Genome-Wide Identification and Expression Analysis of the BTB Domain-Containing Protein Gene Family in Sugar Beet
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Treating Methods
2.2. Expression Analysis of BvBTB1, BvBTB2, and BvBTB3
2.3. Bioinformatic Analysis of BTB Proteins and Genes
2.4. Statistical Analysis
3. Results
3.1. Analysis of Physicochemical Properties of the BTB Protein Family in Sugar Beet
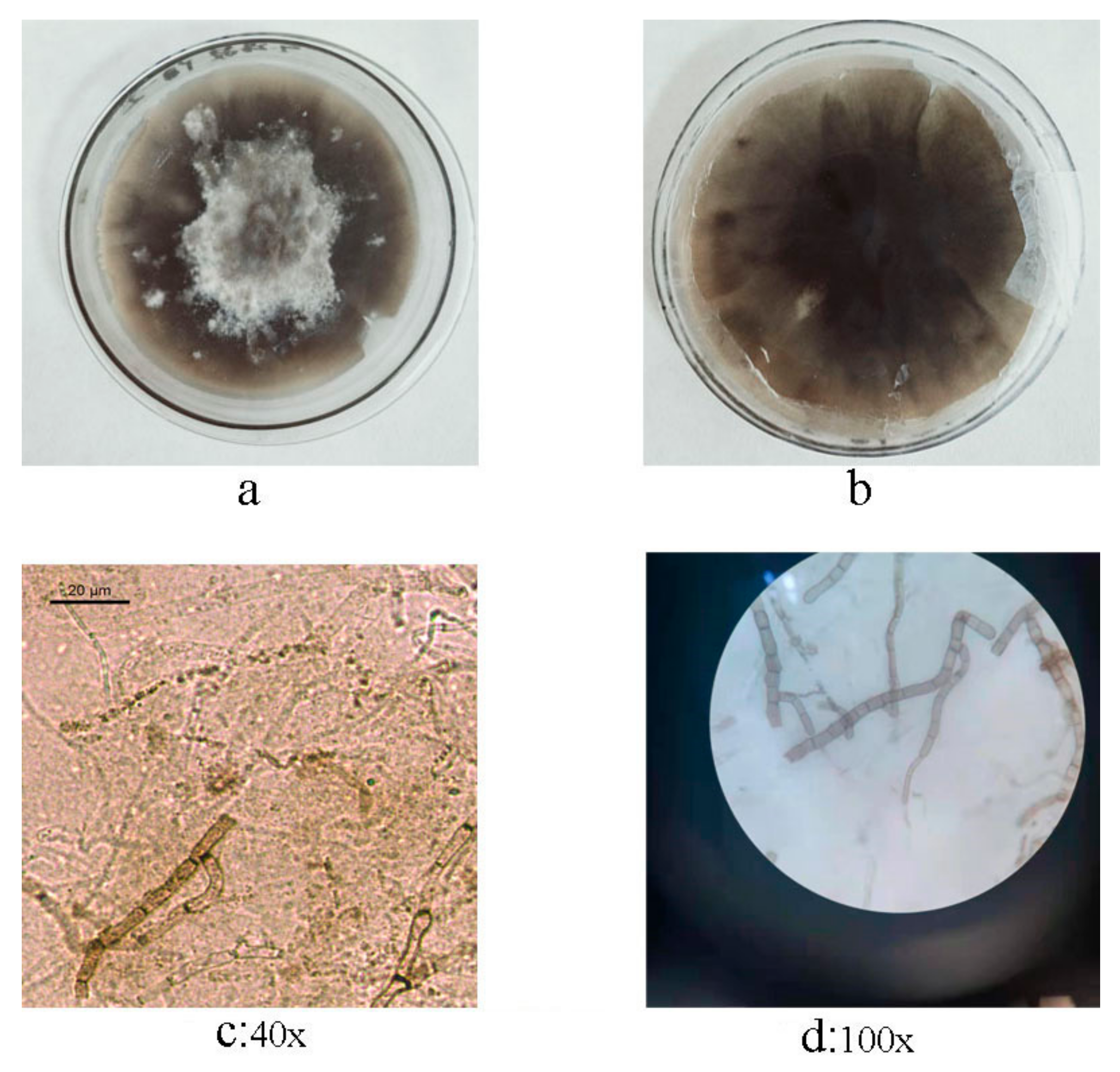
3.2. Isolation and Morphological Identification of C. beticola in Sugar Beet
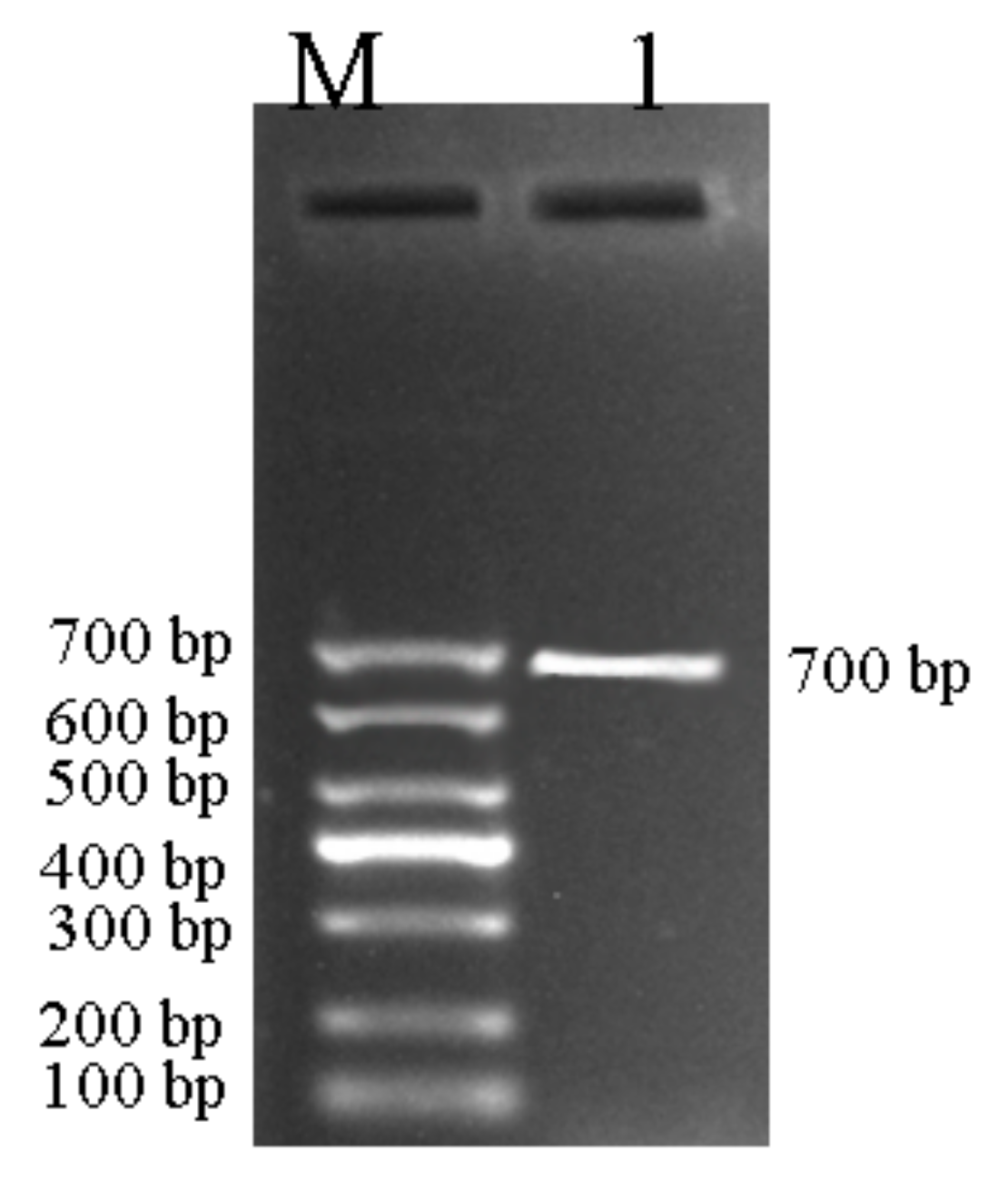
3.3. Confirmation to C. beticola in Sugar Beet
3.4. Screening BvBTB Genes with Significant Differential Expression
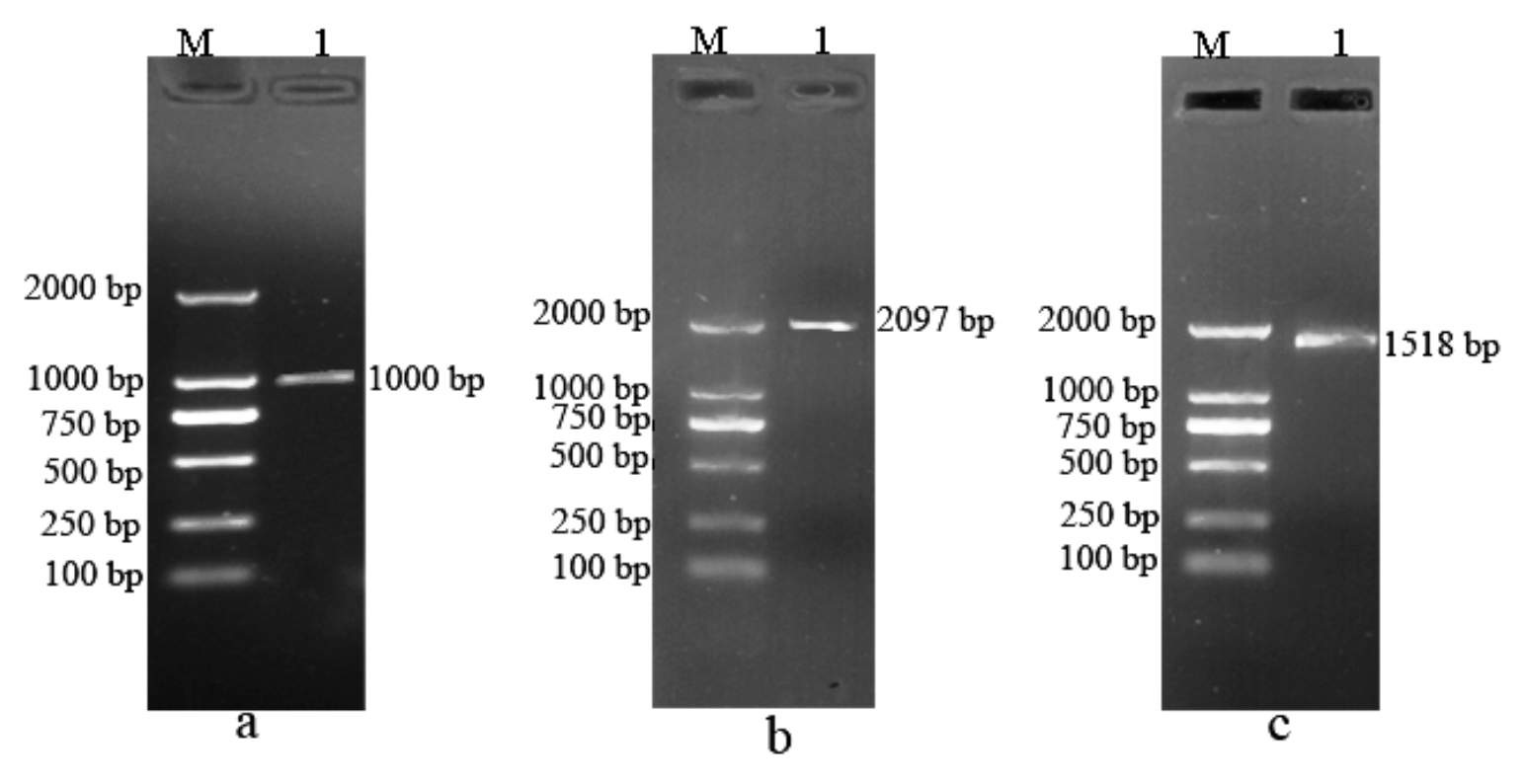
3.5. Agarose Gel Electrophoresis of PCR Amplification of BvBTB1, BvBTB2, and BvBTB3
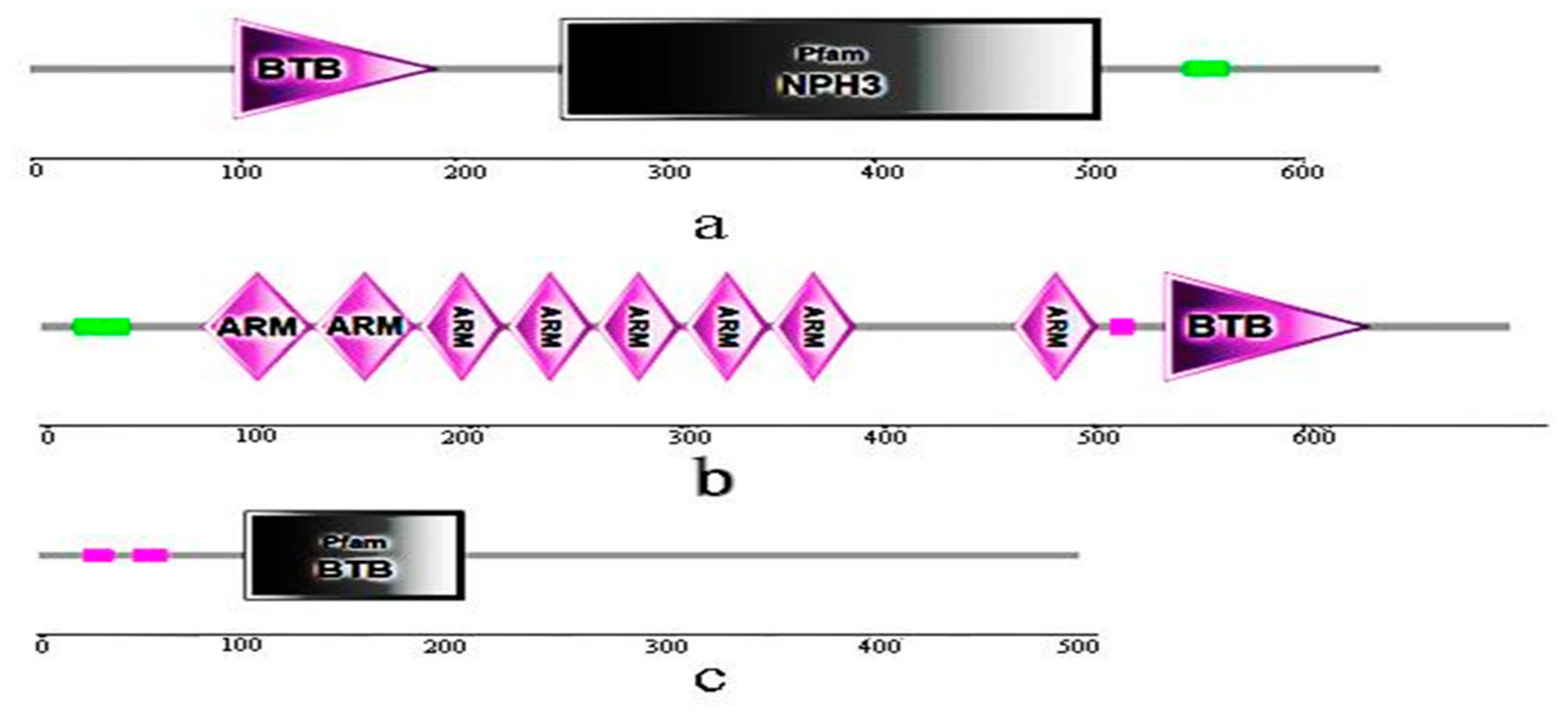
3.6. Conserved Domains of BvBTB1, BvBTB2, and BvBTB3 Proteins
3.7. Analysis of Promoter Elements of BvBTB1, BvBTB2, and BvBTB3
3.8. Phenotypic Analysis of Sugar Beet Resistant and Susceptible Materials at Different Time under Different Spore Concentrations of C. beticola
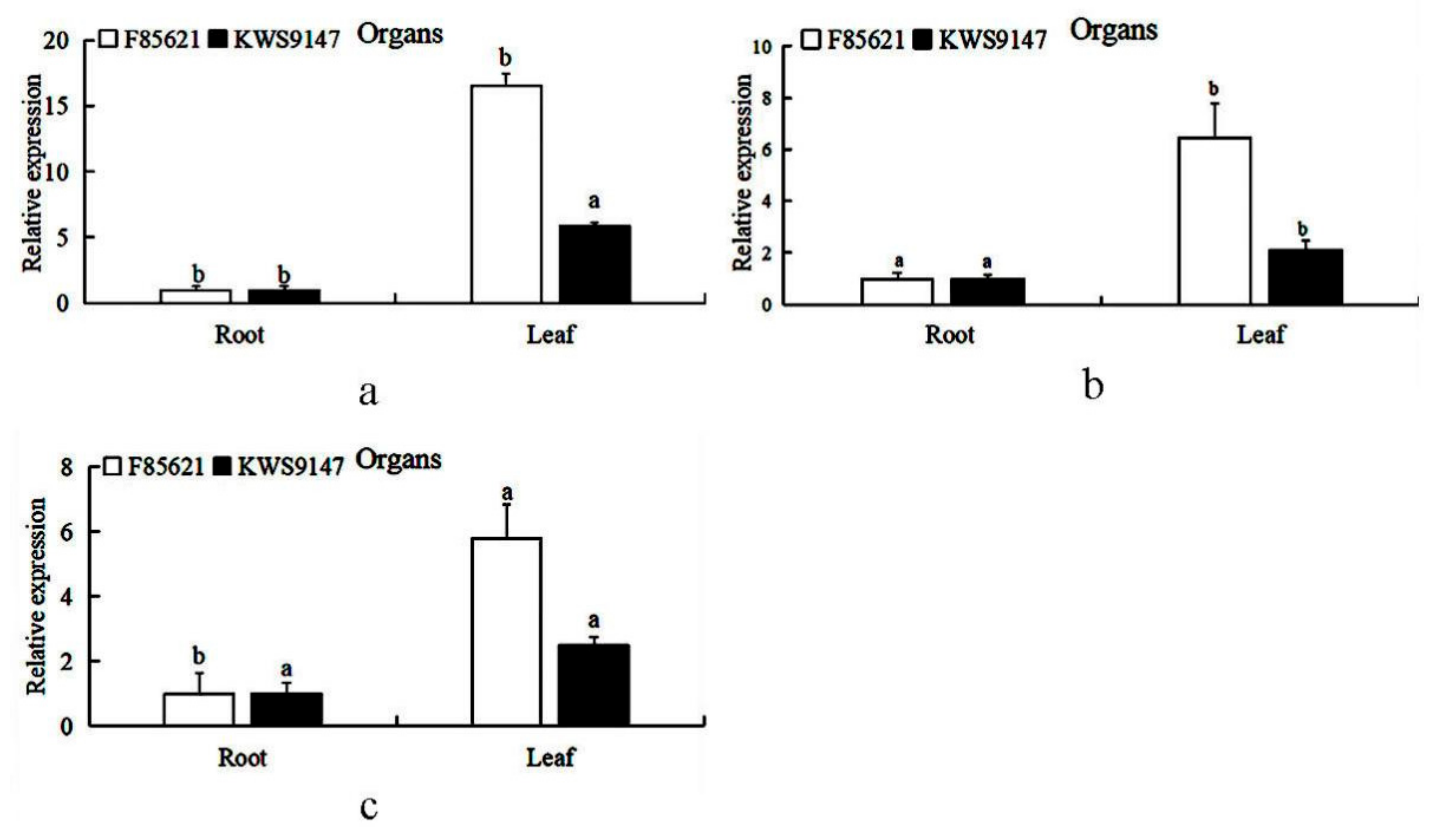
3.9. Tissue Expression Patterns of BvBTB1, BvBTB2, and BvBTB3 in Sugar Beet Resistant Material ‘F85621’ and Susceptible Material ‘KWS9147’
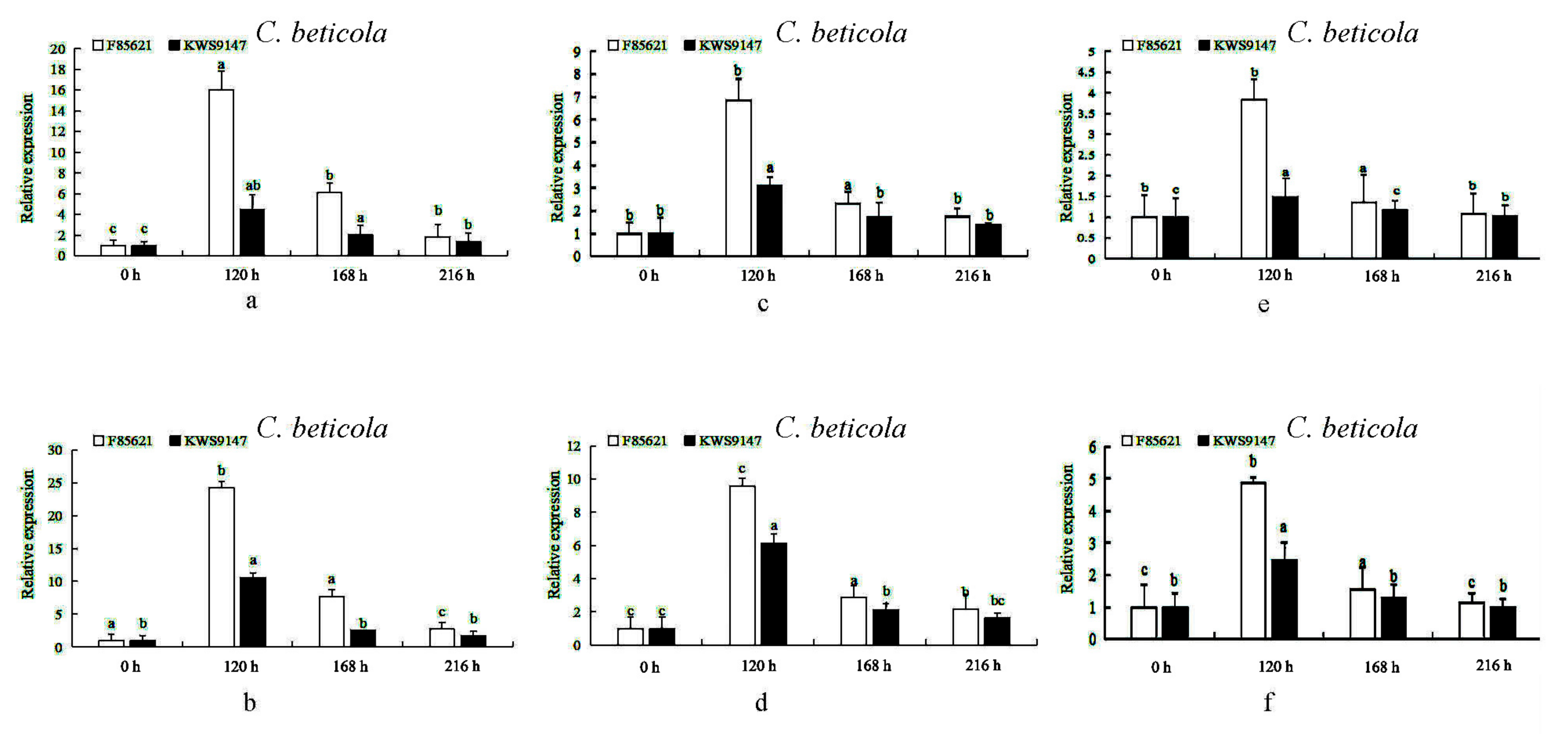
3.10. The Expression Levels of BvBTB1, BvBTB2, and BvBTB3 after High and Low Spore Concentrations of C. beticola Infection at Different Times in Leaves in Sugar Beet Resistant Material ‘F85621’ and Susceptible Material ‘KWS9147’
3.11. The Expression Levels of BvBTB1, BvBTB2, and BvBTB3 after High and Low Spore Concentration of C. beticola Infection at Different Times in Roots in Sugar Beet Resistant Material ‘F85621’and Susceptible Material ‘KWS9147’
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, S.Z.; Yan, X.F.; Zhang, C.Z.; Han, D.; Xu, P.F. Progress in the studies of plant BTB/POZ protein and the disease resistance. Soybean Sci. 2019, 38, 311–316. [Google Scholar]
- Buscaill, P.; Rivas, S. Transcriptional control of plant defence responses. Curr. Opin. Plant Biol. 2014, 20, 35–46. [Google Scholar] [CrossRef]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soosaar, J.L.; Burch-Smith, T.M.; Dinesh-Kumar, S.P. Mechanisms of plant resistance to viruses. Nat. Rev. Microbiol. 2005, 3, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Hong, G.; Li, R.P.; Han, D.; Wang, L.; Xu, P.F.; Zhang, S.Z. GmBvBTB, a novel BTB domain-containing nuclear protein, positively regulates the response of soybean to Phytophthora sojae infection. Mol. Plant Pathol. 2019, 20, 78–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hui, C.; Glazebrook, J.; Clarke, J.D.; Volko, S.; Dong, X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Plant Cell 1997, 88, 57–63. [Google Scholar]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Phys. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedele, M.; Crescenzi, E.; Cerchia, L. The POZ/BTB and AT-Hook Containing zinc finger 1 (PATZ1) transcription regulator: Physiological functions and disease involvement. Int. J. Mol. Sci. 2017, 18, 2524. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Li, X.; Dong, X. Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. USA 1998, 95, 6531–6536. [Google Scholar] [CrossRef] [Green Version]
- Tsuda, K.; Somssich, I.E. Transcriptional networks in plant immunity. New Phytol. 2014, 206, 932–947. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.J.; Luo, D.P.; Luo, X.M.; Feng, J.X. Overexpression of arabidopsis thaliana AtNPR1 gene in rice enhances the rice resistance to bacterial blight and blast diseases. Guihaia 2012, 32, 800–805. [Google Scholar]
- Vidhyasekaran, P. Plant Hormone Signaling Systems in Plant Innate Immunity; Springer: Berlin/Heidelberg, Germany, 2015; Volume 2, pp. 117–122. [Google Scholar]
- Makandar, R.; Essig, J.S.; Schapaugh, M.A.; Trick, H.N.; Shah, J. Genetically engineered resistance to fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol. Plant-Microbe Interact. 2006, 19, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Xu, P.; Wu, J.; Xue, A.G.; Zhang, J.; Li, W.; Chen, C.; Chen, W.; Lv, H. Races of Phytophthora sojae and their virulences on soybean cultivars in Heilongjiang, China. Plant Dis. 2010, 94, 87–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endah, R.; Beyene, G.; Kiggundu, A.; Berg, N.; Schluter, U.; Kunert, K.; Chikwamba, R. Elicitor and Fusariuminduced expression of NPR1-like genes in banana. Plant Physiol. Biochem. 2008, 46, 1007–1014. [Google Scholar] [CrossRef]
- Rasmussen, M.W.; Roux, M.; Petersen, M.; Mundy, J. MAP kinase cascades in Arabidopsis innate immunity. Front. Plant Sci. 2012, 3, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Che, D.D.; Fan, J.P.; Wang, J.G.; Xu, P.; Yang, T.; Liu, S.K. Cloning and analysis of full-length cDNA of PumNPR1 gene from Pyrus ussuriensis Maxim. J. Northeast Agric. Univ. 2008, 15, 12–17. [Google Scholar]
- Zhang, S.B.; Liu, J.B.; Wang, Y.S. Study on the resistance of arbuscular mycorrhizal fungi to Phytophthora capsici of pepper. North. Hortic. 2015, 30, 125–128. [Google Scholar]
- He, Y.M.; Liu, K.K.; Zhang, H.X.; Cheng, G.X.; Gong, Z.H. Contribution of cabpm4, a BTB domain–containing gene, to the response of pepper to Phytophthora capsici infection and abiotic stresses. Agronomy 2019, 9, 417. [Google Scholar] [CrossRef] [Green Version]
- Weiland, J.; Koch, G. Sugar beet leaf spot disease (C. beticola beticola sacc.)†. Mol. Plant Pathol. 2010, 5, 157–166. [Google Scholar] [CrossRef]
- Skaracis, G.N.; Pavli, O.I.; Biancardi, E. C. beticola leaf spot disease of sugar beet. Sugar Tech. 2010, 12, 220–228. [Google Scholar] [CrossRef]
- Vaghefi, N.; Kikkert, J.R.; Hay, F.S.; Carver, G.D.; Koenick, L.B.; Bolton, M.D. Cryptic diversity, pathogenicity, and evolutionary species boundaries in C. beticola populations associated with C. beticola leaf spot of Beta vulgaris. Fungal Biol. 2018, 122, 264–282. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yang, S.Y.; Ma, W.J.; Zhou, J.H.; Ma, W.J. Isolation and identification of the pathogen of walnut brown spot and investigation of its pathogenesis. For. Sci. Res. 2017, 30, 1004–1008. [Google Scholar]
- Berbee, M.L.; Pirseyedi, M.; Hubbard, S. Cochliobolus phylogenetics and the origin of know, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 1999, 91, 964–977. [Google Scholar] [CrossRef]
- Wolf, P.; Verreet, J.A. Factors affecting the onset of C. beticola leaf spot epidemics in sugar beet and establishment of disease-monitoring thresholds. Phytopathology 2005, 95, 269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.T.; Yang, Q.; Hao, J.J.; Wang, Y.; Zhang, Y.Y.; Li, B.Q. Cloning and expression analysis of Maize ZmNPR1 gene. Acta Bot. Boreali-Occident. Sin. 2018, 38, 1817–1822. [Google Scholar]




| Primer | Primer Sequences (5′–3′) | Tm |
|---|---|---|
| BvBTB1 Amplification primer | R: TTAAGAAATGGAGAACCTTCTTCTCCTTGG F: ATGATGAGTGCAACAGCGTTGAACCCT | 62 °C |
| BvBTB2 Amplification primer | R: CTAATTTAGAGCATTAGGCTTGGTAAGTAACCT F: ATGGGGTCCAAGGAGTGGC | 58 °C |
| BvBTB3 Amplification prime | R: GTTACCAAAGATGATCGAGTAGC F: TCTCCCAATGTTACACCACAAC | 54 °C |
| BvBTB1 Fluorescent primer | R: TCGTTGGCTCCACTCCTTAGCTTTA F: GAGTACATGCCTCACAGAACAAGCG | 60 °C |
| BvBTB 2 Fluorescent primer | R: AGTGGACTTGAGAAGCCCAAGAAGT F: AGTTGCTTTGGCTCTTGCTCATCTG | 60 °C |
| BvBTB3 Fluorescent primer | R: TCCATCGATCGGAGAACAAAGCAGA F: ACTCCGATTCTCTCTCCTCCATTGC | 60 °C |
| Gapdh Fluorescent primer | R: GTTGGAACACGGAA AGCC F: TGGAGAGGTGGAAGG | 60 °C |
| ITS1 Amplification primer | TCCTCCGCTTATTGATATGC | 60 °C |
| ITS4 Amplification primer | TCCGTAGGTGAACCTGCGG | 60 °C |
| Gene Subfamily | Gene Accession Number | Protein Accession Number | Amino Acid Number | Isoelectric Point | Average Hydrophobicity | Number of Phosphorylation Sites | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| NPH3 | KMS95472.1 | BVRB—007930 | 1120 | 5.91 | −0.054 | 19 | Nucleus |
| KMT02116.1 | BVRB—9 g 207600 | 539 | 6.68 | −0.128 | 21 | Nucleus | |
| KMT16927.1 | BVRB—2 g 043970 | 615 | 7.50 | −0.351 | 17 | Nucleus | |
| KMT09829.1 | BVRB—6 g 128050 | 625 | 7.54 | −0.342 | 21 | Nucleus | |
| KMT03832.1 | BVRB—8 g 189750 | 621 | 6.73 | −0.256 | 25 | Cytoplasm | |
| KMT08787.1 | BVRB—6 g 135110 | 1076 | 5.99 | −0.722 | 24 | Nucleus | |
| KMT13794.1 | BVRB—4 g 081670 | 559 | 9.10 | −0.258 | 13 | Cytomembrane Nucleus | |
| KMT00896.1 | BVRB—9 g 221680 | 695 | 6.88 | −0.463 | 22 | Cytomembrane | |
| KMT11299.1 | BVRB—5 g 109820 | 581 | 8.18 | −0.172 | 12 | Cytomembrane Nucleus | |
| KMT09814.1 | BVRB—6 g 127900 | 634 | 5.96 | −0.185 | 15 | Cytomembrane | |
| 104899057 | BVRB—7 g 164450 | 636 | 9.07 | −0.292 | 15 | Cytomembrane | |
| KMT05614.1 | BVRB—7 g 167550 | 610 | 6.63 | −0.327 | 23 | Cytomembrane Nucleus | |
| KMT06491.1 | BVRB—7 g 156670 | 617 | 6.00 | −0.230 | 14 | Cytomembrane | |
| KMT03943.1 | BVRB—8 g 187520 | 633 | 5.74 | −0.186 | 13 | Cytomembrane | |
| KMT17316.1 | BVRB—2 g 040100 | 688 | 5.64 | −0.350 | 13 | Cytomembrane Nucleus | |
| KMT09842.1 | BVRB—6 g 128170 | 488 | 5.59 | −0.301 | 11 | Cytomembrane Nucleus | |
| BACK | KMT11993.1 | BVRB—5 g 099550 | 553 | 5.45 | −0.265 | 9 | Cytomembrane |
| KMT20372.1 | BVRB—1 g 003710 | 556 | 5.04 | −0.369 | 10 | Cytomembrane Chloroplast | |
| 104898402 | BVRB—7 g 159950 | 979 | 5.83 | 0.108 | 14 | Nucleus | |
| 104905652 | BVRB—1 g 021120 | 805 | 5.95 | −0.239 | 13 | Cytomembrane | |
| Arm | KMS95782.1 | BVRB—005090 | 1012 | 6.88 | 0.068 | 15 | Cytomembrane Nucleus |
| KMT06143.1 | BVRB—7 g 163130 | 709 | 6.14 | −0.119 | 13 | Nucleus | |
| 104883420 | BVRB—000890 | 698 | 6.13 | −0.084 | 13 | Nucleus | |
| Ank | KMS99096.1 | BVRB—3 g 066940 | 579 | 5.03 | −0.260 | 11 | Nucleus |
| KMS96007.1 | BVRB—003000 | 852 | 5.88 | −0.453 | 15 | Nucleus | |
| KMT15126.1 | BVRB—3 g 062440 | 498 | 6.25 | −0.273 | 12 | Nucleus | |
| KMS96842.1 | BVRB—8 g 199180 | 604 | 5.92 | −0.271 | 13 | Nucleus | |
| MATH | KMT05007.1 | BVRB—7 g 171670 | 398 | 5.72 | −0.161 | 8 | Nucleus |
| KMS97744.1 | BVRB—5 g 124160 | 407 | 7.16 | −0.144 | 5 | Nucleus | |
| KMS97745.1 | BVRB—5 g 124170 | 405 | 5.98 | −0.183 | 10 | Nucleus | |
| KMT18392.1 | BVRB—2 g 025440 | 420 | 5.98 | −0.133 | 9 | Nucleus | |
| TAZ | KMS97248.1 | BVRB—7 g 176960 | 345 | 9.51 | −0.323 | 5 | Nucleus |
| KMT09833.1 | BVRB—6 g 128090 | 388 | 9.28 | −0.321 | 8 | Cytomembrane Nucleus | |
| KMT15541.1 | BVRB—3 g 058930 | 139 | 9.18 | −0.340 | 3 | Nucleus | |
| BTB-only | KMT11639.1 | BVRB—5 g 109530 | 346 | 5.69 | −0.330 | 8 | Cytomembrane |
| KMT12649.1 | BVRB—4 g 091080 | 329 | 5.87 | −0.141 | 8 | Cytomembrane | |
| KMT19646.1 | BVRB—1 g 010330 | 253 | 5.03 | −0.313 | 8 | Cytomembrane Nucleus | |
| KMT18872.1 | BVRB—2 g 029750 | 279 | 5.43 | −0.238 | 5 | Cytomembrane Nucleus | |
| TPR | KMT10233.1 | BVRB—5 g 119930 | 886 | 5.72 | −0.204 | 22 | Nucleus |
| Other | KMS99416.1 | BVRB—2 g 045380 | 484 | 5.35 | −0.281 | 9 | Chloroplast |
| KMT12084.1 | BVRB—5 g 100390 | 462 | 5.38 | −0.152 | 17 | Chloroplast | |
| KMT14083.1 | BVRB—4 g 079080 | 558 | 6.56 | −0.429 | 18 | Chloroplast | |
| KMT08528.1 | BVRB—6 g 138110 | 532 | 7.89 | −0.434 | 16 | Chloroplast Nucleus | |
| KMT12064.1 | BVRB—5 g 100210 | 585 | 6.59 | −0.394 | 20 | Nucleus | |
| KMT05357.1 | BVRB—7 g 174890 | 508 | 5.42 | −0.359 | 16 | Nucleus | |
| KMS93550.1 | BVRB—030350 | 181 | 5.03 | −0.160 | 4 | Cytomembrane Nucleus | |
| KMT12599.1 | BVRB—5 g 098380 | 433 | 4.70 | −0.008 | 13 | Nucleus | |
| KMT12434.1 | BVRB—5 g 103650 | 436 | 4.93 | −0.097 | 12 | Nucleus | |
| KMT00450.1 | BVRB—9 g 217510 | 481 | 6.48 | −0.165 | 24 | Nucleus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Q.; Liu, Y.; Zhao, C.; Wang, X.; Ding, G.; Li, Y.; Chen, L. Genome-Wide Identification and Expression Analysis of the BTB Domain-Containing Protein Gene Family in Sugar Beet. Agronomy 2022, 12, 253. https://doi.org/10.3390/agronomy12020253
Yang Q, Liu Y, Zhao C, Wang X, Ding G, Li Y, Chen L. Genome-Wide Identification and Expression Analysis of the BTB Domain-Containing Protein Gene Family in Sugar Beet. Agronomy. 2022; 12(2):253. https://doi.org/10.3390/agronomy12020253
Chicago/Turabian StyleYang, Qiao, Yu Liu, Chunlei Zhao, Xi Wang, Guangzhou Ding, Yanli Li, and Li Chen. 2022. "Genome-Wide Identification and Expression Analysis of the BTB Domain-Containing Protein Gene Family in Sugar Beet" Agronomy 12, no. 2: 253. https://doi.org/10.3390/agronomy12020253
APA StyleYang, Q., Liu, Y., Zhao, C., Wang, X., Ding, G., Li, Y., & Chen, L. (2022). Genome-Wide Identification and Expression Analysis of the BTB Domain-Containing Protein Gene Family in Sugar Beet. Agronomy, 12(2), 253. https://doi.org/10.3390/agronomy12020253






