Abstract
Wild relatives of cultivated potato are used in breeding to increase the genetic diversity of Solanum tuberosum (AAAA genome) varieties. Wild Mexican allotetraploid species Solanum stoloniferum (AABB genome) was used in breeding for extreme resistance to viruses and late blight. In this study, genomic in situ hybridization (GISH) was used for visualization of introgression of genetic material of the B subgenome of S. stoloniferum into the genome of backcross hybrids. The fertile hexaploid hybrid had 48 chromosomes of the A genome and 24 chromosomes of the B subgenome. Plants of the BC1 generation were pentaploid having the AAAAB genome constitution and three selected BC2 hybrids were aneuploid, containing one to six chromosomes of the B subgenome and 48 chromosomes of the A genome. The B subgenome of S. stoloniferum was inherited in the backcross generations as single chromosomes and in rare cases as recombinant chromosomes. GISH showed that chromosome pairing in the backcross hybrids was predominantly intragenomic. Most chromosomes of the B subgenome remained as univalents in backcross hybrids. Rare homeologous A/B chromosome pairing was detected in all analyzed hybrids. The obtained data indicate that the B subgenome of S. stoloniferum was able to recombine with the A genome.
1. Introduction
Transferring genes/Quantitative Trait Loci conferring desirable traits via interspecific hybridization and classical breeding still remains an effective approach in breeding programs aimed at developing new varieties resistant to biotic and abiotic stresses. This is especially relevant for the common potato Solanum tuberosum L. Many wild relatives of this cultivated species were used in breeding to increase the genetic diversity of the varieties’ gene pool and to introduce alien genes conferring resistance to diseases and pests [1,2,3,4].
Wild potato relatives constitute a complex of di-, tri-, tetra-, penta- and hexaploid species of the section Petota Dumort of the Solanum L. genus having the same basic chromosome number x = 12. About 60% of the 107 wild potato species are diploid, and the remaining are polyploid [5]. Genomic in situ hybridization (GISH) is successfully used for the detection of allopolyploidy and discrimination of different subgenomes of wild allopolyploid species [6,7]. Among them, the wild allotetraploid Mexican potato species Solanum stoloniferum Schltdl. is known as a source of resistance to late blight [8,9], to potato virus Y (PVY) which can lead to the yield losses on susceptible varieties as high as 80% [1,2,10,11,12,13], and is characterized by aphid resistance [14] and by abiotic stress (heat) tolerance [15]. Solanum stoloniferum belongs to the secondary potato gene pool [16]. Interspecific hybridization with cultivated potato is complicated by unilateral interspecific incompatibility (UI) [17,18], differences in endosperm balance numbers (EBNs) [19,20,21,22,23] and male-sterility of interspecific hybrids having cytoplasm of wild species [1,24,25,26].
GISH analysis supports an AABB genome constitution and hybrid origin of S. stoloniferum, with wild Mexican diploid species S. verrucosum (or its progenitor) supported as the A subgenome donor and other diploid Mexican species (for example, S. jamesii Torrey, S. bulbocastanum Dunal) as the donor of B subgenome [6]. GISH confirms the strict allopolyploid nature of S. stoloniferum, which is characterized by the only intragenomic chromosome pairing, but not the intergenomic chromosome associations [6].
Since S. stoloniferum (2n = 4x = 48, AABB genome, EBN = 2) and S. tuberosum (2n = 4x = 48, AAAA genome, EBN = 4) have different EBN values, involvement of this wild species into breeding is based on the use of artificial polyploids, unreduced gametes [27,28,29,30] or using bridge species for crossing with tetraploid S. tuberosum [31]. Introgression is achieved using several breeding schemes. Usually, interspecific hybrids are obtained following pollinations of S. stoloniferum with pollen of fertile diploid clones of S. tuberosum (2n = 2x = 24, AA, EBN = 2). The resulting triploid hybrids (2n = 3x = 36, AAB) are sterile, and the further breeding cycle is based on their polyploidization, leading to the production of fertile hexaploid hybrids [29,30,31]. Because of UI, hybrid seeds are usually obtained when wild species are used as a female parent [17,18].
Another way to involve S. stoloniferum (2n = 4x = 48, EBN = 2) in breeding is the use of selected clones of this wild species which produce unreduced gametes (2n = 4x = 48, EBN = 2) and to cross them with potato diploid lines (2n = 4x = 48, EBN = 2) [19,31,32] or the use in interspecific crosses of the octaploid clones (2n = 8x = 96, EBN = 4) of S. stoloniferum and tetraploid potato varieties (2n = 4x = 48, EBN = 4) [19,27,28,31]. In exceptional cases, hybrid seeds can be obtained in crosses between the species with different EBN [33,34]. Thus, tetraploid interspecific hybrids (AAAB) were obtained after numerous controlled pollinations in crosses of S. stoloniferum (2n = 4x = 48, AABB genome, EBN = 2) with tetraploid S. tuberosum (2n = 4x = 48, AAAA genome, EBN = 4) [20,32,35].
Introgressive lines in different breeding schemes were produced following further pollinations of interspecific hybrids with pollen of fertile varieties or diploid breeding clones. Despite the fact that interspecific hybrids with S. stoloniferum are used in potato breeding, the specificity of the introgression of wild genome and the potential for intergenomic recombination are still unclear.
Recently, Yermishin with colleagues realized several breeding schemes to involve S. stoloniferum into interspecific crosses both as female and as male parent [23,36,37]. Here, we analyzed these hybrids and their BC progenies to investigate the potential of the introgression of genetic material of the B subgenome of S. stoloniferum into the genomes of selected backcross hybrids using GISH and to study the possibility of the homeologous chromosome pairing.
2. Materials and Methods
2.1. Plant Materials
In the previous study of Yermishin with colleagues [23,36], sexual hybrids were produced both with S. stoloniferum as a female parent and as a male parent (Figure 1). Plant material in the present study was represented by selected backcross clones derived from two breeding schemes:
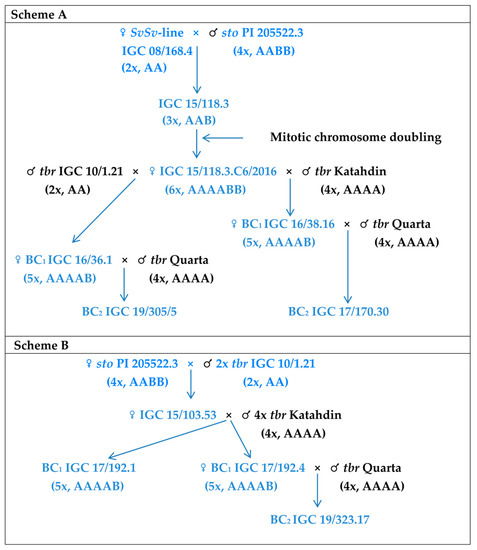
Figure 1.
Crosses used for development of the introgressive forms which were analyzed in the present study. Expected genome composition is indicated in brackets. All pollinators are marked by black color, and maternal hybrid forms by blue color.
- -
- Scheme ‘A’—sterile triploid hybrid was obtained in crosses between the SvSv-diploid line [diploid SvSv-line IGC 08/168.4 (F2 dihaploid S. tuberosum × S. verrucosum) [23,36] and S. stoloniferum, PI 205522 (Figure 1). After colchicine treatment of this triploid hybrid, a fertile hexaploid hybrid IGC 15.118.3C6/2016 was obtained as the result of chromosome doubling;
- -
- Scheme ‘B’—hybrid material was developed in interspecific crosses involved S. stoloniferum as a female parent with pollen of the fertile diploid line IGC 10/1.21 (Figure 1).
These two schemes of interspecific crosses, data on confirming hybridity with molecular markers and morphology, results of pathogen resistance tests and data of chromosome counts made by conventional acetocarmine method were described earlier [23,37].
Backcross progenies in both schemes were generated from mature seeds produced after hand emasculations and repeated pollinations of the interspecific hybrids by the pollen of male fertile varieties Katahdin or Quarta (Figure 1).
Seven introgressive forms were used to prepare mitotic and meiotic chromosome spreads for GISH including: an F1 (chromosome doubling) hybrid 15/118.3; three BC1 clones (IGC 16/38.16, IGC 17/192.1, IGC 17/192.4); three BC2 clones (IGC 17/170.30, IGC 19/305.5, IGC 19/323.17). Backcross hybrids selected for the present study had normal growth and morphology, good pollen fertility, as well as resistance to late blight and to PVY [23,36,37].
2.2. Genomic In Situ Hybridization of the Introgressive Forms
Plant material for GISH assays was grown in a greenhouse. Root tips and flower buds were used to determine the genome composition of the hybrids. Root tips were fixed in Carnoy’s solution (an ethanol–glacial acetic acid = 3:1) after pretreatment in water with ice for 24 h and further stored at −20 °C for chromosome counts. The flower buds were fixed directly in Carnoy’s solution and then stored at −20 °C for meiotic analysis in pollen mother cells (PMC). Chromosome slides were prepared after the enzymatic treatment of anthers and root meristems. The enzyme solution contained 4% cellulase (1.14 U/mg) and 1% pectolyase (0.94 U/mg), the time of incubation was 65–120 min.
Genomic DNA was isolated from green leaves according to the protocol of Bernatzky and Tanksley [38]. For GISH, DNA was isolated from the leaves of the wild A genome diploid species S. verrucosum (PI 545745) and the B genome diploid species S. jamesii (PI 458424).
DNA of both species was labeled using Nick-Translation Digoxigenin-NT Labeling Kit or BIO-NT Labeling Kit (Jena Bioscience (Jena, Germany), PP-310-DIGX and PP-310-BIO16). FITS anti-DIG conjugate (FAB fragments) (Roche 11207741910) and Rhodamine conjugated avidin Red TM-X (Thermo Fisher Scientific (Waltham, MA, USA) S6366) were used in a two-color GISH. Differentially labeled DNA of the A and B genomic species was used in the hybridization mix. GISH was performed according to the standard techniques [39] with slight modifications [6].
An AxioImager M2 epifluorescence microscope with an AxioCamMRm camera and AxioVision Rel 4.8 software was used to analyze the slides and create and process images. In addition, Adobe Photoshop 6.0 was used for image processing.
3. Results
3.1. Genomic In Situ Hybridization of the Introgression Forms
Chromosomes of the A subgenome of S. stoloniferum and the A genome of S. tuberosum are indistinguishable in GISH analysis of the interspecific hybrids. The B subgenome of S. stoloniferum was identified using S. jamesii genomic DNA. Table 1 presents the content of the B subgenome of S. stoloniferum in analyzed hybrid material.

Table 1.
Genomic constitution of the backcross hybrids of cultivated potato with S. stoloniferum.
Fertile hexaploid hybrid IGC 15.118.3C6/2016 derived from the chromosome doubling of the primary F1 sterile triploid hybrid had, as expected, AAAABB genome composition with 48 chromosomes of the A genome and 24 chromosomes of the B subgenome (Figure 2a).
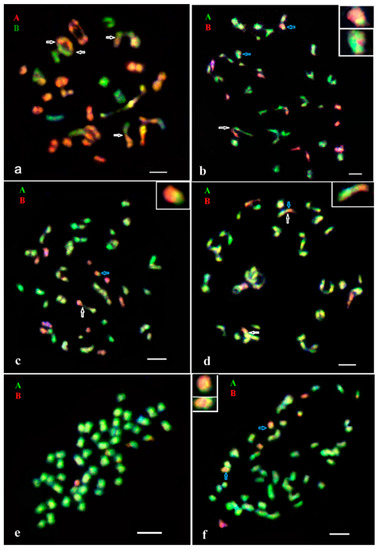
Figure 2.
GISH analysis of chromosome content and homeologous pairing in hybrids of different generations. (a) Diakinesis in PMC of F1 (chromosome doubling) hybrid IGC 15/118.3.C6.2016: 48 chromosomes of the A genome and 24 chromosomes of the B subgenome of S. stoloniferum, three intergenomic chromosome associations were identified (2 IV A/A/B/B and 1 II A/B). (b) Diakinesis in PMC of BC1 hybrid IGC 16/38.16: 48 chromosomes of the A genome (one of them with the B subgenome introgression) and 12 chromosomes of the B subgenome (one of them with the A genome introgression), A/A/B trivalent is detected. (c) Diakinesis in PMC of BC1 hybrid IGC 17/192.4: 48 chromosomes of the A genome and 12 chromosomes of the B subgenome (one of them with the A genome introgression), A/B rod bivalent is observed. (d) Diakinesis in PMC of BC2 hybrid IGC 17/170.30: 48 chromosomes of the A genome and five chromosomes of the B subgenome (one of them with the A genome introgression), two A/B rod bivalents are observed. (e) Somatic chromosomes of BC2 hybrid IGC 19/323.17 hybrid: 48 chromosomes of the A genome and one chromosome of the B subgenome of S. stoloniferum. (f) Somatic chromosomes of BC2 hybrid IGC 19/305.5:48 chromosomes of the A genome and six chromosomes of the B subgenome (one of them with the A genome introgression). The color of the fluorescence of the B subgenome (labeled DNA probe of S. jamesii) and the A genome (DNA probe of S. verrucosum) corresponds to the color of the letters above the images. Blue arrows show recombinant chromosomes. Homeologous pairing between the A genome chromosomes and the B subgenome chromosomes are indicated by white arrows. Scale bar = 5 μm.
GISH revealed the AAAAB constitution of the BC1 hybrid plants. All three BC1 hybrids were pentaploid (2n = 60) having 48 chromosomes of the A genome and 12 of the B subgenome of S. stoloniferum (Table 1, Figure 2b,c).
Three selected BC2 hybrids were aneuploids as expected (Table 1), indicating the loss of the B subgenome chromosomes in the backcross progeny. According to the GISH results, these BC2 hybrids have the chromosome counts of 2n = 48A + 1B (IGC 19/323.17) (Figure 2e); 2n = 48A + 5B (IGC 17/170.30) (Figure 2d); 2n = 48A + 6B (IGC 19/305.5) (Figure 2f).
One-two A/B recombinant chromosomes were observed in four of the six analyzed backcross hybrids (Table 1, Figure 2b–d,f). The GISH signal from the A genome probe was located at the terminal parts of the recombinant chromosomes of the B subgenome. In one hybrid (BC1 IGC 16/38.16) the B subgenome fragment was detected in the terminal part of one A genome chromosome (Table 1, Figure 2d). It is also possible that some of the introgressed fragments were out of range of the GISH resolution.
Detection of the A/B recombinant chromosomes in two BC1 (IGC 16/38.16, IGC 17/192.4) and in two BC2 (IGC 17/170.30, IGC 19/305.5) genotypes indicates that homoeologous recombination occurred in earlier generations (F1 and in BC1 plants) (Table 1, Figure 2a–c). To confirm the occurrence of intergenomic recombination, GISH analysis of meiosis was further performed for corresponding F1 and BC1 plants.
3.2. Chromosome Pairing in Interspecific Hybrids of Different Generations
Meiotic chromosome pairing at diakinesis was studied in three backcross hybrids and in F1 (chromosome doubling) hybrid (Table 2, Figure 2a–d).

Table 2.
Mean chromosome pairing at diakinesis in selected hybrid genotypes analyzed by GISH.
GISH showed that chromosome pairing was predominantly intragenomic, representing the A genome chromosome associations with the mean number of about 20 per cell presented mainly by A/A bivalents (Table 2, Figure 2a–d). The number of the A genome multivalents in all analyzed genotypes was extremely low.
The most chromosomes of the B subgenome of S. stoloniferum were paired in the hexaploid F1 (chromosome doubling) hybrid (AAAABB genome) as expected. Whereas in the BC1 and BC2 hybrids, the most chromosomes of the B subgenome of S. stoloniferum were represented by univalents with the exception of a few chromosomes of the B genome included in various intergenomic associations (Table 2). For example, the pentaploid BC1 hybrids (AAAAB genome) had more than nine univalents of the B subgenome on average per PMC (Table 2).
The homoeologous pairing between chromosomes of A and B genomes was seen at diakinesis relatively seldom, nevertheless, it was detected in all analyzed genotypes (Table 2, Figure 2a–d).
Intergenomic pairing was revealed in the hexaploid F1 (chromosome doubling) hybrid with an average frequency of 0.82 per PMC (0.41 A/B bivalents, 0.18 A/A/B trivalents, 0.12 A/A/B/B quadrivalents) (Table 2, Figure 2a).
In the pentaploid BC1 hybrids intergenomic pairing was observed with an average frequency 2.7 per cell (2.36 A/B bivalents, 0.14 A/A/B trivalents and 0.05 A/A/A/B quadrivalents) (Table 2, Figure 2b,c). The average frequency of homoeologous pairing in the BC2 hybrid was 1.08 per cell (0.96 A/B bivalents and 0.04 A/A/A/B quadrivalents) (Table 2, Figure 2d). Most homeologous A/B bivalents were rod; ring A/B bivalents were observed in single meiocytes.
4. Discussion
The use of S. stoloniferum to improve potato varieties dates back more than 60 years [1]. This wild species was utilized in breeding mainly for transferring extreme resistance to potato virus Y. The genes Rysto and Ry-fsto conferring PVY resistance were introgressed by classical breeding from S. stoloniferum into potato germplasm, which was further used to produce some PVY-resistant European varieties [12,40,41,42,43]. These genes were mapped on chromosome XII, and Rysto was recently isolated from the potato dihaploid clone [44].
Another gene Rpi-sto1 conferring broad-spectrum resistance to late blight, which was mapped on chromosome VIII [8], was also introduced via conventional breeding into several varieties [45,46,47]. Rpi-sto1 is the functional Rb/Rpi-blb1 homologue which was identified in diploid ancestor species of S. stoloniferum [8,48,49]—the B-genome species of the series Bulbocastana, Pinnatisecta (according to system of Hawkes, 1990 [50]) and in the A genome wild species S. verrucosum. However, it is still unknown which of the subgenomes of S. stoloniferum contributes functional alleles of the Rysto, Ry-fsto, Rpi-sto1 into potato varieties. Furthermore, the potential of intergenomic recombination in interspecific hybrids and their progenies is not fully understood.
Few studies were reported in the literature regarding the possibilities of homeologous pairing in interspecific hybrids between S. stoloniferum and the A genome potato species. In conventional cytological meiotic analysis (acetocarmine or acetorcein chromosome staining) multivalents were observed—about two trivalents per meiocyte in triploid hybrids [29,51,52], and rare quadrivalents were detected at metaphase I of tetraploid hybrids [31,32,37]. These results could indicate the possibility of intergenomic pairing.
In recent decades GISH was successfully used to study alien introgression and homeologous pairing in somatic hybrids between potato, S. tuberosum and distantly related non-tuber bearing species belonging to other sections of the genus Solanum: Etuberosum [53,54,55,56,57,58], Lycopersicum [59,60] and the Solanum nigrum complex—also known as Solanum L. section Solanum [61]. In these distant hybrids, the parental genomes were easily discriminated by GISH and alien chromatine introgression was distinguishable in backcross progenies of somatic hybrids (Reviews: [62,63,64,65]). Within the section Petota to which cultivated potato belongs, GISH was able to discriminate parental genomes in somatic hybrids between S. tuberosum (AAAA) and wild diploid species S. bulbocastanum (BB) from the tertiary gene pool [66,67]. In these studies, GISH helped to detect alien chromosome transmission and introgressions in backcross progenies. However, distinguishing chromatin using GISH for more closely related potato species within the section Petota is more difficult or impossible. Thus, parental genomes could not be discriminated through GISH in hybrids between diploid cultivated potato and South American wild diploid species S. commersonii [68], which belongs to the tertiary gene pool of potato [69].
Detection of homeologous chromosome pairing in the present study suggests meiotic recombination as the possible mechanism of alien fragment introgressions in interspecific hybrids of potato with S. stoloniferum and their backcross progenies. To our knowledge, the present research is the first GISH study of the introgression into potato genome A of the chromosomes/chromosomal segments of the subgenome B of S. stoloniferum which belongs to the secondary gene pool. Our results provide evidence that homeologous recombination between chromosomes of the B subgenome of S. stoloniferum and the A genome might occur. In four of the seven analyzed hybrids, intergenomic recombinant chromosomes were observed. Small terminal regions have been recombined into one or two chromosomes of A or B genomes. Besides hybrids carrying A/B recombinant chromosomes, one BC2 hybrid with a single additional alien chromosome was distinguished; this indicates potential for receiving addition lines.
Our results improve the understanding of the introgression of S. stoloniferum genetic material into the potato genome and can help to plan introgression breeding to contribute adaptive traits, including resistance to aphids and tolerance to abiotic stresses from this wild allotetraploid species into the varieties’ gene pool.
In future, it will be of value to observe the transmission of the B subgenome chromosomes and stability of recombinant chromosomes in introgressive lines over several cross generations. We plan to focus our research on GISH and FISH with Chromosome-Specific Cytogenetic DNA Markers (CSCDMs). This approach helped to identify individual chromosomes in interspecific hybrids of Solanum species [55,58,70] as well as in a wide hybrids of other crops having small chromosomes [71,72].
5. Conclusions
New data about the potential of homeologous pairing and intergenomic recombination between chromosomes of the subgenome B of wild Mexican species S. stoloniferum and chromosomes of the A genome of potato were obtained using the GISH method. Genomic in situ hybridization allows visualization of the chromosomes/chromosomal segments of the B subgenome of S. stoloniferum into the backcross progenies. Chromosomes of the B subgenome of S. stoloniferum undergo the process of chromosome elimination during backcrosses with cultivated potato. Recombinant A/B chromosomes were revealed in backcross hybrids. Detection of intergenomic pairing suggests meiotic recombination as the possible mechanism of alien segment introgressions.
Author Contributions
Conceptualization, T.A.G., G.I.P. and A.P.Y.; Methodology, G.I.P.; GISH analysis, G.I.P.; Resources, A.P.Y.; Writing, T.A.G.; Project Administration, T.A.G. All authors have read and agreed to the published version of the manuscript.
Funding
The paper was prepared with assistance provided within the framework of a grant 20-54-00043-bel-a from the Russian Foundation of Basic Research (RFBR).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors express their deep gratitude to N. Fomina for her kind help with preparing the manuscript for submission.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ross, H. Potato breeding–problems and perspectives. In Supplement 13 Advances in Plant Breeding; Parey, P., Ed.; Theoretical and Applied Genetics: Berlin/Heidelberg, Germany, 1986. [Google Scholar]
- Hawkes, J.G. Origins of cultivated potatoes and species relationships. In Potato Genetics; Bradshaw, J.E., Mackay, G.R., Eds.; CAB International: Wallingford, UK, 1994; pp. 3–42. [Google Scholar]
- Bradshaw, J.E.; Ramsay, G. Chapter 1–Potato Origin and Production. In Advances in Potato Chemistry and Technology; Singh, J., Kaur, L., Eds.; Academic Press: San Diego, CA, USA, 2009; pp. 1–26. [Google Scholar] [CrossRef]
- Watanabe, K.; Ortiz, R.; Handayani, T. Potato Germplasm Enhancement with Genetic Resources and Biotechnology Applications and Related Achievements in the Early Years of the International Potato Center (CIP). Jpn. Agric. Res. Q. 2022, 55, 405–418. [Google Scholar] [CrossRef]
- Spooner, D.M.; Ghislain, M.; Simon, R.; Jansky, S.H.; Gavrilenko, T. Systematics, Diversity, Genetics, and Evolution of Wild and Cultivated Potatoes. Bot. Rev. 2014, 80, 283–383. [Google Scholar] [CrossRef]
- Pendinen, G.; Gavrilenko, T.; Jiang, J.; Spooner, D.M. Allopolyploid Speciation of the Mexican Tetraploid Potato Species Solanum stoloniferum and S. hjertingii Revealed by Genomic In Situ Hybridization. Genome 2008, 51, 714–720. [Google Scholar] [CrossRef] [PubMed]
- Pendinen, G.; Spooner, D.M.; Jiang, J.; Gavrilenko, T. Genomic in Situ Hybridization Reveals Both Auto- and Allopolyploid Origins of Different North and Central American Hexaploid Potato (Solanum Sect. Petota) Species. Genome 2012, 55, 407–415. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, M.; Allefs, S.; Berg, R.; Vleeshouwers, V.; Vossen, E.; Vosman, B. Allele Mining in Solanum: Conserved Homologues of Rpi-Blb1 Are Identified in Solanum stoloniferum. Theor. Appl. Genet. 2008, 116, 933–943. [Google Scholar] [CrossRef]
- Karki, H.S.; Jansky, S.H.; Halterman, D.A. Screening of Wild Potatoes Identifies New Sources of Late Blight Resistance. Plant Disease 2021, 105, 368–376. [Google Scholar] [CrossRef]
- Solomon-Blackburn, R.M.; Barker, H. A Review of Host Major-Gene Resistance to Potato Viruses X, Y, A and V in Potato: Genes, Genetics and Mapped Locations. Heredity 2001, 8, 8–16. [Google Scholar] [CrossRef]
- Valkonen, J.P.T.; Wiegmann, K.; Hämäläinen, J.H.; Marczewski, W.; Watanabe, K.N. Evidence for Utility of the Same PCR-Based Markers for Selection of Extreme Resistance to Potato Virus Y Controlled by Rysto of Solanum stoloniferum Derived from Different Sources. Ann. Appl. Biol. 2008, 152, 121–130. [Google Scholar] [CrossRef]
- Valkonen, J.P.T.; Gebhardt, C.; Zimnoch-Guzowska, E.; Watanabe, K. Resistance to Potato Virus Y in Potato. In Potato Virus Y Biodiversity, Pathogenicity, Epidemiology and Management; Lacomme, C., Glais, L., Bellstedt, D.U., Dupuis, B., Karasev, A.V., Jacquot, E., Eds.; Springer: Cham, Germany, 2017; pp. 207–241. [Google Scholar] [CrossRef]
- Ahmadvand, R.; Takács, A.; Taller, J.; Wolf, I.; Polgár, Z. Potato Viruses and Resistance Genes in Potato. Acta Agron. Hung. 2012, 60, 283–298. [Google Scholar] [CrossRef]
- Alvarez, A.E. Resistance Mechanisms of Solanum Species to Myzus Persicae. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 2007. Available online: http://edepot.wur.nl/25052 (accessed on 1 December 2021).
- Handayani, T.; Gilani, S.A.; Watanabe, K.N. Climatic Changes and Potatoes: How Can We Cope with the Abiotic Stresses? Breed. Sci. 2019, 69, 545–563. [Google Scholar] [CrossRef]
- Castañeda-Álvarez, N.P.; de Haan, S.; Juárez, H.; Khoury, C.K.; Achicanoy, H.A.; Sosa, C.C.; Bernau, V.; Salas, A.; Heider, B.; Simon, R.; et al. Ex Situ Conservation Priorities for the Wild Relatives of Potato (Solanum L. Section Petota). PLoS ONE 2015, 10, e0122599. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.A.; Hanneman, R.E. Crossability between Cultivated and Wild Tuber-and Non-Tuber-Bearing Solanums. Euphytica 1999, 109, 51–67. [Google Scholar] [CrossRef]
- Hayes, R.J.; Dinu, I.I.; Thill, C.A. Unilateral and Bilateral Hybridization Barriers in Inter-Series Crosses of 4x 2EBN Solanum stoloniferum, S. pinnatisectum, S. cardiophyllum, and 2x 2EBN S. tuberosum Haploids and Haploid-Species Hybrids. Sex. Plant Reprod. 2005, 17, 303–311. [Google Scholar] [CrossRef]
- Brown, C.R. Characteristics of 2N pollen producing triploid hybrids between Solanum stoloniferum and cultivated diploid potatoes. Am. Potato J. 1988, 65, 75–84. [Google Scholar] [CrossRef]
- Singsit, C.; Hanneman, R.E. Rescuing Abortive Inter-EBN Potato Hybrids through Double Pollination and Embryo Culture. Plant Cell Rep. 1991, 9, 475–478. [Google Scholar] [CrossRef]
- Ortiz, R.; Ehlenfeldt, M.K. The Importance of Endosperm Balance Number in Potato Breeding and the Evolution of Tuber-Bearing Solanum Species. Euphytica 1992, 60, 105–113. [Google Scholar] [CrossRef]
- Cho, H.M.; Kim-Lee, H.Y.; Om, Y.H.; Kim, J.K. Influence of endosperm balance number (EBN) in interploidal and interspecific crosses between Solanum tuberosum dihaploids and wild species. Korean J. Breed. 1997, 29, 154–161. [Google Scholar]
- Yermishin, A.; Levy, A.; Voronkova, E.; Polyukhovich, Y.; Ageeva, A. Overcoming Unilateral Incompatibility in Crosses with Wild Allotetraploid Potato Species Solanum stoloniferum Schldtl. & Bouchet. Euphytica 2017, 213, 249. [Google Scholar] [CrossRef]
- Lössl, A.; Götz, M.; Braun, A.; Wenzel, G. Molecular Markers for Cytoplasm in Potato: Male Sterility and Contribution of Different Plastid-Mitochondrial Configurations to Starch Production. Euphytica 2000, 116, 221–230. [Google Scholar] [CrossRef]
- Song, Y.; Schwarzfischer, A. Development of STS Markers for Selection of Extreme Resistance (RySto) to PVY and Maternal Pedigree Analysis of Extremely Resistant Cultivars. Am. J. Potato Res. 2008, 85, 159–170. [Google Scholar] [CrossRef]
- Anisimova, I.; Gavrilenko, T. Cytoplasmic Male Sterility and Prospects for Its Utilization in Potato Breeding, Genetic Studies and Hybrid Seed Production. Russ. J. Genet. Appl. Res. 2017, 7, 721–735. [Google Scholar] [CrossRef]
- Swaminathan, M.S. Notes on induced polyploids in the tuber-bearing Solanum species and their crossability with S. tuberosum. Am. Potato J. 1951, 28, 472–489. [Google Scholar] [CrossRef]
- Lamm, R. Investigations on Some Tuber-Bearing Solanum Hybrids. Hereditas 1953, 39, 97–112. [Google Scholar] [CrossRef]
- Ramanna, M.S.; Abdalla, M.M.F. Fertility, Late Blight Resistance and Genome Relationship in an Interspecific Hybrid, Solanum Polytrichon Rydb × S. phureja Juz. et Buk. Euphytica 1970, 19, 317–326. [Google Scholar] [CrossRef]
- Adiwilaga, K.D.; Brown, C.R. Use of 2n Pollen-Producing Triploid Hybrids to Introduce Tetraploid Mexican Wild Species Germ Plasm to Cultivated Tetraploid Potato Gene Pool. Appl Genet. 1991, 81, 645–652. [Google Scholar] [CrossRef]
- Bamberg, J.B.; Hanneman, R.E.; Palta, J.P.; Harbage, J.F. Using Disomic 4x(2EBN) Potato Species’ Germplasm via Bridge Species Solanum Commersonii. Genome 1994, 37, 866–870. [Google Scholar] [CrossRef]
- von Wangenheim, K.H. Zur Ursache der Kreuzungsschwierigkeiten zwischen Solanum tuberosum L. und S. acaule Bitt. bzw. S. stoloniferum Schlechtd et Bouche. Z. Pflanz. J. Plant Breed. 1954, 34, 7–48. [Google Scholar]
- Bamberg, J.B.; Hanneman, R.E. Allelism of Endosperm Balance Number (EBN) in Mexican Tuber-Bearing Solanum Species. Appl. Genet. 1990, 80, 161–166. [Google Scholar] [CrossRef]
- Jansky, S.; Hamernik, A. The Introgression of 2× 1EBN Solanum Species into the Cultivated Potato Using Solanum Verrucosum as a Bridge. Genet. Resour. Crops Evol. 2009, 56, 1107–1115. [Google Scholar] [CrossRef]
- Panahandeh, J. Chromosome Pairing in Auto-Allotetraploid (AAAB) Interspecific Hybrid Potatoes. N. Z. J. Crops Hortic. Sci. 2019, 47, 11–18. [Google Scholar] [CrossRef]
- Yermishin, A.; Polyukhovich, Y.; Voronkova, E.; Gukasyan, O. SvSv-Lines Is an Effective Tool for Involvement of the Valuable Genepool of 1 EBN Diploid Potato Species into Breeding. Vavilov J. Genet. Breed. 2017, 21, 42–50. [Google Scholar] [CrossRef][Green Version]
- Antonova, O.Y.; Yermishin, A.P.; Levy, A.V.; Ageeva, A.S.; Voronkova, E.V.; Gavrilenko, T.A. Development of chromosome-specific markers for a study on introgressive hybridization of potato with the wild Mexican allotetraploid species Solanum stoloniferum Schltdl. Plant Biotechnol. Breed. 2019, 2, 24–35. [Google Scholar] [CrossRef]
- Bernatzky, R.; Tanksley, S.D. Genetics of Actin-Related Sequences in Tomato. Appl. Genet. 1986, 72, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Leitch, A.R.; Schwarzacher, T.; Jackson, D.P. In Situ Hybridisation, A Practical Guide; BIOS Scientific Publishers: Oxford, UK, 1994. [Google Scholar]
- Van Berloo, R.; Hutten, R.C.B.; van Eck, H.J.; Visser, R.G.F. An Online Potato Pedigree Database Resource. Potato Res. 2007, 50, 45–57. [Google Scholar] [CrossRef]
- Song, Y.-S.; Hepting, L.; Schweizer, G.; Hartl, L.; Wenzel, G.; Schwarzfischer, A. Mapping of Extreme Resistance to PVY (RySto) on Chromosome XII Using Anther-Culture-Derived Primary Dihaploid Potato Lines. Appl. Genet. 2005, 111, 879–887. [Google Scholar] [CrossRef]
- Flis, B.; Hennig, J.; Strzelczyk-Żyta, D.; Gebhardt, C.; Marczewski, W. The Ry-Fstogene from Solanum stoloniferum for Extreme Resistant to Potato Virus Y Maps to Potato Chromosome XII and Is Diagnosed by PCR Marker GP122718 in PVY Resistant Potato Cultivars. Mol. Breed. 2005, 15, 95–101. [Google Scholar] [CrossRef]
- Cernák, I.; Decsi, K.; Nagy, S.; Wolf, I.; Polgár, Z.; Gulyás, G.; Hirata, Y.; Taller, J. Development of a Locus-Specific Marker and Localization of the Rysto Gene Based on Linkage to a Catalase Gene on Chromosome XII in the Tetraploid Potato Genome. Breed. Sci. 2008, 58, 309–314. [Google Scholar] [CrossRef][Green Version]
- Grech-Baran, M.; Witek, K.; Szajko, K.; Witek, A.I.; Morgiewicz, K.; Wasilewicz-Flis, I.; Jakuczun, H.; Marczewski, W.; Jones, J.D.; Hennig, J. Extreme Resistance to Potato Virus Y in Potato Carrying the Rysto Gene Is Mediated by a TIR-NLR Immune Receptor. bioRxiv 2018, 445031. [Google Scholar] [CrossRef]
- Gavrilenko, T.A.; Klimenko, N.S.; Antonova, O.Y.; Lebedeva, V.A.; Evdokimova, Z.Z.; Gadjiyev, N.M.; Apalikova, O.V.; Alpatyeva, N.V.; Kostina, L.I.; Zoteyeva, N.M.; et al. Molecular screening of potato varieties bred in the northwestern zone of the Russian Federation. Vavilov J. Genet. Breed. 2018, 22, 35–45. [Google Scholar] [CrossRef]
- Antonova, O.Y.; Klimenko, N.S.; Evdokimova, Z.Z.; Kostina, L.I.; Gavrilenko, T.A. Finding RB/Rpi-blb1/Rpi-sto1-like sequences in conventionally bred potato varieties. Vavilov J. Genet. Breed. 2018, 22, 693–702. [Google Scholar] [CrossRef]
- Kochetov, A.V.; Afonnikov, D.A.; Shmakov, N.; Vasiliev, G.V.; Antonova, O.Y.; Shatskaya, N.V.; Glagoleva, A.Y.; Ibragimova, S.M.; Khiutti, A.; Afanasenko, O.S.; et al. NLR Genes Related Transcript Sets in Potato Cultivars Bearing Genetic Material of Wild Mexican Solanum Species. Agronomy 2021, 11, 2426. [Google Scholar] [CrossRef]
- Lokossou, A.A.; Rietman, H.; Wang, M.; Krenek, P.; van der Schoot, H.; Henken, B.; Hoekstra, R.; Vleeshouwers, V.G.A.A.; van der Vossen, E.A.G.; Visser, R.G.F.; et al. Diversity, Distribution, and Evolution of Solanum bulbocastanum Late Blight Resistance Genes. Mol. Plant Microbe. Interact. 2010, 23, 1206–1216. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Halterman, D. Identification and Characterization of RB-Orthologous Genes from the Late Blight Resistant Wild Potato Species Solanum verrucosum. Physiol. Mol. Plant Pathol. 2006, 4–6, 230–239. [Google Scholar] [CrossRef]
- Hawkes, J.G. The Potato Evolution, Biodiversity and Genetic Resources; Belhaven Press a Division of Pinter Publishers: London, UK, 1990. [Google Scholar]
- Magoon, M.L.; Hougas, R.W.; Cooper, D.C. Chromosome Pairing at Different Ploidy Levels in the Tuber-Bearing Solanums. J. Genet. 1960, 57, 279–297. [Google Scholar] [CrossRef]
- Marks, G.E. Cytogenetic Studies in Tuberous Solanum Species. New Phytol. 1965, 64, 293–306. [Google Scholar] [CrossRef]
- Dong, F.; Novy, R.G.; Helgeson, J.P.; Jiang, J. Cytological Characterization of Potato–Solanum etuberosum Somatic Hybrids and Their Backcross Progenies by Genomic in Situ Hybridization. Genome 1999, 42, 987–992. [Google Scholar] [CrossRef]
- Dong, F.; McGrath, J.M.; Helgeson, J.P.; Jiang, J. The Genetic Identity of Alien Chromosomes in Potato Breeding Lines Revealed by Sequential GISH and FISH Analyses Using Chromosome-Specific Cytogenetic DNA Markers. Genome 2001, 44, 729–734. [Google Scholar] [CrossRef]
- Dong, F.; Tek, A.L.; Frasca, A.B.L.; McGrath, J.M.; Wielgus, S.M.; Helgeson, J.P.; Jiang, J. Development and Characterization of Potato-Solanum brevidens Chromosomal Addition/Substitution Lines. Cytogenet. Genome Res. 2005, 109, 368–372. [Google Scholar] [CrossRef]
- Gavrilenko, T.; Larkka, J.; Pehu, E.; Rokka, V.M. Identification of Mitotic Chromosomes of Tuberous and Non-Tuberous Solanum Species (Solanum tuberosum and Solanum brevidens) by GISH in Their Interspecific Hybrids. Genome 2002, 45, 442–449. [Google Scholar] [CrossRef][Green Version]
- Gavrilenko, T.; Thieme, R.; Heimbach, U.; Thieme, T. Somatic hybrids of Solanum etuberosum (+) dihaploid Solanum tuberosum and their backcrossing progenies: Relationships of genome dosage with tuber development. Euphytica 2003, 131, 323–332. [Google Scholar] [CrossRef]
- Gavrilenko, T.A.; Pendinen, G.I.; Rokka, V.-M.; Antonova, O.; Thieme, R. Homeologous Chromosome Pairing in Distant Allohaploid Hybrids of the Genus Solanum. Russ. J. Genet. Appl. Res. 2015, 5, 182–190. [Google Scholar] [CrossRef]
- Garriga-Calderé, F.; Huigen, D.J.; Jacobsen, E.; Ramanna, M.S. Prospects for Introgressing Tomato Chromosomes into the Potato Genome: An Assessment through GISH Analysis. Genome 1999, 42, 282–288. [Google Scholar] [CrossRef]
- Ali, S.; Ramanna, M.; Jacobsen, E.; Visser, R. Establishment of a Complete Series of a Monosomic Tomato Chromosome Addition Lines in the Cultivated Potato Using RFLP and GISH Analyses. Appl. Genet. 2001, 103, 687–695. [Google Scholar] [CrossRef]
- Horsman, K.; Gavrilenko, T.; Bergervoet, M.; Huigen, D.J.; Joe, A.T.W.; Jacobsen, E. Alteration of the genomic composition of Solanum nigrum (+) potato backcross derivatives by somatic hybridization: Selection of fusion hybrids by DNA measurements. Plant Breed. 2001, 120, 201–207. [Google Scholar] [CrossRef]
- Gavrilenko, T. Chapter 10–Potato Cytogenetics. In Potato Biology and Biotechnology; Vreugdenhil, D., Bradshaw, J., Gebhardt, C., Govers, F., Mackerron, D.K.L., Taylor, M.A., Ross, H.A., Eds.; Elsevier Science B.V.: Amsterdam, The Netherlands, 2007; pp. 203–216. [Google Scholar]
- Gavrilenko, T. Application of molecular cytogenetics in fundamental and applied research of potato (Review, Chapter 9). In Genetics, Genomics and Breeding of Potato, 1st ed.; Bradeen, J., Kole, C., Eds.; Science Publishers: New York, NY, USA, 2011; pp. 184–207. [Google Scholar]
- Gaiero, P.; Speranza, P.; de Jong, H. Introgressive Hybridization in Potato Revealed by Novel Cytogenetic and Genomic Technologies. Am. J. Potato Res. 2018, 95, 607–621. [Google Scholar] [CrossRef]
- Gaiero, P.; Torres, G.A.; Iovene, M. Cytogenetics of Potato and Tomato Wild Relatives. In The Wild Solanums Genomes; Compendium of Plant Genomes; Carputo, D., Aversano, R., Ercolano, M.R., Eds.; Springer International Publishing: Cham, Germany, 2021; pp. 11–33. [Google Scholar] [CrossRef]
- Iovene, M.I.; Savarese, S.S.; Cardi, T.C.; Frusciante, L.F.; Scotti, N.S.; Simon, P.W.S.W.; Carputo, D.C. Nuclear and Cytoplasmic Genome Composition of Solanum bulbocastanum (+) S. tuberosum Somatic Hybrids. Genome 2007, 50, 443–450. [Google Scholar] [CrossRef]
- Rakosy-Tican, E.; Thieme, R.; König, J.; Nachtigall, M.; Hammann, T.; Denes, T.-E.; Kruppa, K.; Molnár-Láng, M. Introgression of Two Broad-Spectrum Late Blight Resistance Genes, Rpi-Blb1 and Rpi-Blb3, From Solanum bulbocastanum Dun Plus Race-Specific R Genes into Potato Pre-Breeding Lines. Front. Plant Sci. 2020, 11, 699. [Google Scholar] [CrossRef]
- Gaiero, P.; Mazzella, C.; Vilaró, F.; Speranza, P.; de Jong, H. Pairing Analysis and in Situ Hybridisation Reveal Autopolyploid-like Behaviour in Solanum commersonii × S. tuberosum (Potato) Interspecific Hybrids. Euphytica 2017, 213, 137. [Google Scholar] [CrossRef]
- Haan, S.; Rodriguez, F. Potato Origin and Production; Academic Press: Cambridge, MA, USA, 2016; pp. 1–32. [Google Scholar]
- Tek, A.L.; Stevenson, W.R.; Helgeson, J.P.; Jiang, J. Transfer of tuber soft rot and early blight resistance from Solanum brevidens into cultivated potato. Appl. Genet. 2004, 109, 249–254. [Google Scholar] [CrossRef]
- Zhan, Z.; Nwafor, C.C.; Hou, Z.; Gong, J.; Zhu, B.; Jiang, Y.; Zhou, Y.; Wu, J.; Piao, Z.; Tong, Y.; et al. Cytological and morphological analysis of hybrids between Brassicoraphanus, and Brassica napus for introgression of clubroot resistant trait into Brassica napus L. PLoS ONE 2017, 12, e0177470. [Google Scholar] [CrossRef]
- Šimoníková, D.; Cížková, J.; Zoulová, V.; Christelová, P.; Hribová, E. Advances in the Molecular Cytogenetics of Bananas, Family Musaceae. Plants 2022, 11, 482. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).