 [27], respectively.
2.5. Genome Annotation
Assembled de novo plastomes were annotated using an online tool GESEQ [28] with default settings.
3. Results
3.1. Sequencing Results
For Alaska, Argo, and Shah potato varieties, more than 30 Gb of whole-genome sequencing data were available as these varieties had been sequenced for our earlier study [29]. The Legenda variety was also sequenced separately, and ~7 Gb of whole-genome data were available for this study. The other 24 varieties were simultaneously sequenced in multiplex using a single flowcell, resulting in 12 Gb of data. After filtering by the quality and read length, the reads were used to assemble the plastome. In addition to filtering, the reads related to the plastome of Solanum tuberosum (MT120865) were extracted before the assembly of the varieties Alaska, Argo, Shah, and Legenda. Additional details are presented in Table S1. A large amount of data might hamper the assembly. However, plastome assembly from whole-genome data was possible using even modest computing capacities for the other varieties. For the barcoded samples, the proportion of reads belonging to the plastome was 8.8–22%, which provided 50–386x sequencing coverage of individual sequencing samples with 86.2–94.2% belonging to the plastome reads, with more than 2000× coverage. At the same time, the average mapping quality was ~46 according to the mapping quality score, and the average error rate was ~0.0025.
3.2. Phylogenetic Identification
One of the earlier studies on the type of plastid genome type showed that the T-type dominates among the Russian potato varieties and is present in 40 of those, while 23 varieties belong to the W-type [30]. This study notes that more than 1000 breeding varieties descend from Rough Purple Chili, which has a T-type plastome.
For phylogenetic identification and plastome classification, the resolution of nanopore sequencing is sufficient. Despite a higher error rate of ONT long-reads than Illumina short-reads [31], a wide range bacterial genome studies based only on ONT long-reads demonstrated the accuracy of ONT technology for the phylogenetic tree construction [32,33,34], and applicability for in situ surveillance protocols [35]. Even the variety Otrada, whose sequencing depth was only 50, showed 99.97% identity to MT120865 (S. tuberosum). Therefore, we used all de novo plastome assemblies for phylogenetic identification. Plastomes with a confirmed type were taken as reference-based sequences of the potato plastid genome [6]: W-type—Solanum tuberosum (MT120865) and S. commersonii (NC_028069); W2-type—S. bukasovii (MT120867); T-type—S. tuberosum (DQ231562); C-type—S. tuberosum ssp. andigenum (MT120861), S. ahanhuiri (MT120857) and S. juzepczukii (MT120863); A-type—S. chaucha (MT120864) and S. tuberosum ssp. andigenum (MT120862); S-type—S. curtilobum (MT120866), S. bukasovii (MT120860), S. stenotomum subsp. stenotomum (MT120859), and S. stenotomum subsp. goniocalyx (MT120855 и MT120856). Other known sequences were also added to the analysis: S. tuberosum (KM489056), S. chacoense (MH021455, MK398247), S. aucale (NC_041551, MK036506), S. demissium (NC_041552, MK036508), S. stoloniferum (MF471373), and S. bulbocastanum (NC_007943, MH021439). To avoid the false clades, we manually flipped SSC regions in those assembled genomes, where required, in accordance with the reference sequences.
In the resulting cladogram (Figure 1) and phylogram (Figure S1), two types characteristic of our collection could be distinguished. The W-type clade included 16 plastomes from our collection, where the variety 15-22-4 was closer to S. stoloniferum, and varieties Baron, Start, 14-4-1, and 16-4-3 were closer to S. demissium. The T-type clade was comprised of 10 plastomes. No plastomes belonging to the other types mentioned above were found. However, two of our varieties, Legenda and 15-27-1, formed a common clade with sequences belonging to the wild species S. chacoense.
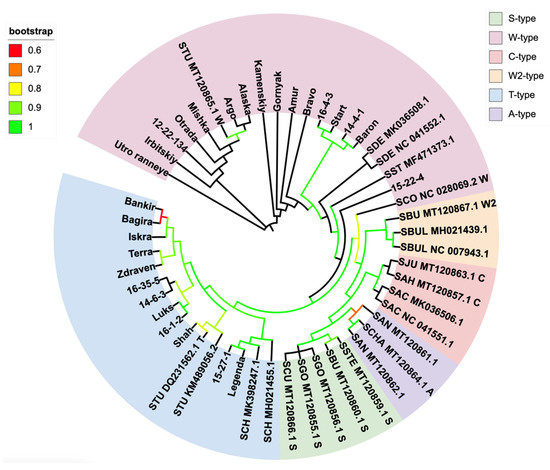
Figure 1.
Cladogram of reference plastid genomes and de novo assemblies of plastomes of breeding varieties developed at the Ural Research Institute of Agriculture. Low support is defined as bootstrap <60. STU—Solanum tuberosum, SCO—S. commersonii, SST—S. stoloniferum, SDE—S. demissium, SCH– S. chacoense, SSTE—S. stenotomum subsp. stenotomum, SBU—S. bukasovii, SGO—S. stenotomum subsp. goniocalyx, SCU—S. curtilobum, SAN—S. tuberosum ssp. andigenum, SCHA—S. chaucha, SAC—S. aucale, SAH—S. ahanhuiri, SJU—S. juzepczukii, SBUL—S. bulbocastanum.
3.3. Long-Reads Confirm the Presence of Two Isoforms of the Plastid Genome in Potatoes
It has long been known that the plastid genome is present in two isoforms differing in the orientation of SSC region [36], which has been confirmed for a wide range of terrestrial plants [37]. For varieties Alaska, Argo, and Shah, a sufficient number of long-reads were available, covering the LSC, SSC, and IR regions in unambiguous orientation. We estimated plastome isoforms using the Cp-hap [37]. The results are presented in Table 1.

Table 1.
Variability of the plastid genome in three Ural Research Institute of Agriculture potato varieties.
3.4. Nanopore Sequencing Cannot Cope with Homopolymer Repeats
We compared our plastome assemblies with the closest reference-based sequences obtained at the stage of phylogenetic identification. The detected structural variations are presented in Table 2. The proportion of SNPs in the detected variants was insignificant, while indels were more abundant, with a higher number of deletions than insertions. Additionally, in our study, all indels were located in the region of homopolymer repeats with a length of more than 5 nucleotides. The examples of Alaska, Argo, and Shah showed that the highest read depth does not guarantee correct assembly. Although the number of deletions in these varieties was 2–4 times lower than the average, the number of insertions was 2–20 times higher than in other assemblies relative to the reference. Numerous errors in homopolymeric regions led to changes in the reading frames of genes or nonsense mutations, which in turn made it impossible to annotate the plastome obtained by de novo assembly from long-reads (Figure 2).

Table 2.
The number of SNPs and indels in plastomes of potato varieties developed at the Ural Research Institute of Agriculture.
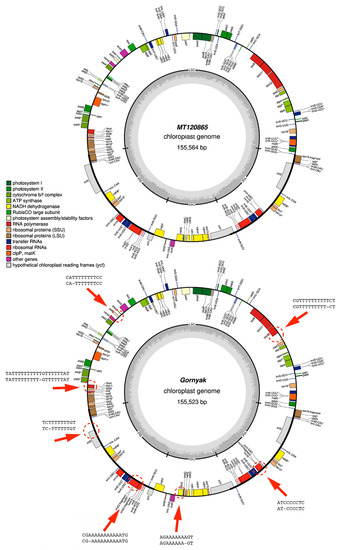
Figure 2.
Annotated plastome sequences Solanum tuberosum (MT120865) and our variety Gornyak. Red arrows and dotted circles indicate the homopolymer regions in genes with indels. Next to the arrows, the sequences at the top, are related to MT120865, and those at the bottom are related to Gornyak.
It was previously reported that homopolymer sequences longer than five nucleotides contain up to 80% deletions and up to 62% insertions [38]. The lack of indels in other sites of our de novo assemblies can be explained by the software updates: basecalling—Guppy (with the release of new versions, accuracy increases [39]), genome polishing—Medaka, as well as the use of the Trycycler, which combined the results of Flye, Unicycler, and Raven in our study.
Due to the high error rate in nanopore sequencing, it is better to use hybrid assembly based on combining ONT long-reads with Illumina short-reads [38]. For example, a combination of 20× long-read coverage and 20× short-read coverage can be sufficient for a de novo assembly with few or no errors [40].
4. Discussion
Illumina technology provides a good solution for phylogeny inference and annotation of the potato plastome [41]. However, its higher costs and excessive performance can be redundant for the plastome type identification. In the present study, we demonstrated that nanopore sequencing is suitable for the phylogenetic identification of the plastomes. Combining de novo assemblies using Trycycler and polishing the resulting consensus assembly with Medaka allowed obtaining plastomes suitable for the construction of the phylogenetic tree and determination of the plastome type. By multiplexing up to 24 samples per run, enough data could be obtained for 150× plastome coverage from whole-genome sequencing libraries. Therefore, ONT provides the cheapest sequencing method for subsequent phylogenetic identification, which can be used to determine the plastome type and identify the potato species rapidly.
In potatoes, as in many other plants, the plastome comes in two variants with a non-inverted and inverted SSC region.
Despite the annual technological improvements of the sequencing depth and nanopore read quality and the software used in the analysis, genome assembly based on long-read data alone cannot replace hybrid genome assembly (ONT + Illumina). Numerous indels in the homopolymer-rich regions prevent correct annotation, leading to the appearance of nonsense mutations in assemblies and gene loss. Thus, the short-read technology or the Sanger sequencing are more suitable for the development of new markers [42,43], k-mers analysis [44], or gene annotation [45,46] of the plastome. In the same way, short-reads are more accurate and more applicable in studies of evolutionary relations [47] than ONT technology.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy12040846/s1, Table S1: Sequencing results. Figure S1: Phylogram of reference plastid genomes and de novo assemblies of plastomes of breeding varieties developed at the Ural Research Institute of Agriculture. Low support is defined as bootstrap < 60. STU—Solanum tuberosum, SCO—S. commersonii, SST—S. stoloniferum, SDE—S. demissium, SCH–S. chacoense, SSTE—S. stenotomum subsp. stenotomum, SBU—S. bukasovii, SGO—S. stenotomum subsp. goniocalyx, SCU—S. curtilobum, SAN—S. tuberosum ssp. andigenum, SCHA—S. chaucha, SAC—S. aucale, SAH—S. ahanhuiri, SJU—S. juzepczukii, SBUL—S. bulbocastanum.
Author Contributions
Conceptualization, all authors; methodology, all authors; validation, G.A.L.; investigation, G.A.L.; writing—original draft preparation, G.A.L.; writing—review and editing, E.P.S.; visualization, G.A.L.; supervision, E.P.S.; funding acquisition, E.P.S. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Ministry of Science and Higher Education of the Russian Federation under agreement No. 0773-2020-0022, which provides a grant in the form of subsidies from the Federal budget of the Russian Federation. The grant was provided within the framework of the State Target “Development of competitive, high-yielding, world-class varieties of grain, leguminous, forage, fruit and berry crops and potatoes based on promising genetic resources that are resistant to bio- and abiotic factors”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets of basecalled and filtered reads supporting the results of this article are available on NCBI: https://www.ncbi.nlm.nih.gov/bioproject/807056 (accessed on 4 January 2022), and de novo assembled plastomes: https://drive.google.com/file/d/1C2A628FaurL_-roLKkDezDEsBaVvkL5J/view?usp=sharing (accessed on 4 January 2022).
Acknowledgments
The authors are grateful to Mikryukov V.S. for his help on the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jansen, R.K.; Raubeson, L.A.; Boore, J.L.; de Pamphilis, C.W.; Chumley, T.W.; Haberle, R.C.; Wyman, S.K.; Alverson, A.J.; Peery, R.; Herman, S.J.; et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 2005, 395, 348–384. [Google Scholar] [PubMed]
- Thode, V.A.; Lohmann, L.G. Comparative Chloroplast Genomics at Low Taxonomic Levels: A Case Study Using Amphilophium (Bignonieae, Bignoniaceae). Front. Plant Sci. 2019, 10, 796. [Google Scholar] [CrossRef]
- Chung, H.-J.; Jung, J.D.; Park, H.-W.; Kim, J.-H.; Cha, H.W.; Min, S.R.; Jeong, W.-J.; Liu, J.R. The complete chloroplast genome sequences of Solanum tuberosum and comparative analysis with Solanaceae species identified the presence of a 241-bp deletion in cultivated potato chloroplast DNA sequence. Plant Cell Rep. 2006, 25, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.-S.; Cheon, K.-S.; Hong, S.-Y.; Cho, J.-H.; Im, J.-S.; Mekapogu, M.; Yu, Y.S.; Park, T.H. Complete chloroplast genome sequences of Solanum commersonii and its application to chloroplast genotype in somatic hybrids with Solanum tuberosum. Plant Cell Rep. 2016, 35, 2113–2123. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.; Yang, C.; Liu, J.; Jin, S.; Harijati, N.; Hu, Z.; Diao, Y.; Zhao, L. Comparative analysis of complete chloroplast genome sequences of four major Amorphophallus species. Sci. Rep. 2019, 9, 809. [Google Scholar] [CrossRef] [PubMed]
- Achakkagari, S.R.; Kyriakidou, M.; Tai, H.H.; Anglin, N.L.; Ellis, D.; Stromvik, M.V. Complete plastome assemblies from a panel of 13 diverse potato taxa. PLoS ONE 2020, 15, e0240124. [Google Scholar] [CrossRef] [PubMed]
- Spooner, D.M.; Ghislain, M.; Simon, R.; Jansky, S.H.; Gavrilenko, T. Systematics, Diversity, Genetics, and Evolution of Wild and Cultivated Potatoes. Bot. Rev. 2014, 80, 283–383. [Google Scholar] [CrossRef]
- Huang, B.; Ruess, H.; Liang, Q.; Colleoni, C.; Spooner, D.M. Analyses of 202 plastid genomes elucidate the phylogeny of Solanum section Petota. Sci. Rep. 2019, 9, 4454. [Google Scholar] [CrossRef]
- Hosaka, K. Who is the mother of the potato?—Restriction endonuclease analysis of chloroplast DNA of cultivated potatoes. Theor. Appl. Genet. 1986, 72, 606–618. [Google Scholar] [CrossRef]
- Hosaka, K. Successive domestication and evolution of the Andean potatoes as revealed by chloroplast DNA restriction endonuclease analysis. Theor. Appl. Genet. 1995, 90, 356–363. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Available online: https://community.nanoporetech.com/posts/guppy-v6-0-1-patch-release (accessed on 1 December 2021).
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef] [PubMed]
- Sedlazeck, F.J.; Rescheneder, P.; Smolka, M.; Fang, H.; Nattestad, M.; von Haeseler, A.; Schatz, M.C. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 2018, 15, 461–468. [Google Scholar] [CrossRef] [Green Version]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaser, R.; Šikić, M. Time- and memory-efficient genome assembly with Raven. Nat. Comput. Sci. 2021, 1, 332–336. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Cerdeira, L.T.; Hawkey, J.; Méric, G.; Vezina, B.; Wyres, K.L.; Holt, K.E. Trycycler: Consensus long-read assemblies for bacterial genomes. Genome Biol. 2021, 22, 266. [Google Scholar] [CrossRef]
- Available online: https://github.com/nanoporetech/medaka (accessed on 25 December 2021).
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef]
- Available online: https://github.com/connor-lab/msa2vcf (accessed on 4 January 2022).
- Available online: https://github.com/RealTimeGenomics/rtg-tools (accessed on 4 January 2022).
- Available online: https://github.com/iliuh/FMAlign (accessed on 15 January 2022).
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucl. Acids Res. 2021, 49, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://chlorobox.mpimp-golm.mpg.de/geseq.html (accessed on 17 January 2022).
- Lihodeevskiy, G.A.; Shanina, E.P. Structural Variations in the Genome of Potato Varieties of the Ural Selection. Agronomy 2021, 11, 1703. [Google Scholar] [CrossRef]
- Gavrilenko, T.A.; Antonova, O.Y.; Kostina, L.I. Study of the genetic diversity of potato varieties using PCR analysis of DNA organelles. Genetika 2007, 43, 1550–1555. [Google Scholar] [PubMed]
- Rang, F.J.; Kloosterman, W.P.; de Ridder, J. From squiggle to basepair: Computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 2018, 19, 90. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Erickson, D.L.; Meng, J. Benchmarking Long-Read Assemblers for Genomic Analyses of Bacterial Pathogens Using Oxford Nanopore Sequencing. Int. J. Mol. Sci. 2020, 21, 9161. [Google Scholar] [CrossRef]
- Tanaka, M.; Mino, S.; Ogura, Y.; Hayashi, T.; Sawabe, T. Availability of Nanopore Sequences in the Genome Taxonomy for Vibrionaceae Systematics: Rumoiensis Clade Species as a Test Case. PeerJ. 2018, 6, e5018. [Google Scholar] [CrossRef]
- Bokma, J.; Vereecke, N.; De Bleecker, K.; Callens, J.; Ribbens, S.; Nauwynck, H.; Haesebrouck, F.; Theuns, S.; Boyen, F.; Pardon, B. Phylogenomic analysis of Mycoplasma bovis from Belgian veal, dairy and beef herds. Vet. Res. 2020, 51, 121. [Google Scholar] [CrossRef]
- Baeza, J.A. Yes, we can use it: A formal test on the accuracy of low-pass nanopore long-read sequencing for mitophylogenomics and barcoding research using the Caribbean spiny lobster Panulirus argus. BMC Genom. 2020, 21, 882. [Google Scholar] [CrossRef]
- Palmer, J.D. Chloroplast DNA exists in two orientations. Nature 1983, 301, 92–93. [Google Scholar] [CrossRef]
- Wang, W.; Lanfear, R. Long-Reads Reveal That the Chloroplast Genome Exists in Two Distinct Versions in Most Plants. Genome Biol. Evol. 2019, 11, 3372–3381. [Google Scholar] [CrossRef] [PubMed]
- Scheunert, A.; Dorfner, M.; Lingl, T.; Oberprieler, C. Can we use it? On the utility of de novo and reference-based assembly of Nanopore data for plant plastome sequencing. PLoS ONE 2020, 15, e0226234. [Google Scholar]
- Delahaye, C.; Nicolas, J. Sequencing DNA with nanopores: Troubles and biases. PLoS ONE 2021, 16, e0257521. [Google Scholar] [CrossRef]
- Wang, W.; Schalamun, M.; Morales-Suarez, A.; Kainer, D.; Schwessinger, B.; Lanfear, R. Assembly of chloroplast genomes with long- and short-read data: A comparison of approaches using Eucalyptus pauciflora as a test case. BMC Genom. 2018, 19, 977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Achakkagari, S.R.; Tai, H.H.; Davidson, C.; Jong, H.; Strömvik, M.V. The complete plastome sequences of nine diploid potato clones. Mitochondrial DNA B Resour. 2021, 6, 811–813. [Google Scholar] [CrossRef]
- Gargano, D.; Scotti, N.; Vezzi, A.; Bilardi, A.; Valle, G.; Grillo, S.; Cozzolino, S.; Cardi, T. Genome-wide analysis of plastome sequence variation and development of plastidial CAPS markers in common potato and related Solanum species. Genet. Resour. Crop Evol. 2012, 59, 419–430. [Google Scholar] [CrossRef]
- Yang, Y.; Dang, Y.; Li, Q.; Lu, J.; Li, X.; Wang, Y. Complete chloroplast genome sequence of poisonous and medicinal plant Datura stramonium: Organizations and implications for genetic engineering. PLoS ONE 2014, 9, e110656. [Google Scholar] [CrossRef]
- Raime, K.; Remm, M. Method for the Identification of Taxon-Specific k-mers from Chloroplast Genome: A Case Study on Tomato Plant (Solanum lycopersicum). Front Plant Sci. 2018, 9, 6. [Google Scholar] [CrossRef]
- Yan, L.; Lai, X.; Li, X.; Wei, C.; Tan, X.; Zhang, Y. Analyses of the complete genome and gene expression of chloroplast of sweet potato [Ipomoea batata]. PLoS ONE 2015, 10, e0124083. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. The chloroplast genome sequence of bittersweet (Solanum dulcamara): Plastid genome structure evolution in Solanaceae. PLoS ONE 2018, 13, e0196069. [Google Scholar] [CrossRef]
- Tamburino, R.; Sannino, L.; Cafasso, D.; Cantarella, C.; Orrù, L.; Cardi, T.; Cozzolino, S.; D’Agostino, N.; Scotti, N. Cultivated Tomato (Solanum lycopersicum L.) Suffered a Severe Cytoplasmic Bottleneck during Domestication: Implications from Chloroplast Genomes. Plants 2020, 9, 1443. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).