Meta-Analysis of Genetic Factors for Potato Starch Phosphorylation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Starch Isolation
2.3. DNA Isolation and Genotyping
2.4. Phosphorus Content Analysis
2.5. PCA and Population Structure Analysis
2.6. Partial Least Squares (PLS) Analysis
2.7. Association Analysis
2.8. Meta-Analysis
2.9. Basic Local Alignment Tool (BLAST)
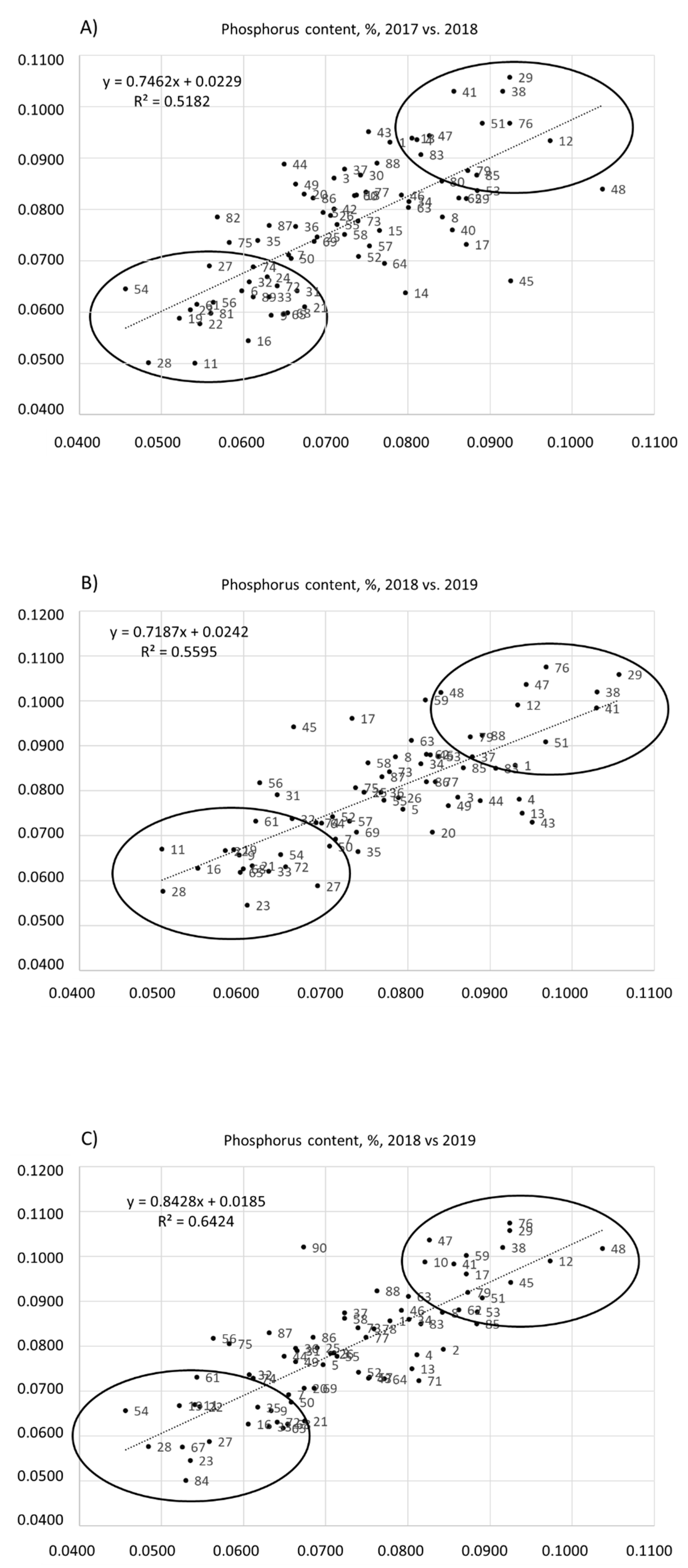
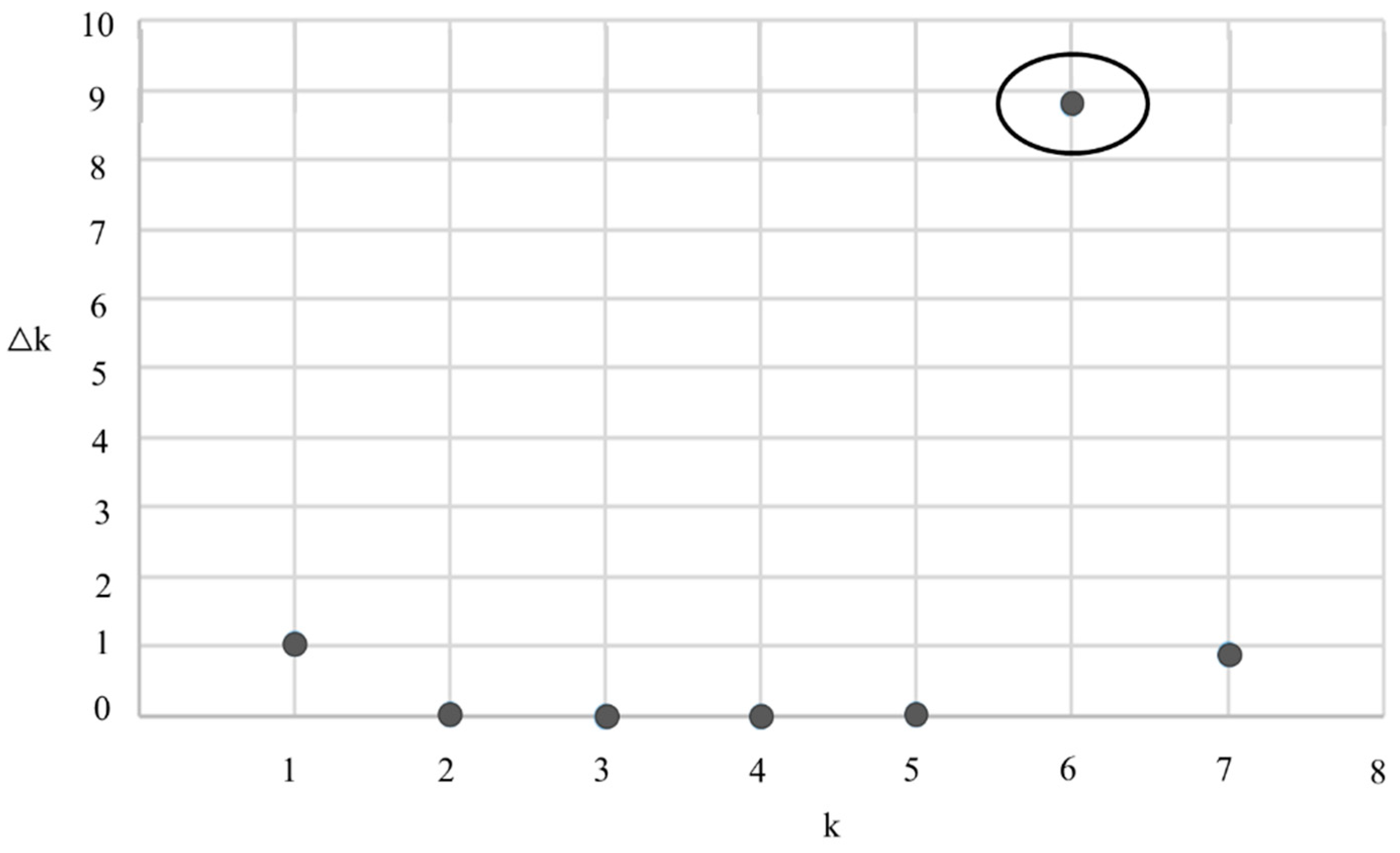
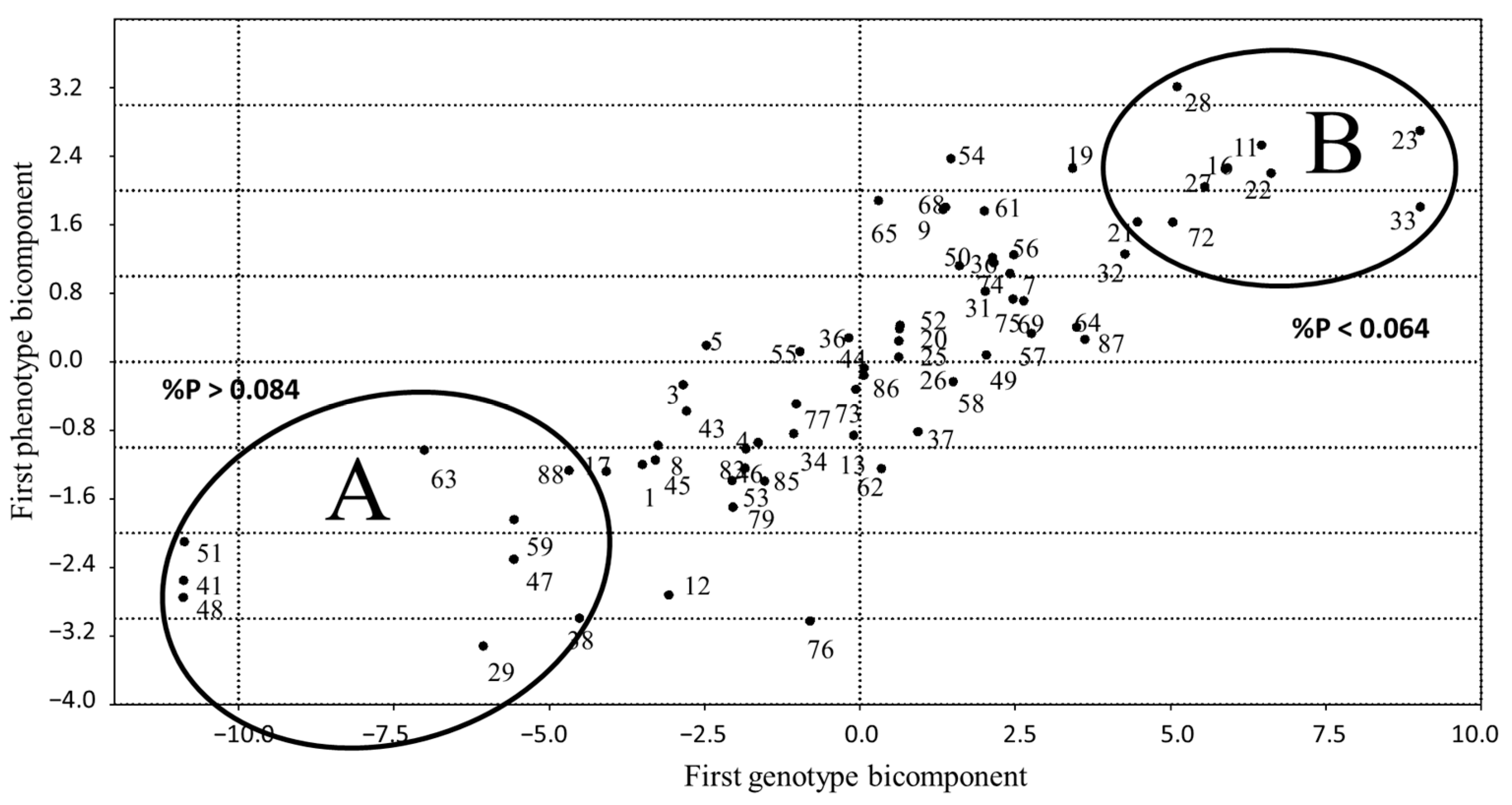
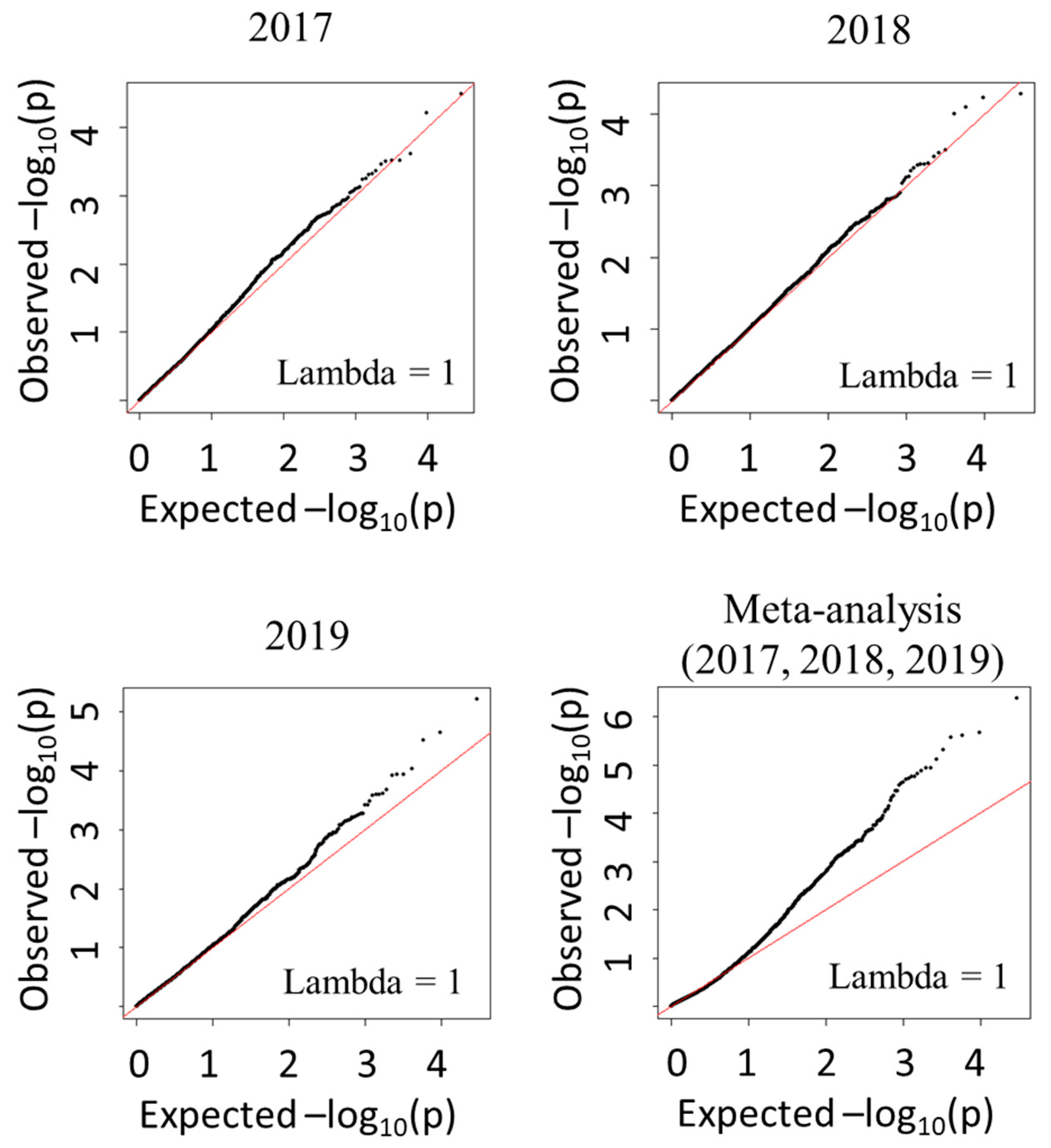
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khlestkin, V.K.; Peltek, S.E.; Kolchanov, N.A. Review of direct chemical and biochemical transformations of starch. Carbohydr. Polym. 2018, 181, 460–476. [Google Scholar] [CrossRef] [PubMed]
- Khlestkin, V.K.; Rozanova, I.V.; Efimov, V.M.; Khlestkina, E.K. Starch phosphorylation associated SNPs found by genome-wide association studies in the potato (Solanum tuberosum L.). BMC Genet. 2019, 20 (Suppl. S1), 29. [Google Scholar] [CrossRef] [Green Version]
- Law, M.; Jackson, D.; Turner, R.; Rhodes, K.; Viechtbauer, W. Two new methods to fit models for network meta-analysis with random inconsistency effects. BMC Med. Res. Methodol. 2016, 16, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, H.-J.; Li, X.-Q.; Kalinga, D.; Lim, S.-T.; Yada, R.; Liu, Q. Physicochemical properties of dry matter and isolated starch from potatoes grown in different locations in Canada. Food Res. Int. 2014, 57, 89–94. [Google Scholar] [CrossRef]
- Khlestkin, V.K.; Erst, T.V.; Rozanova, I.V.; Efimov, V.M.; Khlestkina, E.K. Genetic loci determining potato starch yield and granule morphology revealed by genome-wide association study (GWAS). PeerJ 2020, 8, e10286. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Palentological statistics software package for education and data analysis. Paleontol. Electron. 2001, 4, 9. [Google Scholar]
- Gower, J.C. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 1966, 53, 325–338. [Google Scholar] [CrossRef]
- Polunin, D.A.; Shtaiger, I.A.; Efimov, V.M. Development of the software package jacobi 4 for multidementional analysis of microchip data. Bull. Novosib. State Univ. Ser. Inf. Technol. 2014, 2, 90–98. (In Russian) [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genet 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Cinar, O.; Viechtbauer, W. The poolr Package for Combining Independent and Dependent p Values. J. Stat. Softw. 2022, 101, 1–42. [Google Scholar] [CrossRef]
- van den Berg, S.; Vandenplas, J.; van Eeuwijk, F.A.; Lopes, M.S.; Veerkamp, R.F. Significance testing and genomic inflation factor using high-density genotypes or whole-genome sequence data. J. Anim. Breed. Genet. 2019, 136, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.; Muñoz-Amatriaín, M.; Hokin, S.A.; Cisse, N.; Roberts, P.A.; Farmer, A.D.; Xu, S.; Close, T.J. A genome-wide association and meta-analysis reveal regions associated with seed size in cowpea [Vigna unguiculata (L.) Walp]. Theor. Appl. Genet. 2019, 132, 3079–3087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonova, I.N.; Skolotneva, E.S.; Salina, E.A. Genome-wide association study of leaf rust resistance in Russian spring wheat varieties. BMC Plant Biol. 2020, 20, 135. [Google Scholar] [CrossRef] [PubMed]
- Mengist, M.F. Investigating the Genetics and Physiological Basis of Differences in Cadmium and Zinc Concentrations in Tubers of Potato (Solanum tuberosum L.): Implications for Food Safety and Biofortification; University College Cork: Cork, Ireland, 2018. [Google Scholar]
- Hourcadec, D.; Garcia, A.D.; Post, T.W.; Taillon-Miller, P.; Holers, V.M.; Wagner, L.M.; Bora, N.S.; Atkinson, J.P. Analysis of the human regulators of complement activation (RCA) gene cluster with yeast artificial chromosomes (YACs). Genomics 1992, 12, 289–300. [Google Scholar] [CrossRef]
- Osbourn, A.E.; Field, B. Operons. Cell. Mol. Life Sci. 2009, 66, 3755–3775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uitdewilligen, J.G.A.M.L.; Wolters, A.M.A.; van Eck, H.J.; Visser, R.G.F. Allelic variation for alpha-Glucan Water Dikinase is associated with starch phosphate content in tetraploid potato. Plant Mol. Biol. 2022, 108, 469–480. [Google Scholar] [CrossRef] [PubMed]


| SNP | p-Value (2017) | p-Value (2018) | p-Value (2019) | p-Value Meta-Analysis |
|---|---|---|---|---|
| solcap_snp_c2_55899 | 6.14 × 10−5 | 7.97 × 10−5 | 7.00 × 10−4 | 4.23 × 10−7 |
| PotVar0091465 | 2.46 × 10−4 | 4.86 × 10−4 | 3.83 × 10−4 | 2.16 × 10−6 |
| PotVar0001436 | 7.86 × 10−4 | 5.98 × 10−5 | 1.20 × 10−3 | 2.46 × 10−6 |
| solcap_snp_c2_50231 | 3.49 × 10−4 | 9.93 × 10−5 | 1.87 × 10−3 | 2.69 × 10−6 |
| 2017 | 2018 | 2019 | |
|---|---|---|---|
| 2017 | 1 | ||
| 2018 | 0.72 * | 1 | |
| 2019 | 0.80 * | 0.75 * | 1 |
| Axis | Sing.Val. | % Covar |
|---|---|---|
| 1 | 5.5948 | 91.503 |
| 2 | 1.3609 | 5.4137 |
| 3 | 1.027 | 3.0835 |
| Year | First Bicomponent | Second Bicomponent | Third Bicomponent | |||
|---|---|---|---|---|---|---|
| Phenotype | Genotype | Phenotype | Genotype | Phenotype | Genotype | |
| 2017 | 0.93 * | 0.78 * | 0.14 | 0.09 | 0.36 * | 0.27 * |
| 2018 | 0.89 * | 0.74 * | 0.47 * | 0.32 * | 0.11 | 0.06 |
| 2019 | 0.94 * | 0.79 * | 0.22 | 0.21 | 0.20 | 0.21 |
| N | SNP | Ch | Position | Threshold | p-Value | Polymorphysm | Minor Allele | Minor Allele Frequency | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Current Study | Article [2] | Current Study, Meta-Analysis | Article [2] | |||||||
| 1 | solcap_snp_c2_55899 | 5 | 9,639,637 | Bonferroni | FDR | 4.23 × 10−7 | 9.01 × 10−6 | T/G | G | 0.30 |
| 2 | PotVar0091465 | 5 | 9,834,091 | Bonferroni | Below FDR | 2.16 × 10−6 | -- | A/G | G | 0.34 |
| 3 | PotVar0001436 | 5 | 20,274,370 | Bonferroni | Below FDR | 2.46 × 10−6 | -- | T/C | C | 0.38 |
| 4 | solcap_snp_c2_50231 | 5 | 36,834,630 | Bonferroni | FDR | 2.69 × 10−6 | 2.70 × 10−5 | T/C | C | 0.33 |
| N | SNP | Gene Code | Gene Statistics | Protein Name | Protein Statistics |
|---|---|---|---|---|---|
| 1 | solcap_snp_c2_50231 | -- | -- | -- | -- |
| 2 | solcap_snp_c2_55899 | PGSC0003DMG401016989 | Exons: 2, Coding exons: 1, Transcript length: 2961 bps, Translation length: 212 residues | LescPth4, or protein serine/threonine kinase | Ave. residue weight: 113.766 g/mol Charge: 0.0 Isoelectric point: 6.4888 Molecular weight: 24,118.36 g/mol Number of residues: 212 aa |
| PGSC0003DMG402016989 | Exons: 1, Coding exons: 1, Transcript length: 1372 bps, Translation length: 68 residues | Protein kinase family protein | Ave. residue weight: 115.902 g/mol Charge: 2.5 Isoelectric point: 8.0567 Molecular weight: 7,881.32 g/mol Number of residues: 68 aa | ||
| PGSC0003DMG400016991 | Exons: 1, Coding exons: 1, Transcript length: 1101 bps, Translation length: 205 residues | LescPth2, protein kinase | Ave. residue weight: 113.319 g/mol Charge: −5.0 Isoelectric point: 5.4433 Molecular weight: 36,375.41 g/mol Number of residues: 321 aa | ||
| PGSC0003DMG400016992 | Exons: 1, Coding exons: 1, Transcript length: 1448 bps, Translation length: 120 residues | LescPth2, protein kinase | Ave. residue weight: 113.414 g/mol Charge: 7.0 Isoelectric point: 10.3705 Molecular weight: 15,991.43 g/mol Number of residues: 141 aa | ||
| 3 | PotVar0091465 | PGSC0003DMG400007677 | Exons: 34, Coding exons: 33, Transcript length: 5008 bps, Translation length: 1464 residues | GWD, or Starch-granule-bound R1 protein | Ave. residue weight: 111.535 g/mol Charge: −10.0 Isoelectric point: 5.8464 Molecular weight: 163,287.07 g/molNumber of residues: 1464 aa |
| 4 | PotVar0001436 | -- | -- | -- | -- |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khlestkin, V.; Erst, T.; Igoshin, A.; Rozanova, I.; Khlestkina, E. Meta-Analysis of Genetic Factors for Potato Starch Phosphorylation. Agronomy 2022, 12, 1343. https://doi.org/10.3390/agronomy12061343
Khlestkin V, Erst T, Igoshin A, Rozanova I, Khlestkina E. Meta-Analysis of Genetic Factors for Potato Starch Phosphorylation. Agronomy. 2022; 12(6):1343. https://doi.org/10.3390/agronomy12061343
Chicago/Turabian StyleKhlestkin, Vadim, Tatyana Erst, Alexander Igoshin, Irina Rozanova, and Elena Khlestkina. 2022. "Meta-Analysis of Genetic Factors for Potato Starch Phosphorylation" Agronomy 12, no. 6: 1343. https://doi.org/10.3390/agronomy12061343
APA StyleKhlestkin, V., Erst, T., Igoshin, A., Rozanova, I., & Khlestkina, E. (2022). Meta-Analysis of Genetic Factors for Potato Starch Phosphorylation. Agronomy, 12(6), 1343. https://doi.org/10.3390/agronomy12061343








