Complete Chloroplast Genome Sequence of a New Variety of Brasenia schreberi: Genome Characteristics, Comparative Analysis, and Phylogenetic Relationships
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Plant Material
2.2. Physiological Indicators of the Leaves from Three Varieties
2.3. DNA Extraction and Genome Sequencing
2.4. Genome Sequence Assembly and Annotation
2.5. Chloroplast Genome Data
2.5.1. SSR and IRS Analysis
2.5.2. KaKs Calculation and Nucleotide Diversity Analysis
2.5.3. IR Boundary Analysis
2.6. Comparative Analysis of cpDNA Genomes and Related Species
2.6.1. Comparative Analysis on the Structure of Chloroplast Related Species and Chloroplast Sequence Homology Analysis
2.6.2. Genetic Distance and Phylogenetic Analysis
3. Results
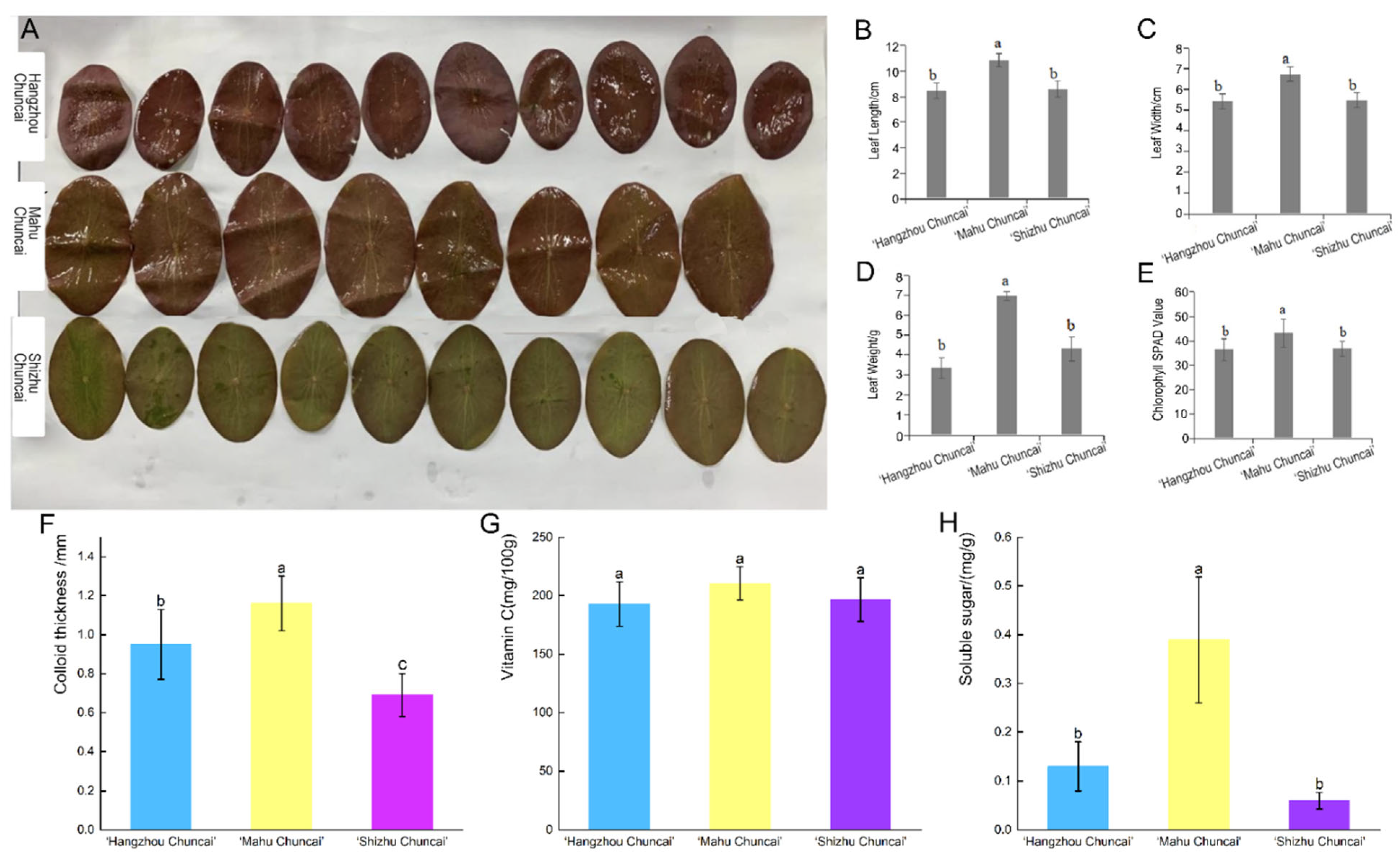
3.1. Comparison of Morphology and Nutrition in Different Varieties of B. schreberi
3.2. Chloroplast Genome of ‘Mahu Chuncai’
3.3. Codon Usage Pattern Analysis
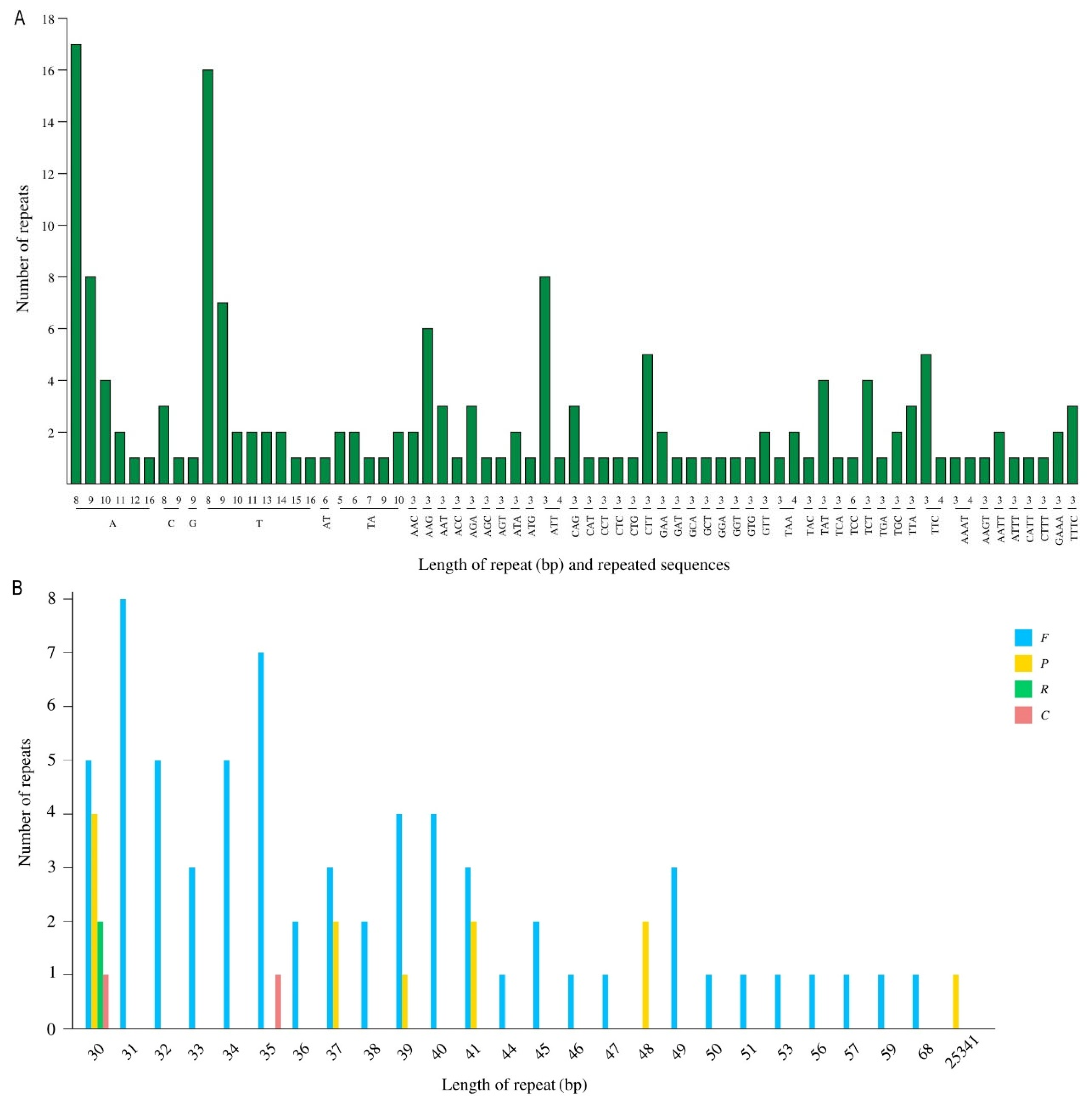
3.4. Analysis of Repeat Sequences
3.5. KaKs Analysis
3.6. Analysis of Nucleotide Diversity (Pi)
3.7. Contraction and Expansion of IR Regions
3.8. Genome Comparison and Collinearity Analysis
3.9. Phylogenetic Relationships
4. Discussion
4.1. Analysis of the Chloroplast Genome of B. schreberi
4.2. Evolutionary and Phylogenetic Relatedness
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, C.; Na, H.R.; Choi, H.K. Conservation genetics of endangered Brasenia schreberi based on RAPD and AFLP markers. J. Plant Biol. 2008, 51, 260–268. [Google Scholar] [CrossRef]
- Li, Z.Z.; Gichira, A.W.; Wang, Q.F.; Chen, J.M. Genetic diversity and population structure of the endangered basal angiosperm Brasenia schreberi (Cabombaceae) in China. Peer J. 2018, 6, e5296. [Google Scholar] [CrossRef] [PubMed]
- Elakovich, S.D.; Wooten, J.W. An examination of the phytotoxicity of the water shield, Brasenia schreberi. J. Chem. Ecol. 1987, 13, 1935–1940. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Li, J.; Pan, F.; Fu, J.; Zhou, W.; Lu, S.; Li, P.; Zhou, C. Environmental factors influencing mucilage accumulation of the endangered Brasenia schreberi in China. Sci. Rep. 2018, 8, 17955. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Liu, Y.; Yang, Y.; Chen, Z.; Li, J.; Luo, J. Mechanism of biological liquid superlubricity of Brasenia schreberi mucilage. Langmuir 2014, 30, 3811–3816. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yi, C.; Zhang, C.; Pan, F.; Xie, C.; Zhou, W.; Zhou, C. Effects of light quality on leaf growth and photosynthetic fluorescence of Brasenia schreberi seedlings. Heliyon 2021, 7, e06082. [Google Scholar] [CrossRef]
- Feng, S.; Luan, D.; Ning, K.; Shao, P.; Sun, P. Ultrafiltration isolation, hypoglycemic activity analysis and structural characterization of polysaccharides from Brasenia schreberi. Int. J. Biol. Macromol. 2019, 135, 141–151. [Google Scholar] [CrossRef]
- Yang, F.; Xiao, K.; Pan, H.; Liu, J. Chloroplast: The Emerging Battlefield in Plant-Microbe Interactions. Front. Plant Sci. 2021, 12, 637853. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Fan, Y.; Jin, Y.; Ding, M.; Tang, Y.; Cheng, J.; Zhang, K.; Zhou, M. The complete chloroplast genome sequences of eight Fagopyrum species: Insights into genome evolution and phylogenetic relationships. Front. Plant Sci. 2021, 12, 799904. [Google Scholar] [CrossRef]
- Tian, X.; Zheng, J.; Hu, S.; Yu, J. The rice mitochondrial genomes and their variations. Plant Physiol. 2006, 140, 401–410. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Li, W.H.; Sharp, P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad Sci. USA 1987, 84, 9054–9058. [Google Scholar] [CrossRef]
- Drouin, G.; Daoud, H.; Xia, J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 2008, 49, 827–831. [Google Scholar] [CrossRef]
- Wicke, S.; Schneeweiss, G.M.; dePamphilis, C.W.; Müller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef]
- Ren, T.; Li, Z.X.; Xie, D.F.; Gui, L.J.; Peng, C.; Wen, J.; He, X.J. Plastomes of eight Ligusticum species: Characterization, genome evolution, and phylogenetic relationships. BMC Plant Biol. 2020, 20, 519. [Google Scholar] [CrossRef]
- Cai, Z.; Guisinger, M.; Kim, H.G.; Ruck, E.; Blazier, J.C.; McMurtry, V.; Kuehl, J.V.; Boore, J.; Jansen, R.K. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J. Mol. Evol. 2008, 67, 696–704. [Google Scholar] [CrossRef]
- Shinozaki, K.; Ohme, M.; Tanaka, M.; Wakasugi, T.; Hayashiuda, N.; Matsubayashi, T.; Zaita, N.; Chunwongse, J.; Obokata, J.; Yamaguchi-Shinosaki, K.; et al. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986, 5, 2043–2049. [Google Scholar] [CrossRef]
- Ohyama, K.; Fukuzawa, H.; Kohchi, T.; Shirai, H.; Sano, T.; Sano, S.; Umesono, K.; Shiki, Y.; Takeuchi, M.; Chang, Z.; et al. Chloroplast gene organization deduced from complete sequence of liverwort, Marchantia polymorpha chloroplast DNA. Nature 1986, 322, 572–574. [Google Scholar] [CrossRef]
- Bull, K.R.; Rimmer, A.J.; Siggs, O.M.; Miosge, L.A.; Roots, C.M.; Enders, A.; Bertram, E.M.; Crockford, T.L.; Whittle, B.; Potter, P.K.; et al. Unlocking the bottleneck in forward genetics using whole-genome sequencing and identity by descent to isolate causative mutations. PLoS Genet. 2013, 9, e1003219. [Google Scholar] [CrossRef]
- Muhammad, B.L.; Ki, J.S. Hybrid origin of the invasive Spartina anglica inferred from chloroplast and nuclear ITS phylogenies. Aquat. Botany 2021, 178, 103484. [Google Scholar] [CrossRef]
- Kolter, A.; Gemeinholzer, B. Plant DNA barcoding necessitates marker-specific efforts to establish more comprehensive reference databases. Genome 2021, 64, 265–298. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.F.; Zhu, G.F.; Li, D.M.; Wang, X.J. Complete chloroplast genome sequence and phylogenetic analysis of Spathiphyllum Parrish. PLoS ONE 2019, 14, e0224038. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Sun, Y.B.; Huang, L.L.; Xu, Y.C.; Zhao, C.Y.; Liu, X.F. The complete chloroplast genome sequence of Camellia chuongtsoensis. Mitochondrial DNA B Resour. 2021, 6, 247–249. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Jung, J.; Na, H.R.; Kim, S.W.; Li, W.; Kadono, Y.; Shin, H.; Choi, H.-K. Population Genetic Structure of the Endangered Brasenia schreberi in South Korea Based on Nuclear Ribosomal Spacer and Chloroplast DNA Sequences. J. Plant Biol. 2012, 55, 81–91. [Google Scholar] [CrossRef]
- Yang, Y.; Li, J.W.; Wang, N.; Zou, X.X.; Zou, S.Q. The complete chloroplast genome sequence of Brasenia schreberi (Cabombaceae). Mitochondrial DNA. Mitochondrial DNA Part B 2019, 4, 3842–3843. [Google Scholar] [CrossRef] [PubMed]
- Gruenstaeudl, M.; Nauheimer, L.; Borsch, T. Plastid genome structure and phylogenomics of Nymphaeales: Conserved gene order and new insights into relationships. Plant Syst. Evol. 2017, 303, 1251–1270. [Google Scholar] [CrossRef]
- Li, M.Y.; Li, J.; Zhang, R.; Lin, Y.X.; Xiong, A.S.; Tan, G.F.; Luo, Y.; Zhang, Y.; Chen, Q.; Wang, Y.; et al. Combined Analysis of the Metabolome and Transcriptome to Explore Heat Stress Responses and Adaptation Mechanisms in Celery (Apium graveolens L.). Int. J. Mol. Sci. 2022, 23, 3367. [Google Scholar] [CrossRef]
- Hanania, U.; Velcheva, M.; Sahar, N.; Perl, A. An improved method for isolating high-quality DNA from Vitis vinifera nuclei. Plant Mol. Biol. Rep. 2004, 22, 173–177. [Google Scholar] [CrossRef]
- Menezes, A.P.A.; Resende-Moreira, L.C.; Buzatti, R.S.O.; Nazareno, A.G.; Carlsen, M.; Lobo, F.P.; Kalapothakis, E.; Lovato, M.B. Chloroplast genomes of Byrsonima species (Malpighiaceae): Comparative analysis and screening of high divergence sequences. Sci. Rep. 2018, 8, 2210. [Google Scholar] [CrossRef]
- Wang, H.; Ma, Z.H.; Mao, J.; Chen, B.H. Genome-wide identification and expression analysis of the EXO70 gene family in grape (Vitis vinifera L.). Peer J. 2021, 9, e11176. [Google Scholar] [CrossRef]
- Li, M.; Song, Y.F.; Sylvester, S.P.; Sylvester, S.P.; Wang, X.R. Comparative analysis of the complete plastid genomes in Prunus subgenus Cerasus (Rosaceae): Molecular structures and phylogenetic relationships. PLoS ONE 2022, 17, e0266535. [Google Scholar]
- Nei, M.; Li, W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Xie, D.F.; Yu, Y.; Deng, Y.Q.; Li, J.; Liu, H.Y.; Zhou, S.D.; He, X.J. Comparative analysis of the chloroplast genomes of the Chinese endemic genus Urophysa and their contribution to chloroplast phylogeny and adaptive evolution. Int. J. Mol. Sci. 2018, 19, 1847. [Google Scholar] [CrossRef]
- Cavagnaro, P.F.; Senalik, D.A.; Yang, L.; Simon, P.W.; Harkins, T.T.; Kodira, C.D.; Huang, S.; Weng, Y. Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genomics 2010, 11, 569. [Google Scholar] [CrossRef]
- Gu, C.; Ma, L.; Wu, Z.; Chen, K.; Wang, Y. Comparative analyses of chloroplast genomes from 22 Lythraceae species: Inferences for phylogenetic relationships and genome evolution within Myrtales. BMC Plant Biol. 2019, 19, 281. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, Z.; Huang, S.; An, W.; Li, J.; Zheng, X. Comprehensive analysis of Rhodomyrtus tomentosa chloroplast genome. Plants 2019, 8, 89. [Google Scholar] [CrossRef]
- Nguyen, H.Q.; Nguyen, T.; Doan, T.N.; Nguyen, T.; Phạm, M.H.; Le, T.L.; Sy, D.T.; Chu, H.H.; Chu, H.M. Complete chloroplast genome of novel Adrinandra megaphylla Hu species: Molecular structure, comparative and phylogenetic analysis. Sci. Rep. 2021, 11, 11731. [Google Scholar] [CrossRef]
- Xiong, Y.; Xiong, Y.; Jia, S.; Ma, X. The complete chloroplast genome sequencing and comparative analysis of reed canary grass (Phalaris arundinacea) and hardinggrass (P. aquatica). Plants 2020, 9, 748. [Google Scholar] [CrossRef]
- Liu, H.; Su, Z.; Yu, S.; Liu, J.; Yin, X.; Zhang, G.; Liu, W.; Li, B. Genome comparison reveals mutation hotspots in the chloroplast genome and phylogenetic relationships of Ormosia Species. Biomed Res. Int. 2019, 2019, 7265030. [Google Scholar] [CrossRef]
- Dong, W.; Liu, J.; Yu, J.; Wang, L.; Zhou, S. Highly Variable Chloroplast Markers for Evaluating Plant Phylogeny at Low Taxonomic Levels and for DNA Barcoding. PLoS ONE 2012, 7, e35071. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Shi, T.; Luo, W.; Ni, X.; Iqbal, S.; Ni, Z.; Huang, X.; Yao, D.; Shen, Z.; Gao, Z. Comparative analysis of the complete chloroplast genome among Prunus mume, P. armeniaca, and P. salicina. Hortic. Res. 2019, 6, 89. [Google Scholar] [CrossRef] [PubMed]
- Hurst, G.D.D.; Jiggins, F.M. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: The effects of inherited symbionts. Proc. Biol. Sci. 2005, 272, 1525–1534. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shen, Z.; Meng, X.; Zhang, L.; Liu, Z.; Liu, M.; Zhang, F.; Zhao, J. Codon usage patterns across seven Rosales species. BMC Plant Biol. 2022, 22, 65. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zheng, H.; Xu, A.; Yan, D.; Jiang, Z.; Qi, Q.; Sun, J. Analysis of codon usage bias of envelope glycoprotein genes in nuclear polyhedrosis virus (NPV) and its relation to evolution. BMC Genom. 2016, 17, 677. [Google Scholar] [CrossRef] [PubMed]
- Pintó, R.M.; Bosch, A. The codon usage code for cotranslational folding of viral capsids. Genome Biol. Evol. 2021, 13, evab089. [Google Scholar] [CrossRef]
- Li, M.Y.; Zhang, R.; Li, J.; Zheng, K.; Xiao, J.; Zheng, Y. Analyses of chloroplast genome of Eutrema japonicum provide new insights into the evolution of Eutrema species. Agronomy 2021, 11, 2546. [Google Scholar] [CrossRef]
- Sheng, J.J.; Yan, M.; Wang, J.; Zhao, L.L.; Zhou, F.S.; Hu, Z.L.; Jin, S.R.; Diao, Y. The complete chloroplast genome sequences of five miscanthus species, and comparative analyses with other grass plastomes. Ind. Crops Prod. 2021, 162, 113248. [Google Scholar] [CrossRef]
- Zhu, B.; Feng, Q.; Yu, J.; Yu, Y.; Zhu, X.; Wang, Y.; Guo, J.; Hu, X.; Cai, M. Chloroplast genome features of an important medicinal and edible plant: Houttuynia cordata (Saururaceae). PLoS ONE 2020, 15, e0239823. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, L.; Yan, T.L.; Liu, Q. Complete chloroplast genome sequences of Praxelis (Eupatorium catarium Veldkamp), an important invasive species. Gene 2014, 549, 58–69. [Google Scholar] [CrossRef]
- Plöchinger, M.; Torabi, S.; Rantala, M.; Tikkanen, M.; Suorsa, M.; Jensen, P.E.; Aro, E.M.; Meurer, J. The Low Molecular Weight Protein PsaI Stabilizes the Light-Harvesting Complex II Docking Site of Photosystem, I. Plant Physiol. 2016, 172, 450–463. [Google Scholar] [CrossRef]
- Wen, F.; Wu, X.; Li, T.; Jia, M.; Liu, X.; Liao, L. The complete chloroplast genome of Stauntonia chinensis and compared analysis revealed adaptive evolution of subfamily Lardizabaloideae species in China. BMC Genomics 2021, 22, 161. [Google Scholar] [CrossRef]
- Wang, L.; Wang, J.; He, C. Characterization and comparison of chloroplast genomes from two sympatric Hippophae species (Elaeagnaceae). J. For. Res. 2021, 32, 307–318. [Google Scholar] [CrossRef]
- Du, Y.P.; Bi, Y.; Yang, F.P.; Zhang, M.F.; Chen, X.Q.; Xue, J.; Zhang, X.H. Complete chloroplast genome sequences of Lilium: Insights into evolutionary dynamics and phylogenetic analyses. Sci. Rep. 2017, 7, 5751. [Google Scholar] [CrossRef]
- Dong, W.; Xu, C.; Cheng, T.; Lin, K.; Zhou, S. Sequencing angiosperm plastid genomes made easy: A complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol. Evol. 2013, 5, 989–997. [Google Scholar] [CrossRef]
- Wang, X.; Dorjee, T.; Chen, Y.; Gao, F.; Zhou, Y. The complete chloroplast genome sequencing analysis revealed an unusual IRs reduction in three species of subfamily Zygophylloideae. PLoS ONE 2022, 17, e0263253. [Google Scholar] [CrossRef]
- Wu, Z.; Gui, S.; Quan, Z.; Pan, L.; Wang, S.; Ke, W.; Liang, D.; Ding, Y. A precise chloroplast genome of Nelumbo nucifera (Nelumbonaceae) evaluated with Sanger, Illumina MiSeq, and PacBio RS II sequencing platforms: Insight into the plastid evolution of basal eudicots. BMC Plant Biol. 2014, 14, 289. [Google Scholar] [CrossRef]
- Noutsos, C.; Richly, E.; Leister, D. Generation and evolutionary fate of insertions of organelle DNA in the nuclear genomes of flowering plants. Genome Res. 2005, 15, 616–628. [Google Scholar] [CrossRef]
- Mukherjee, P.K.; Mukherjee, D.; Maji, A.K.; Rai, S.; Heinrich, M. The sacred lotus (Nelumbo nucifera)-phytochemical and therapeutic profile. J. Pharm. Pharmacol. 2009, 61, 407–422. [Google Scholar] [CrossRef]







| Category | Gene Group | Gene Name |
|---|---|---|
| Photosynthesis | Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of NADH dehydrogenase | NdhA *, ndhB *(2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Subunits of cytochrome b/f complex | petA, petB *, petD *, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF *, atpH, atpI | |
| Large subunit of rubisco | rbcL | |
| Subunits photochlorophyllide reductase | - | |
| Self-replication | Proteins of large ribosomal subunit | rpl14, rpl16 *, rpl2 *(2), rpl20, rpl22, rpl23(2), rpl32, rpl33, rpl36 |
| Proteins of small ribosomal subunit | rps11, rps12 **(2), rps14, rps15, rps16 *, rps18, rps19, rps2, rps3, rps4, rps7(2), rps8 | |
| Subunits of RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | |
| Ribosomal RNAs | rrn16(2), rrn23(2), rrn4.5(2), rrn5(2) | |
| Transfer RNAs | trnA-UGC *(2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC *, trnG-UCC, trnH-GUG, trnI-CAU(2), trnI-GAU *(2), trnK-UUU *, trnL-CAA(2), trnL-UAA *, trnL-UAG, trnM-CAU(2), trnN-GUU(2), trnP-UGG, trnQ-UUG, trnR-ACG(2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC(2), trnV-UAC *, trnW-CCA, trnY-GUA | |
| Other genes | Maturase | matK |
| Protease | clpP ** | |
| Envelope membrane protein | cemA | |
| Acetyl-CoA carboxylase | accD | |
| c-type cytochrome synthesis gene | ccsA | |
| Translation initiation factor | infA | |
| other | - | |
| Genes of unknown function | Conserved hypothetical chloroplast ORF | orf42(2), ycf1(2), ycf2(2), ycf3 **, ycf4 |
| Groups | Each Gene | All Genes | ||||
|---|---|---|---|---|---|---|
| KaKs > 1 | KaKs = 1 | KaKs < 1 | Ka | Ks | KaKs | |
| ‘Mahu Chuncai’ vs KT705316.2 | 0 | 0 | 1 | 0.01417 | 0.02794 | 0.51 |
| ‘Mahu Chuncai’ vs KT705317.3 | 1 | 0 | 66 | 1.03607 | 4.31501 | 0.24 |
| ‘Mahu Chuncai’ vs KY392763.1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ‘Mahu Chuncai’ vs MG720559.1 | 1 | 0 | 62 | 0.99297 | 4.34761 | 0.23 |
| ‘Mahu Chuncai’ vs MG967470.1 | 1 | 0 | 65 | 1.03967 | 4.50068 | 0.23 |
| ‘Mahu Chuncai’ vs MN315507.1 | 0 | 0 | 1 | 0.89994 | 2.34814 | 0.38 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Li, M.; Ma, J.; He, M.; Zheng, Y. Complete Chloroplast Genome Sequence of a New Variety of Brasenia schreberi: Genome Characteristics, Comparative Analysis, and Phylogenetic Relationships. Agronomy 2022, 12, 2972. https://doi.org/10.3390/agronomy12122972
Sun Y, Li M, Ma J, He M, Zheng Y. Complete Chloroplast Genome Sequence of a New Variety of Brasenia schreberi: Genome Characteristics, Comparative Analysis, and Phylogenetic Relationships. Agronomy. 2022; 12(12):2972. https://doi.org/10.3390/agronomy12122972
Chicago/Turabian StyleSun, Yue, Mengyao Li, Junying Ma, Maolin He, and Yangxia Zheng. 2022. "Complete Chloroplast Genome Sequence of a New Variety of Brasenia schreberi: Genome Characteristics, Comparative Analysis, and Phylogenetic Relationships" Agronomy 12, no. 12: 2972. https://doi.org/10.3390/agronomy12122972
APA StyleSun, Y., Li, M., Ma, J., He, M., & Zheng, Y. (2022). Complete Chloroplast Genome Sequence of a New Variety of Brasenia schreberi: Genome Characteristics, Comparative Analysis, and Phylogenetic Relationships. Agronomy, 12(12), 2972. https://doi.org/10.3390/agronomy12122972







